Analysis of the Robotic-Based In Situ Bioprinting Workflow for the Regeneration of Damaged Tissues through a Case Study
Abstract
1. Introduction
2. Materials and Methods
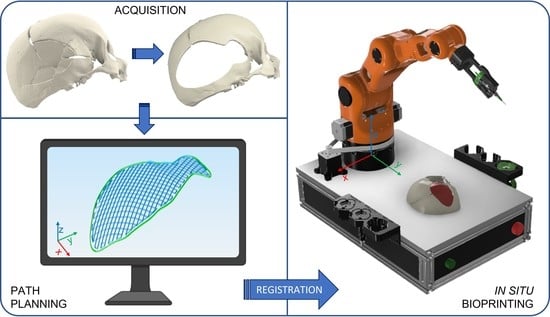
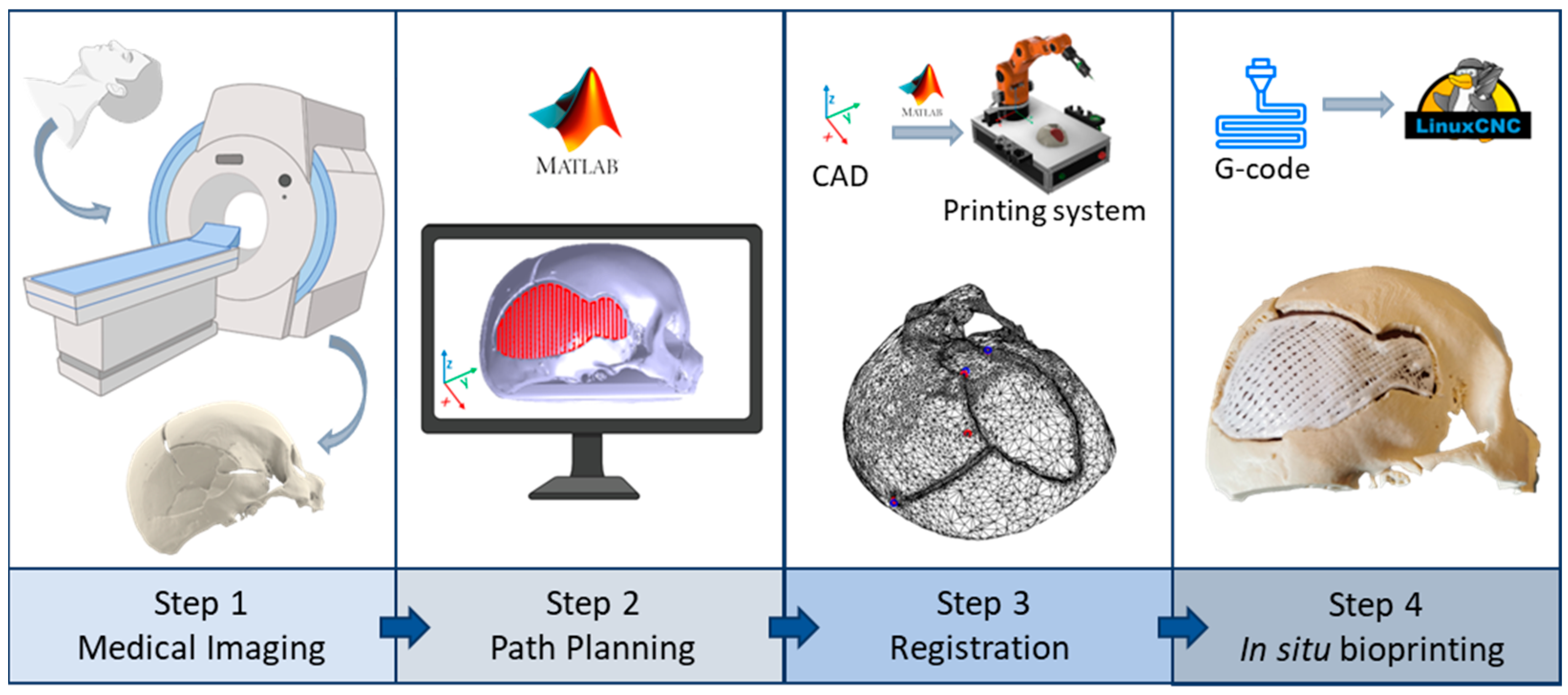
2.1. Robotic-Based In Situ Bioprinting Workflow
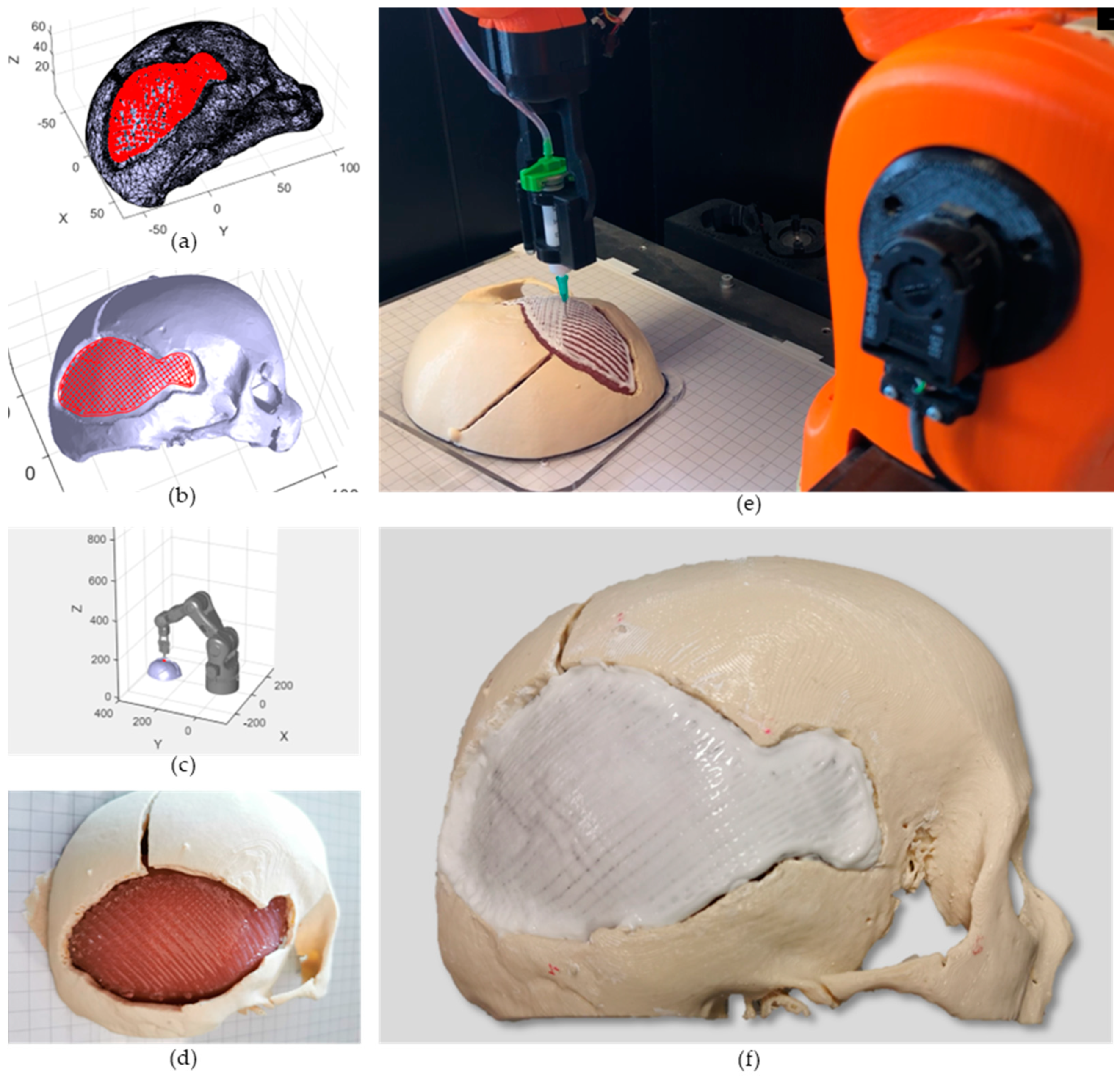
- Acquisition of the 3D digital model of the damaged area: the geometry of the anatomical portion to be reconstructed can be obtained with medical imaging techniques (e.g., computed tomography (CT), magnetic resonance imaging (MRI)) to ensure high accuracy and resolution of the model;
- Printing path planning: based on the acquired geometry the printing pattern to restore the defect is planned. Depending on the slope of the surface on which the material has to be deposited, it is possible to use two different approaches to planning the trajectories: (i) with slopes greater than 45°, the extruder must be kept perpendicular to the surface in order to achieve an optimal deposition; (ii) on the other hand, it is sufficient to keep the extruder vertical and deposit the filler following the profile of the surface. The coordinates of the points of the planned path are referred to the computer-aided design/computed-aided manufacturing (CAD/CAM) software reference frame (e.g., in this work Matlab® is used);
- Registration of the printing path in the robot workspace: the path planning is generally defined in the pre-operative scanner reference frame, which does not coexist in space and time with the robot one, which includes the patient. The transformation matrix between the two reference frames can be computed by acquiring anatomical or artificial landmarks in both reference frames (e.g., Matlab® and IMAGObot workspace);
- Biomaterial in situ deposition: following the registration of the planned printing pattern on the patient in the operating area, using the computed transformation matrix, the in situ bioprinting process is carried out by depositing the biomaterial directly on the damaged tissue. The registered pattern is converted into a G-code and sent to the printing system control software (e.g., LinuxCNC for the IMAGObot platform).
2.2. Overview of the IMAGObot Platform
- Projection method: a single layer pattern, loaded as g-code, can be projected on the substrate mesh and the end-effector path is computed constraining its orientation to be perpendicular to the printing surface;
2.3. Case Study Implementation
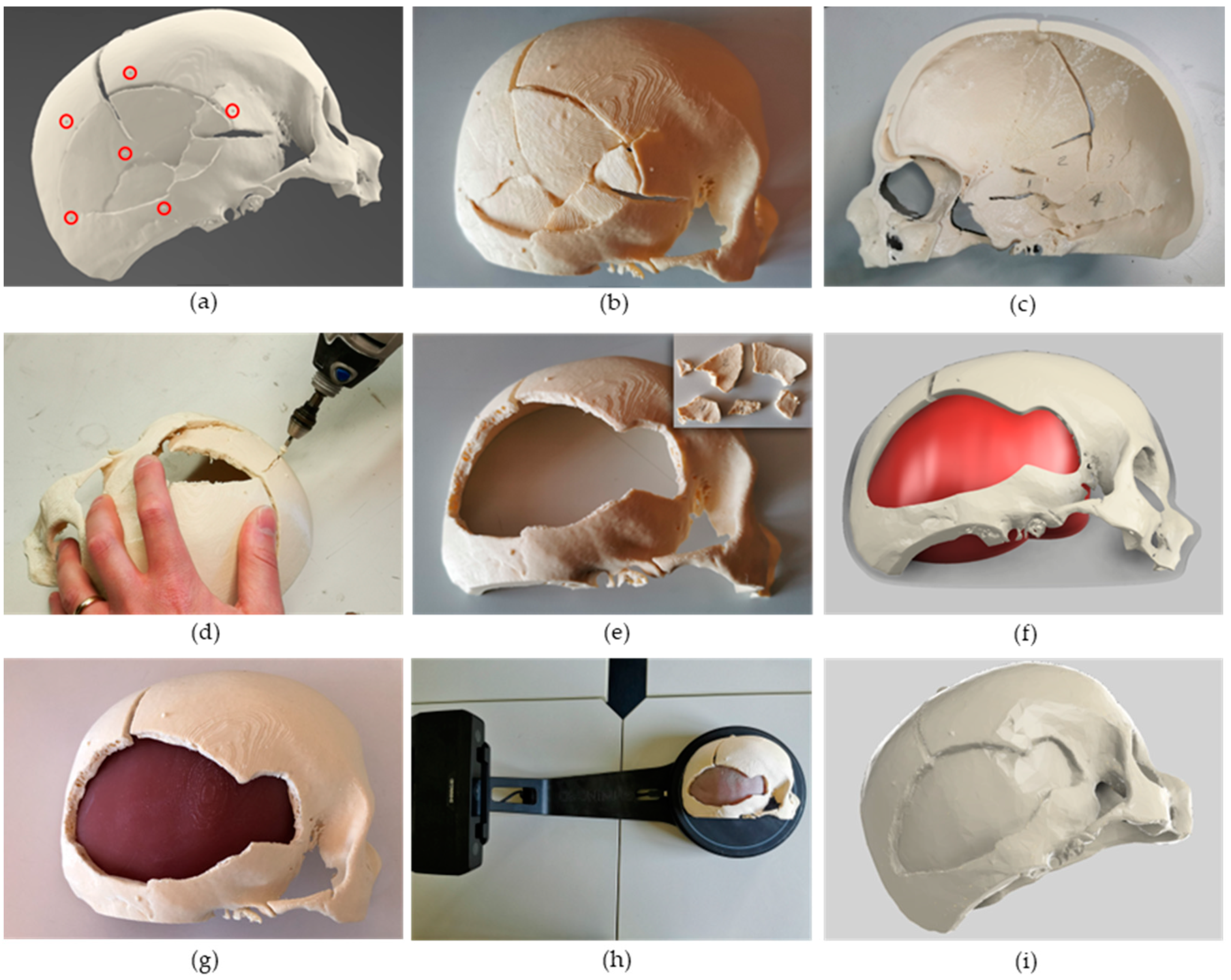
2.3.1. Case Study Selection and CT Scan Processing
2.3.2. Phantom Design and Fabrication
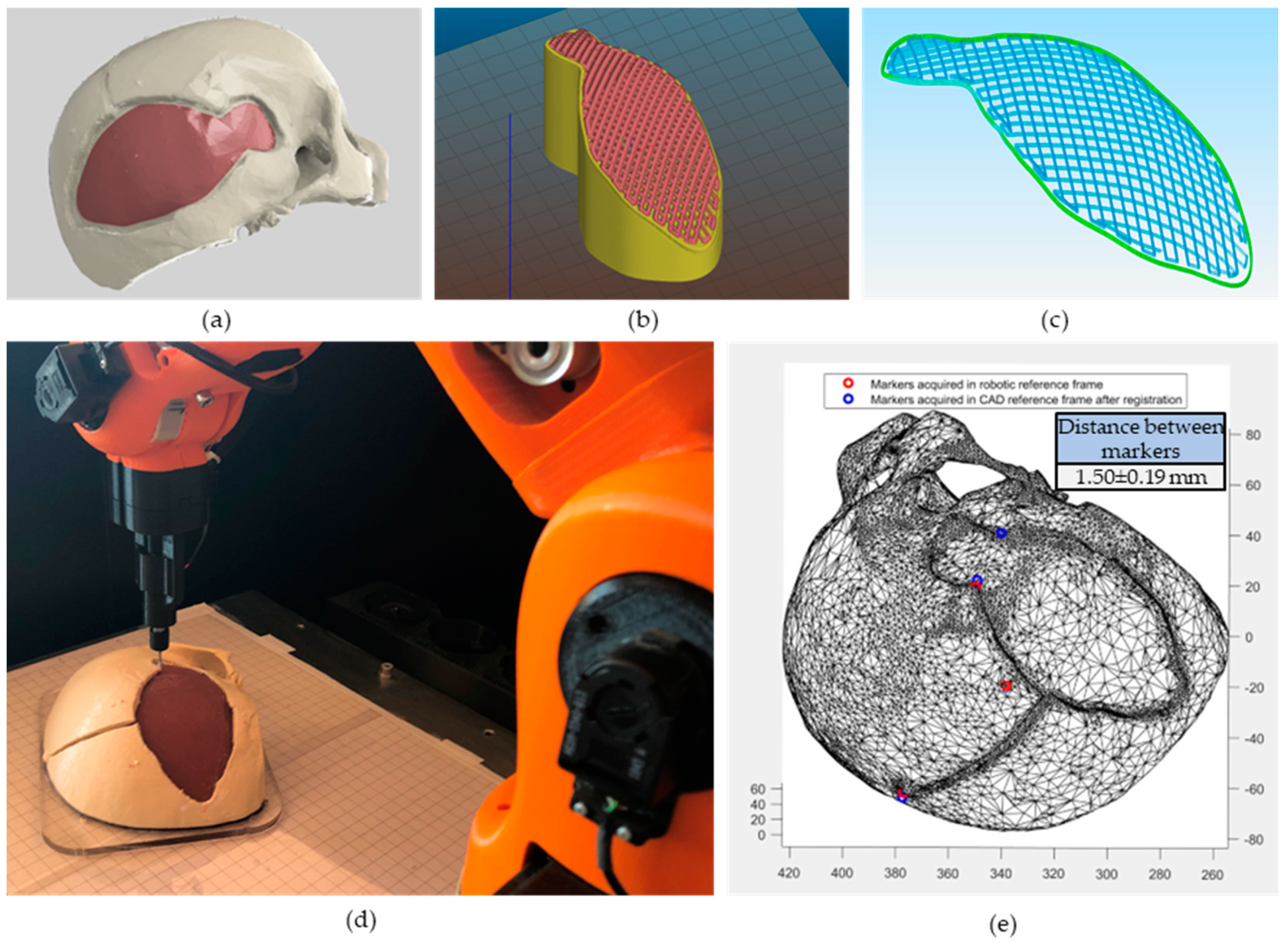
2.4. Printing Path Planning
2.4.1. Non-Planar Slicing
2.4.2. Registration Using Fiducial Markers
2.5. In Situ Bioprinting
3. Results and Discussion
3.1. Cranial Phantom Fabrication
3.2. Path Planning
3.3. In Situ Bioprinting
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Boyle, W.J.; Simonet, W.S.; Lacey, D.L. Osteoclast differentiation and activation. Nature 2003, 423, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Murugan, R.; Ramakrishna, S. Development of nanocomposites for bone grafting. Compos. Sci. Technol. 2005, 65, 2385–2406. [Google Scholar] [CrossRef]
- Marsell, R.; Einhorn, T.A. Emerging Bone Healing Therapies. J. Orthop. Trauma 2010, 24, S4–S8. [Google Scholar] [CrossRef] [PubMed]
- Schemitsch, E.H. Size Matters: Defining Critical in Bone Defect Size! J. Orthop. Trauma 2017, 31, S20–S22. [Google Scholar] [CrossRef]
- Ma, P.X. Scaffolds for tissue fabrication. Mater. Today 2004, 7, 30–40. [Google Scholar] [CrossRef]
- Marsell, R.; Einhorn, T.A. The biology of fracture healing. Injury 2011, 42, 551–555. [Google Scholar] [CrossRef]
- Calori, G.; Mazza, E.; Colombo, M.; Ripamonti, C. The use of bone-graft substitutes in large bone defects: Any specific needs? Injury 2011, 42, S56–S63. [Google Scholar] [CrossRef]
- Li, L.; Yu, F.; Shi, J.; Shen, S.; Teng, H.; Yang, J.; Wang, X.; Jiang, Q. In situ repair of bone and cartilage defects using 3D scanning and 3D printing. Sci. Rep. 2017, 7, 9416. [Google Scholar] [CrossRef]
- Lee, J.C.; Volpicelli, E.J. Bioinspired Collagen Scaffolds in Cranial Bone Regeneration: From Bedside to Bench. Adv. Healthc. Mater. 2017, 6, 1700232. [Google Scholar] [CrossRef]
- Abhay, S.; Haines, S.J. Repairing Holes in the Head: A History of Cranioplasty. Neurosurgery 1997, 40, 588–603. [Google Scholar]
- Williams, L.; Fan, K.; Bentley, R. Custom-made titanium cranioplasty: Early and late complications of 151 cranioplasties and review of the literature. Int. J. Oral Maxillofac. Surg. 2015, 44, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Thakur, B.; Haq, I.; Hettige, S.; Martin, A.J. Complications of titanium cranioplasty—A retrospective analysis of 174 patients. Acta Neurochir. 2014, 156, 989–998. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.S.; Luoma, A.M.V.; Wilson, S.R.; Kitchen, N. Titanium cranioplasty and the prediction of complications. Br. J. Neurosurg. 2012, 26, 832–837. [Google Scholar] [CrossRef] [PubMed]
- Moiduddin, K.; Darwish, S.; Al-Ahmari, A.; ElWatidy, S.; Mohammad, A.; Ameen, W. Structural and mechanical characterization of custom design cranial implant created using additive manufacturing. Electron. J. Biotechnol. 2017, 29, 22–31. [Google Scholar] [CrossRef]
- Berretta, S.; Evans, K.; Ghita, O. Additive manufacture of PEEK cranial implants: Manufacturing considerations versus accuracy and mechanical performance. Mater. Des. 2018, 139, 141–152. [Google Scholar] [CrossRef]
- Tevlin, R.; McArdle, A.; Atashroo, D.; Walmsley, G.; Senarath-Yapa, K.; Zielins, E.; Paik, K.; Longaker, M.; Wan, D. Biomaterials for Craniofacial Bone Engineering. J. Dent. Res. 2014, 93, 1187–1195. [Google Scholar] [CrossRef]
- Spetzger, U.; Vougioukas, V.; Schipper, J. Materials and techniques for osseous skull reconstruction. Minim. Invasive Ther. Allied Technol. 2010, 19, 110–121. [Google Scholar] [CrossRef]
- Shah, A.M.; Jung, H.; Skirboll, S. Materials used in cranioplasty: A history and analysis. Neurosurg. Focus 2014, 36, E19. [Google Scholar] [CrossRef]
- Moroni, L.; Boland, T.; Burdick, J.A.; De Maria, C.; Derby, B.; Forgacs, G.; Groll, J.; Li, Q.; Malda, J.; Mironov, V.A.; et al. Biofabrication: A Guide to Technology and Terminology. Trends Biotechnol. 2017, 36, 384–402. [Google Scholar] [CrossRef]
- Ashammakhi, N.; Ahadian, S.; Pountos, I.; Hu, S.-K.; Tellisi, N.; Bandaru, P.; Ostrovidov, S.; Dokmeci, M.R.; Khademhosseini, A. In situ three-dimensional printing for reparative and regenerative therapy. Biomed. Microdevices 2019, 21, 42. [Google Scholar] [CrossRef]
- Singh, S.; Choudhury, D.; Yu, F.; Mironov, V.; Naing, M.W. In situ bioprinting—Bioprinting from benchside to bedside? Acta Biomater. 2020, 101, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Murphy, S.V.; Atala, A. 3D bioprinting of tissues and organs. Nat. Biotechnol. 2014, 32, 773–785. [Google Scholar] [CrossRef] [PubMed]
- Szpalski, C.; Barr, J.; Wetterau, M.; Saadeh, P.B.; Warren, S.M. Cranial bone defects: Current and future strategies. Neurosurg. Focus 2010, 29, E8. [Google Scholar] [CrossRef] [PubMed]
- Keriquel, V.; Guillemot, F.; Arnault, I.; Guillotin, B.; Miraux, S.; Amédée, J.; Fricain, J.-C.; Catros, S. In vivo bioprinting for computer- and robotic-assisted medical intervention: Preliminary study in mice. Biofabrication 2010, 2, 014101. [Google Scholar] [CrossRef]
- Keriquel, V.; Oliveira, H.; Rémy, M.; Ziane, S.; Delmond, S.; Rousseau, B.; Rey, S.; Catros, S.; Amédée, J.; Guillemot, F.; et al. In situ printing of mesenchymal stromal cells, by laser-assisted bioprinting, for in vivo bone regeneration applications. Sci. Rep. 2017, 7, 1778. [Google Scholar] [CrossRef]
- Dwivedi, R.; Mehrotra, D. 3D bioprinting and craniofacial regeneration. J. Oral Biol. Craniofacial Res. 2020, 10, 650–659. [Google Scholar] [CrossRef]
- Mostafavi, A.; Abdullah, T.; Russell, C.S.; Mostafavi, E.; Williams, T.J.; Salah, N.; Alshahrie, A.; Harris, S.; Basri, S.M.M.; Mishra, Y.K.; et al. In situ printing of scaffolds for reconstruction of bone defects. Acta Biomater. 2021, 127, 313–326. [Google Scholar] [CrossRef]
- Moncal, K.K.; Gudapati, H.; Godzik, K.P.; Heo, D.N.; Kang, Y.; Rizk, E.; Ravnic, D.J.; Wee, H.; Pepley, D.F.; Ozbolat, V.; et al. Intra-Operative Bioprinting of Hard, Soft, and Hard/Soft Composite Tissues for Craniomaxillofacial Reconstruction. Adv. Funct. Mater. 2021, 31, 2010858. [Google Scholar] [CrossRef]
- Wu, Y.; Ravnic, D.J.; Ozbolat, I.T. Intraoperative Bioprinting: Repairing Tissues and Organs in a Surgical Setting. Trends Biotechnol. 2020, 38, 594–605. [Google Scholar] [CrossRef]
- Moncal, K.K.; Aydın, R.S.T.; Godzik, K.P.; Acri, T.M.; Heo, D.N.; Rizk, E.; Wee, H.; Lewis, G.S.; Salem, A.K.; Ozbolat, I.T. Controlled Co-delivery of pPDGF-B and pBMP-2 from intraoperatively bioprinted bone constructs improves the repair of calvarial defects in rats. Biomaterials 2022, 281, 121333. [Google Scholar] [CrossRef]
- Kérourédan, O.; Hakobyan, D.; Rémy, M.; Ziane, S.; Dusserre, N.; Fricain, J.-C.; Delmond, S.; Thébaud, N.B.; Devillard, R. In situ prevascularization designed by laser-assisted bioprinting: Effect on bone regeneration. Biofabrication 2019, 11, 045002. [Google Scholar] [CrossRef] [PubMed]
- Fortunato, G.M.; Rossi, G.; Bonatti, A.F.; De Acutis, A.; Mendoza-Buenrostro, C.; Vozzi, G.; De Maria, C. Robotic platform and path planning algorithm for in situ bioprinting. Bioprinting 2021, 22, e00139. [Google Scholar] [CrossRef]
- Fortunato, G.M.; Batoni, E.; Bonatti, A.F.; Vozzi, G.; De Maria, C. Surface reconstruction and tissue recognition for robotic-based in situ bioprinting. Bioprinting 2022, 26, e00195. [Google Scholar] [CrossRef]
- Fortunato, G.M.; Bonatti, A.F.; Micalizzi, S.; Chiesa, I.; Batoni, E.; De Acutis, A.; De Maria, C.; Vozzi, G. In Situ Bioprinting—Current Applications and Future Challenges. Addit. Manuf. Biomed. Appl. 2022, 23, 225–236. [Google Scholar] [CrossRef]
- Fortunato, G.M.; Bonatti, A.F.; Batoni, E.; Macaluso, R.; Vozzi, G.; De Maria, C. Motion compensation system for robotic based in situ bioprinting to balance patient physiological movements. Bioprinting 2022, 28, e00248. [Google Scholar] [CrossRef]
- Staroveški, T.; Brezak, D.; Udiljak, T. LinuxCNC—The enhanced machine controller: Application and an overview. Teh. Vjesn. 2013, 20, 1103–1110. [Google Scholar]
- Cendrero, A.M.; Fortunato, G.M.; Munoz-Guijosa, J.M.; De Maria, C.; Lantada, A.D. Benefits of Non-Planar Printing Strategies Towards Eco-Efficient 3D Printing. Sustainability 2021, 13, 1599. [Google Scholar] [CrossRef]
- Ahlers, D.; Wasserfall, F.; Hendrich, N.; Zhang, J. 3D Printing of Nonplanar Layers for Smooth Surface Generation. In Proceedings of the 2019 IEEE 15th International Conference on Automation Science and Engineering (CASE), Vancouver, BC, Canada, 22–26 August 2019; pp. 1737–1743. [Google Scholar] [CrossRef]
- Teasdale, G.; Jennett, B. Assessment of coma and impaired consciousness. A practical scale. Lancet Lond. Engl. 1974, 2, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Stein, S.C. The Evolution of Modern Treatment for Depressed Skull Fractures. World Neurosurg. 2019, 121, 186–192. [Google Scholar] [CrossRef]
- Ferrari, V.; Cappelli, C.; Megali, G.; Pietrabissa, A. anatomy driven approach for generation of 3D models from multi-phase CT images. Int. J. Comput. Assist. Radiol. Surg. 2008, 3, 271–273. [Google Scholar]
- Foundation B Blender Free and Open Source 3D Creation Suite. Available online: https://www.blender.org/ (accessed on 6 February 2023).
- Mazgajczyk, E.; Ścigała, K.; Czyż, M.; Jarmundowicz, W.; Będziński, R. Mechanical properties of cervical dura mater. Acta Bioeng. Biomech. 2012, 14, 51–58. [Google Scholar] [PubMed]
- Moreira-Gonzalez, A.; Papay, F.E.; Zins, J.E. Calvarial Thickness and Its Relation to Cranial Bone Harvest. Plast. Reconstr. Surg. 2006, 117, 1964–1971. [Google Scholar] [CrossRef] [PubMed]
- Eric, E. Slicer_NonPlanar_Slicing. Available online: https://github.com/DrEricEbert/Slic3r_NonPlanar_Slicing (accessed on 6 February 2023).
- Arun, K.S.; Huang, T.S.; Blostein, S.D. Least-Squares Fitting of Two 3-D Point Sets. IEEE Trans. Pattern Anal. Mach. Intell. 1987, PAMI-9, 698–700. [Google Scholar] [CrossRef] [PubMed]
- Chiesa, I.; De Maria, C.; Lapomarda, A.; Fortunato, G.M.; Montemurro, F.; Di Gesù, R.; Tuan, R.S.; Vozzi, G.; Gottardi, R. Endothelial cells support osteogenesis in an in vitro vascularized bone model developed by 3D bioprinting. Biofabrication 2020, 12, 025013. [Google Scholar] [CrossRef]
- Bonatti, A.F.; Chiesa, I.; Vozzi, G.; De Maria, C. Open-source CAD-CAM simulator of the extrusion-based bioprinting process. Bioprinting 2021, 24, e00172. [Google Scholar] [CrossRef]
- O’connell, C.D.; Onofrillo, C.; Duchi, S.; Li, X.; Zhang, Y.; Tian, P.; Lu, L.; Trengove, A.; Quigley, A.; Gambhir, S.; et al. Evaluation of sterilisation methods for bio-ink components: Gelatin, gelatin methacryloyl, hyaluronic acid and hyaluronic acid methacryloyl. Biofabrication 2019, 11, 035003. [Google Scholar] [CrossRef]
- Takumi, I.; Akimoto, M. One-stage reconstruction using a vascularized calvarial flap for intractable scalp ulcers in relation with cranial implants without removing the whole prosthesis. Neurosurg. Rev. 2009, 32, 363–368. [Google Scholar] [CrossRef]


| Rheological Properties | Mechanical Properties |
|---|---|
| Herschel-Bulkley model | Pa |
| Flow index range (n): 0.02 | Pa·s |
| Consistency factor: 10 Pa·sn | Poisson modulus in range: 0.49 |
| Yield stress: 500 Pa | Density: 1000 kg/m3 |
| Duration | Accuracy | |
|---|---|---|
| Preparation of the surgical area (removal of bone fragments) | 15 min | N.A. |
| Acquisition of the 3D model (Computed Tomography) | 5 min | 0.4 mm |
| Path planning | 10 min | <0.1 mm |
| Registration | 5 min | ~1.5 mm |
| In situ bioprinting | 1 h 40 min | ~1.5 mm |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fortunato, G.M.; Sigismondi, S.; Nicoletta, M.; Condino, S.; Montemurro, N.; Vozzi, G.; Ferrari, V.; De Maria, C. Analysis of the Robotic-Based In Situ Bioprinting Workflow for the Regeneration of Damaged Tissues through a Case Study. Bioengineering 2023, 10, 560. https://doi.org/10.3390/bioengineering10050560
Fortunato GM, Sigismondi S, Nicoletta M, Condino S, Montemurro N, Vozzi G, Ferrari V, De Maria C. Analysis of the Robotic-Based In Situ Bioprinting Workflow for the Regeneration of Damaged Tissues through a Case Study. Bioengineering. 2023; 10(5):560. https://doi.org/10.3390/bioengineering10050560
Chicago/Turabian StyleFortunato, Gabriele Maria, Sofia Sigismondi, Matteo Nicoletta, Sara Condino, Nicola Montemurro, Giovanni Vozzi, Vincenzo Ferrari, and Carmelo De Maria. 2023. "Analysis of the Robotic-Based In Situ Bioprinting Workflow for the Regeneration of Damaged Tissues through a Case Study" Bioengineering 10, no. 5: 560. https://doi.org/10.3390/bioengineering10050560
APA StyleFortunato, G. M., Sigismondi, S., Nicoletta, M., Condino, S., Montemurro, N., Vozzi, G., Ferrari, V., & De Maria, C. (2023). Analysis of the Robotic-Based In Situ Bioprinting Workflow for the Regeneration of Damaged Tissues through a Case Study. Bioengineering, 10(5), 560. https://doi.org/10.3390/bioengineering10050560