Secretory Expression and Application of Antilipopolysaccharide Factor 3 in Chlamydomonas reinhardtii
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Plasmid Construction and Algae Transformation
2.3. Genomic PCR and RT-PCR Analysis
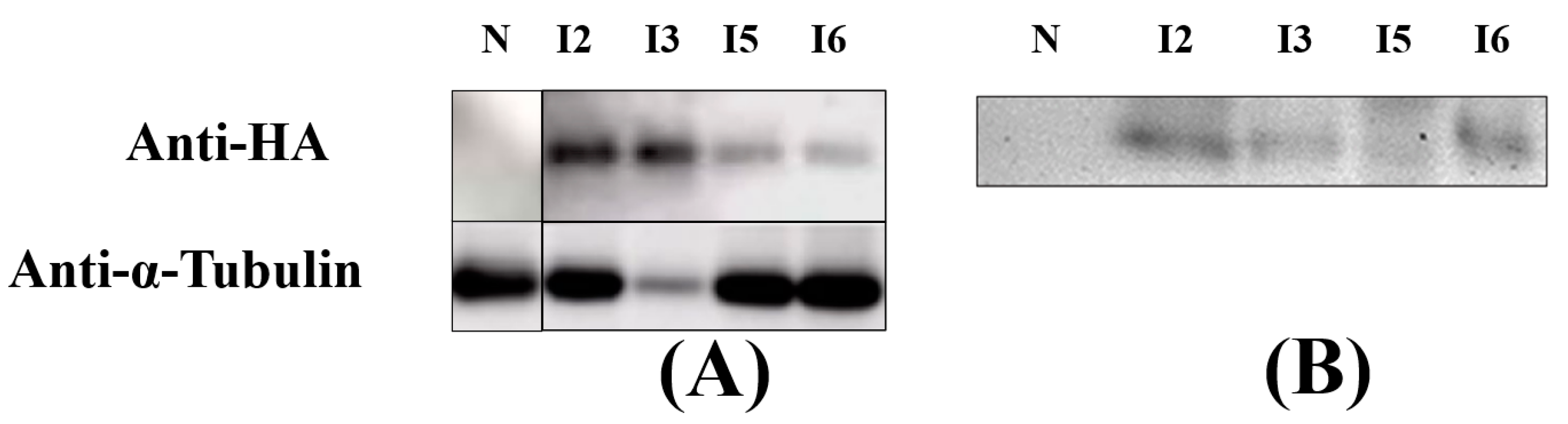
2.4. Protein Extraction and Immunoblot Analysis
2.5. Antibacterial Test
2.6. Minimum Inhibition Concentration (MIC) Assay
2.7. Statistical Analysis
3. Results
3.1. Design of ALFPm3 Expression Cassette
3.2. Confirmation of Transgenic C. reinhardtii
3.3. Secretion of IBP1-ALFPm3 Fusion Protein
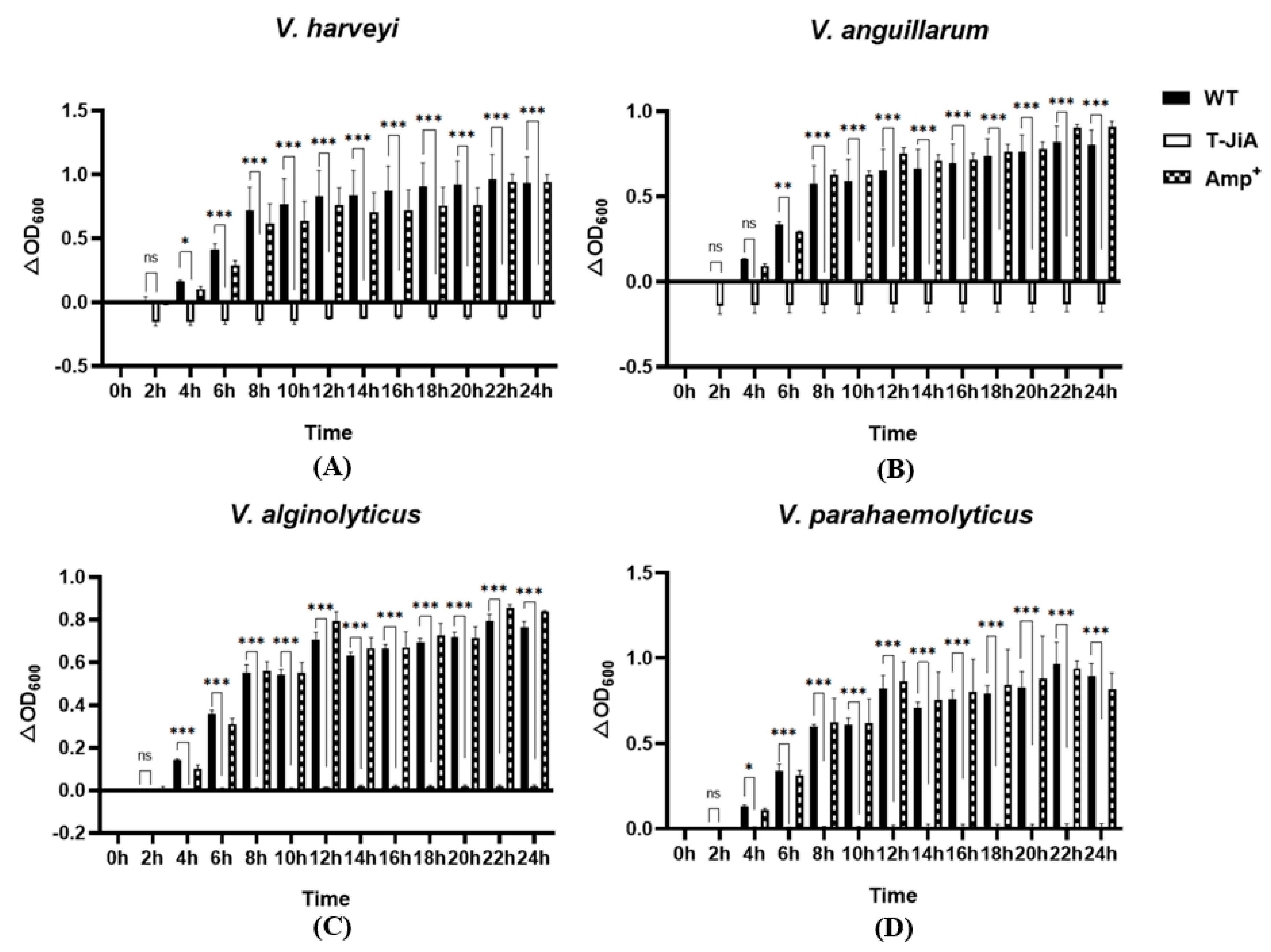
3.4. Extracellular Protein Extracts from T-JiA Showed High Antibacterial Activity
3.5. The MIC of Extracellular Protein Extracts from T-JiA
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Durand, G.A.; Raoult, D.; Dubourg, G. Antibiotic discovery: History, methods and perspectives. Int. J. Antimicrob. Agents 2019, 53, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Montesinos, I.; Dominguez-Guasch, A.; Gomez-Zorrislla, S.; Duran-Jorda, X.; Siverio-Pares, A.; Arenas-Miras, M.M.; Montero, M.M.; Sorli Redo, L.; Grau, S.; Horcajada, J.P. Clinical and economic burden of community-onset multidrug-resistant infections requiring hospitalization. J. Infect. 2020, 80, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Blair, J.M.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Valdez-Miramontes, C.E.; De Haro-Acosta, J.; Aréchiga-Flores, C.F.; Verdiguel-Fernández, L.; Rivas-Santiago, B. Antimicrobial peptides in domestic animals and their applications in veterinary medicine. Peptides 2021, 142, 170576. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.Y.; Chowdhury, M.; Huang, Y.D.; Yu, X.Q. Insect antimicrobial peptides and their applications. Appl. Microbiol. Biotechnol. 2014, 98, 5807–5822. [Google Scholar] [CrossRef] [PubMed]
- Slavokhotova, A.A.; Shelenkov, A.A.; Andreev, Y.A.; Odintsova, T.I. Hevein-Like Antimicrobial Peptides of Plants. Biochemistry 2017, 82, 1659–1674. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.; Brown, K.L.; Mookherjee, N. Host defence peptides from invertebrates--emerging antimicrobial strategies. Immunobiology 2006, 211, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, J.; Sun, Y.; Sun, L. A Crustin from Hydrothermal Vent Shrimp: Antimicrobial Activity and Mechanism. Mar. Drugs 2021, 19, 176. [Google Scholar] [CrossRef]
- Tanaka, S.; Nakamura, T.; Morita, T.; Iwanaga, S. Limulus anti-LPS factor: An anticoagulant which inhibits the endotoxin mediated activation of Limulus coagulation system. Biochem. Biophys. Res. Commun. 1982, 105, 717–723. [Google Scholar] [CrossRef]
- Zhou, L.; Li, G.; Jiao, Y.; Huang, D.; Li, A.; Chen, H.; Liu, Y.; Li, S.; Li, H.; Wang, C. Molecular and antimicrobial characterization of a group G anti-lipopolysaccharide factor (ALF) from Penaeus monodon. Fish Shellfish. Immunol. 2019, 94, 149–156. [Google Scholar] [CrossRef]
- Jaree, P.; Tassanakajon, A.; Somboonwiwat, K. Effect of the anti-lipopolysaccharide factor isoform 3 (ALFPm3) from Penaeus monodon on Vibrio harveyi cells. Dev. Comp. Immunol. 2012, 38, 554–560. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Huang, R.; Wang, C.; Hu, Q.; Li, H.; Li, X. Expression of Anti-Lipopolysaccharide Factor Isoform 3 in Chlamydomonas reinhardtii Showing High Antimicrobial Activity. Mar. Drugs 2021, 19, 239. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, G.; Silvis, M.R.; Talkington, K.C.; Budzik, J.M.; Dodd, C.E.; Paluba, J.M.; Oki, E.A.; Trotta, K.L.; Licht, D.J.; Jimenez-Morales, D.; et al. Ceragenins and Antimicrobial Peptides Kill Bacteria through Distinct Mechanisms. mBio 2022, 13, e0272621. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.M.; Panda, A.K. Solubilization and refolding of bacterial inclusion body proteins. J. Biosci. Bioeng. 2005, 99, 303–310. [Google Scholar] [CrossRef]
- Tang, T.; Liu, J.; Li, S.; Li, H.; Liu, F. Recombinant expression of an oriental river prawn anti-lipopolysaccharide factor gene in Pichia pastoris and its characteristic analysis. Fish Shellfish. Immunol. 2020, 98, 414–419. [Google Scholar] [CrossRef]
- Puxbaum, V.; Mattanovich, D.; Gasser, B. Quo vadis? The challenges of recombinant protein folding and secretion in Pichia pastoris. Appl. Microbiol. Biotechnol. 2015, 99, 2925–2938. [Google Scholar] [CrossRef]
- Scaife, M.A.; Nguyen, G.; Rico, J.; Lambert, D.; Helliwell, K.E.; Smith, A.G. Establishing Chlamydomonas reinhardtii as an industrial biotechnology host. Plant. J. 2015, 82, 532–546. [Google Scholar] [CrossRef]
- Liu, Y.-X.; Li, Z.-F.; Lv, Y.-J.; Dong, B.; Fan, Z.-C. Chlamydomonas reinhardtii-expressed multimer of Bacteriocin LS2 potently inhibits the growth of bacteria. Process. Biochem. 2020, 95, 139–147. [Google Scholar] [CrossRef]
- Hadiatullah, H.; Wang, H.; Liu, Y.-X.; Fan, Z.-C. Chlamydomonas reinhardtii-derived multimer Mytichitin-CB possesses potent antibacterial properties. Process. Biochem. 2020, 96, 21–29. [Google Scholar] [CrossRef]
- Xue, B.; Dong, C.-M.; Hu, H.-H.; Dong, B.; Fan, Z.-C. Chlamydomonas reinhardtii-expressed multimer of ToAMP4 inhibits the growth of bacteria of both Gram-positive and Gram-negative. Process. Biochem. 2020, 91, 311–318. [Google Scholar] [CrossRef]
- Dong, B.; Cheng, R.Q.; Liu, Q.Y.; Wang, J.; Fan, Z.C. Multimer of the antimicrobial peptide Mytichitin-A expressed in Chlamydomonas reinhardtii exerts a broader antibacterial spectrum and increased potency. J. Biosci. Bioeng. 2018, 125, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Rasala, B.A.; Lee, P.A.; Shen, Z.; Briggs, S.P.; Mendez, M.; Mayfield, S.P. Robust expression and secretion of Xylanase1 in Chlamydomonas reinhardtii by fusion to a selection gene and processing with the FMDV 2A peptide. PLoS ONE 2012, 7, e43349. [Google Scholar] [CrossRef] [PubMed]
- Owji, H.; Nezafat, N.; Negahdaripour, M.; Hajiebrahimi, A.; Ghasemi, Y. A comprehensive review of signal peptides: Structure, roles, and applications. Eur. J. Cell Biol. 2018, 97, 422–441. [Google Scholar] [CrossRef] [PubMed]
- Molino, J.V.D.; de Carvalho, J.C.M.; Mayfield, S.P. Comparison of secretory signal peptides for heterologous protein expression in microalgae: Expanding the secretion portfolio for Chlamydomonas reinhardtii. PLoS ONE 2018, 13, e0192433. [Google Scholar] [CrossRef] [PubMed]
- Raymond, J.A.; Janech, M.G.; Fritsen, C.H. Novel Ice-Binding Proteins from a Psychrophilic Antarctic Alga (Chlamydomonadaceae, Chlorophyceae)(1). J. Phycol. 2009, 45, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Juliane, N.; Ning, S.; Yinghong, L.; Ralph, B. Genetic transformation of the model green alga Chlamydomonas reinhardtii. Methods Mol. Biol. 2012, 847, 35–47. [Google Scholar]
- Fischer, N.; Rochaix, J.D. The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii. Mol. Genet. Genom. 2001, 265, 888–894. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Z. RAPD: A database of recombinantly-produced antimicrobial peptides. FEMS Microbiol. Lett. 2008, 289, 126–129. [Google Scholar] [CrossRef]
- Corrales-Garcia, L.L.; Serrano-Carreon, L.; Corzo, G. Improving the heterologous expression of human beta-defensin 2 (HBD2) using an experimental design. Protein Expr. Purif. 2020, 167, 105539. [Google Scholar] [CrossRef]
- Bryksa, B.C.; MacDonald, L.D.; Patrzykat, A.; Douglas, S.E.; Mattatall, N.R. A C-terminal glycine suppresses production of pleurocidin as a fusion peptide in Escherichia coli. Protein Expr. Purif. 2006, 45, 88–98. [Google Scholar] [CrossRef]
- Gong, G.L.; Wei, Y.; Wang, Z.Z. Functional expression, purification, and antimicrobial activity of a novel antimicrobial peptide MLH in Escherichia coli. Prep. Biochem. Biotechnol. 2018, 48, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Ishida, H.; Nguyen, L.T.; Gopal, R.; Aizawa, T.; Vogel, H.J. Overexpression of Antimicrobial, Anticancer, and Transmembrane Peptides in Escherichia coli through a Calmodulin-Peptide Fusion System. J. Am. Chem. Soc. 2016, 138, 11318–11326. [Google Scholar] [CrossRef] [PubMed]
- Yi, T.; Sun, S.; Huang, Y.; Chen, Y. Prokaryotic expression and mechanism of action of alpha-helical antimicrobial peptide A20L using fusion tags. BMC Biotechnol. 2015, 15, 69. [Google Scholar] [CrossRef]
- Cao, J.; de la Fuente-Nunez, C.; Ou, R.W.; Torres, M.T.; Pande, S.G.; Sinskey, A.J.; Lu, T.K. Yeast-Based Synthetic Biology Platform for Antimicrobial Peptide Production. ACS Synth. Biol. 2018, 7, 896–902. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Dong, S.; Zhu, Y.; Jin, H.; Gao, M.; Duan, Z.; Zheng, Z.; Shi, Z.; Li, Z. Effective and stable porcine interferon-alpha production by Pichia pastoris fed-batch cultivation with multi-variables clustering and analysis. Bioprocess Biosyst. Eng. 2010, 33, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Gregory, J.A.; Li, F.; Tomosada, L.M.; Cox, C.J.; Topol, A.B.; Vinetz, J.M.; Mayfield, S. Algae-produced Pfs25 elicits antibodies that inhibit malaria transmission. PLoS ONE 2012, 7, e37179. [Google Scholar] [CrossRef]
- Barahimipour, R.; Neupert, J.; Bock, R. Efficient expression of nuclear transgenes in the green alga Chlamydomonas: Synthesis of an HIV antigen and development of a new selectable marker. Plant Mol. Biol. 2016, 90, 403–418. [Google Scholar] [CrossRef]
- Schroda, M. Good News for Nuclear Transgene Expression in Chlamydomonas. Cells 2019, 8, 1534. [Google Scholar] [CrossRef]
- Liang, Z.C.; Liang, M.H.; Jiang, J.G. Transgenic microalgae as bioreactors. Crit. Rev. Food Sci. Nutr. 2020, 60, 3195–3213. [Google Scholar] [CrossRef]
- Gu, H.J.; Sun, Q.L.; Jiang, S.; Zhang, J.; Sun, L. First characterization of an anti-lipopolysaccharide factor (ALF) from hydrothermal vent shrimp: Insights into the immune function of deep-sea crustacean ALF. Dev. Comp. Immunol. 2018, 84, 382–395. [Google Scholar] [CrossRef]
- Rosa, R.D.; Vergnes, A.; de Lorgeril, J.; Goncalves, P.; Perazzolo, L.M.; Saune, L.; Romestand, B.; Fievet, J.; Gueguen, Y.; Bachere, E.; et al. Functional divergence in shrimp anti-lipopolysaccharide factors (ALFs): From recognition of cell wall components to antimicrobial activity. PLoS ONE 2013, 8, e67937. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Gao, Y.; Wang, X.; Shen, W.; Wang, J.; Zhou, X.; Cai, M.; Zhang, Y. Peroxisome-targeted and tandem repeat multimer expressions of human antimicrobial peptide LL37 in Pichia pastoris. Prep. Biochem. Biotechnol. 2017, 47, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Meng, D.M.; Zhao, J.F.; Ling, X.; Dai, H.X.; Guo, Y.J.; Gao, X.F.; Dong, B.; Zhang, Z.Q.; Meng, X.; Fan, Z.C. Recombinant expression, purification and antimicrobial activity of a novel antimicrobial peptide PaDef in Pichia pastoris. Protein Expr. Purif. 2017, 130, 90–99. [Google Scholar] [CrossRef] [PubMed]
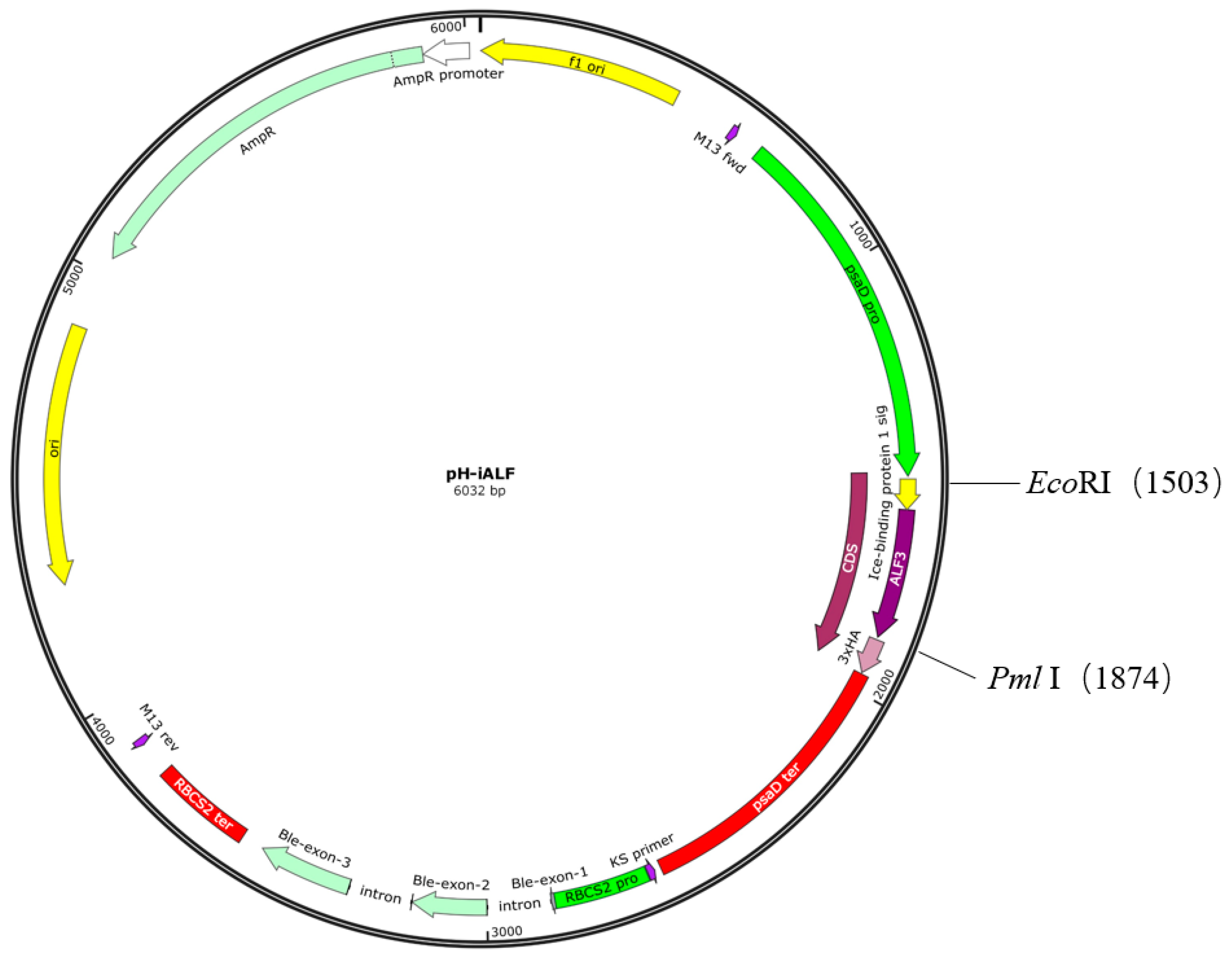
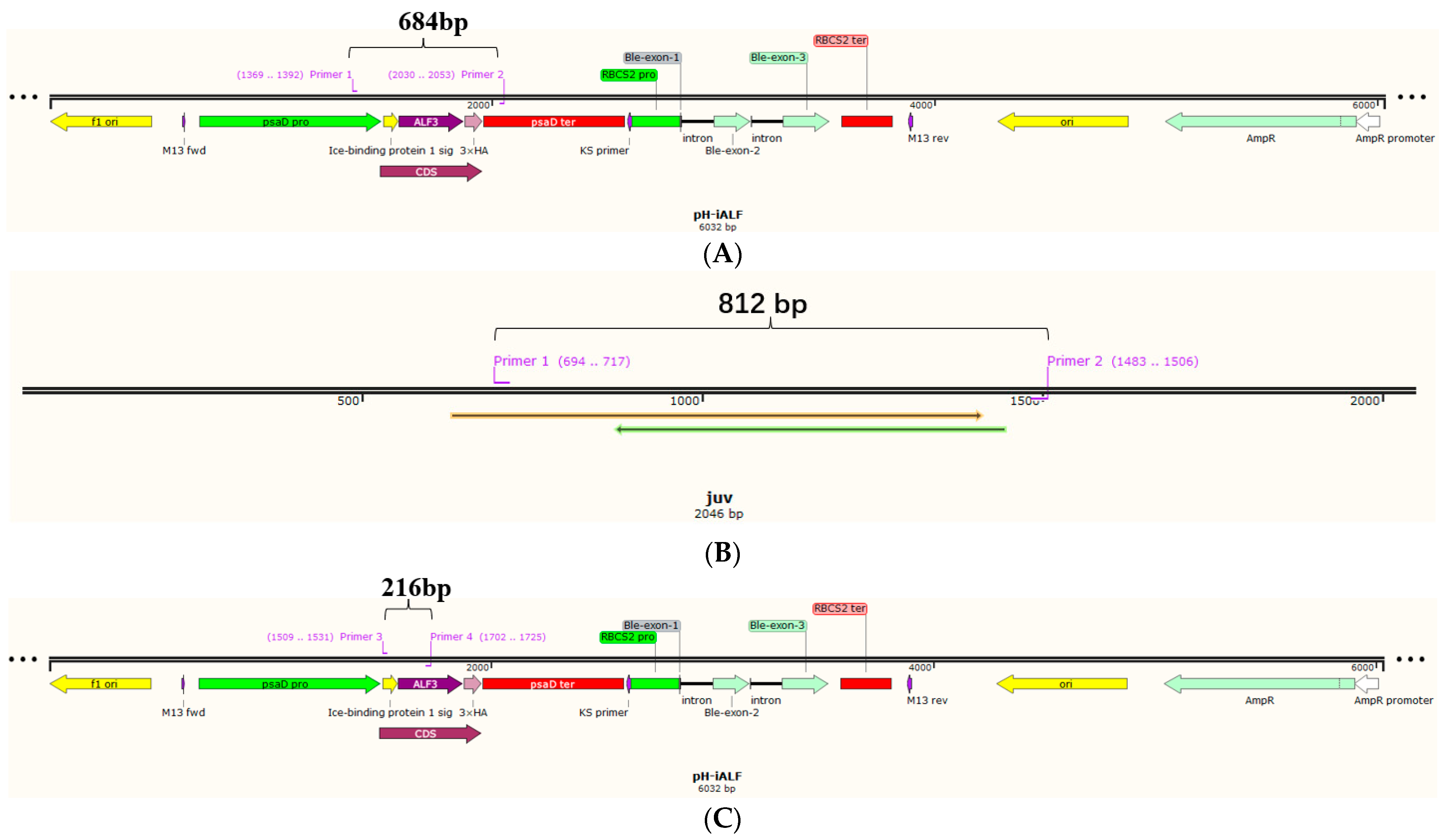


| Name | Sequence (5′-3′) | Target Gene | Expected Product (bp) |
|---|---|---|---|
| psaD-P | GGGAATTGGAGGTACGACCGAGAT | IBP1-ALFPm3 | 684 |
| psaD-T | AGCTCCGATCCCGTATCAATCAGC | ||
| Fi | ATGCCGTCGAGCAGCATGAAGCT | IBP1-ALFPm3 | 216 |
| ALF3R2 | ACATGCGGCCCTTGTAGTACACCT | ||
| actin-F | ACCCCGTGCTGCTGACTG | β-actin | 351 |
| actin-R | ACGTTGAAGGTCTCGAACA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ou, Y.; Zhuang, H.; Chen, R.; Huang, D.; Wang, C. Secretory Expression and Application of Antilipopolysaccharide Factor 3 in Chlamydomonas reinhardtii. Bioengineering 2023, 10, 564. https://doi.org/10.3390/bioengineering10050564
Ou Y, Zhuang H, Chen R, Huang D, Wang C. Secretory Expression and Application of Antilipopolysaccharide Factor 3 in Chlamydomonas reinhardtii. Bioengineering. 2023; 10(5):564. https://doi.org/10.3390/bioengineering10050564
Chicago/Turabian StyleOu, Yaohui, Huilin Zhuang, Ruoyu Chen, Danqiong Huang, and Chaogang Wang. 2023. "Secretory Expression and Application of Antilipopolysaccharide Factor 3 in Chlamydomonas reinhardtii" Bioengineering 10, no. 5: 564. https://doi.org/10.3390/bioengineering10050564
APA StyleOu, Y., Zhuang, H., Chen, R., Huang, D., & Wang, C. (2023). Secretory Expression and Application of Antilipopolysaccharide Factor 3 in Chlamydomonas reinhardtii. Bioengineering, 10(5), 564. https://doi.org/10.3390/bioengineering10050564






