WBM-DLNets: Wrapper-Based Metaheuristic Deep Learning Networks Feature Optimization for Enhancing Brain Tumor Detection
Abstract
1. Introduction
2. Methods and Materials
2.1. Brain MRI Dataset
2.2. Preprocessing
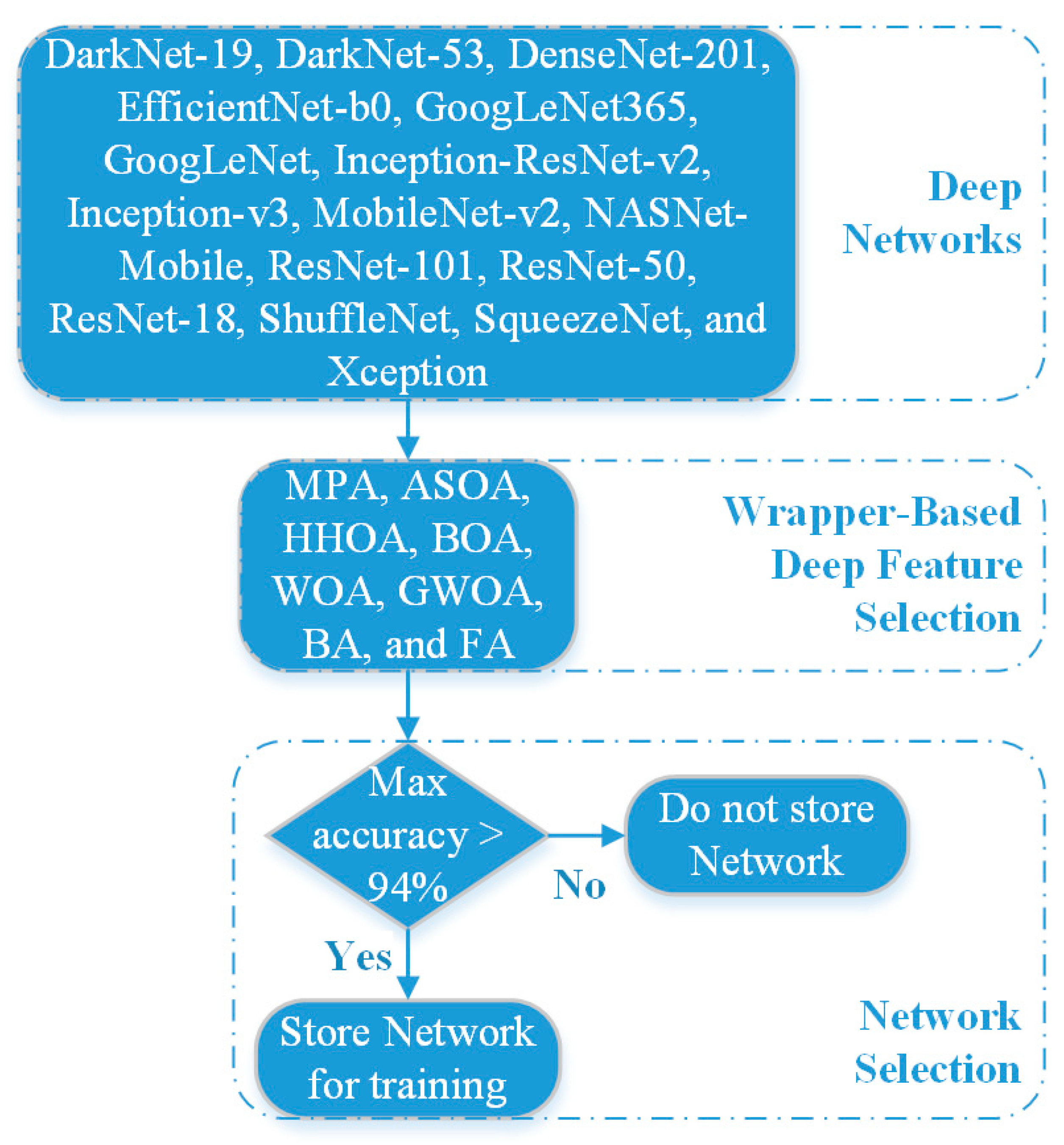
2.3. Deep Feature Extraction
2.4. Wrapper-Based Feature Selection Approach
2.4.1. Marine Predators Algorithm (MPA)
2.4.2. Atom Search Optimization Algorithm (ASOA)
2.4.3. Harris Hawks Optimization Algorithm (HHOA)
2.4.4. Butterfly Optimization Algorithm (BOA)
2.4.5. Whale Optimization Algorithm (WOA)
2.4.6. Grey Wolf Optimization Algorithm (GWOA)
2.4.7. Bat Algorithm (BA)
2.4.8. Firefly Algorithm (FA)
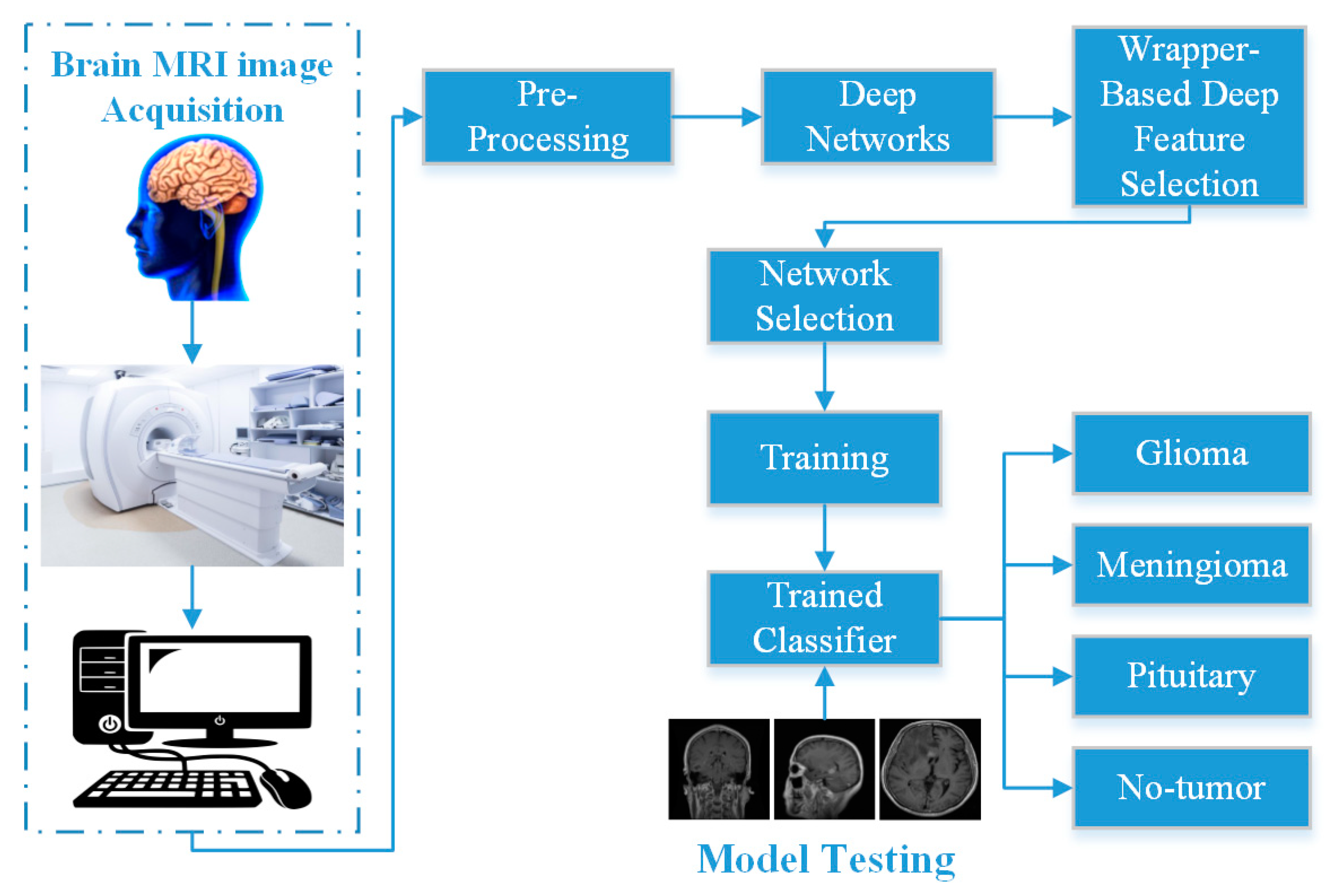
3. Proposed WBM-DLNets Framework for Brain Tumor Detection
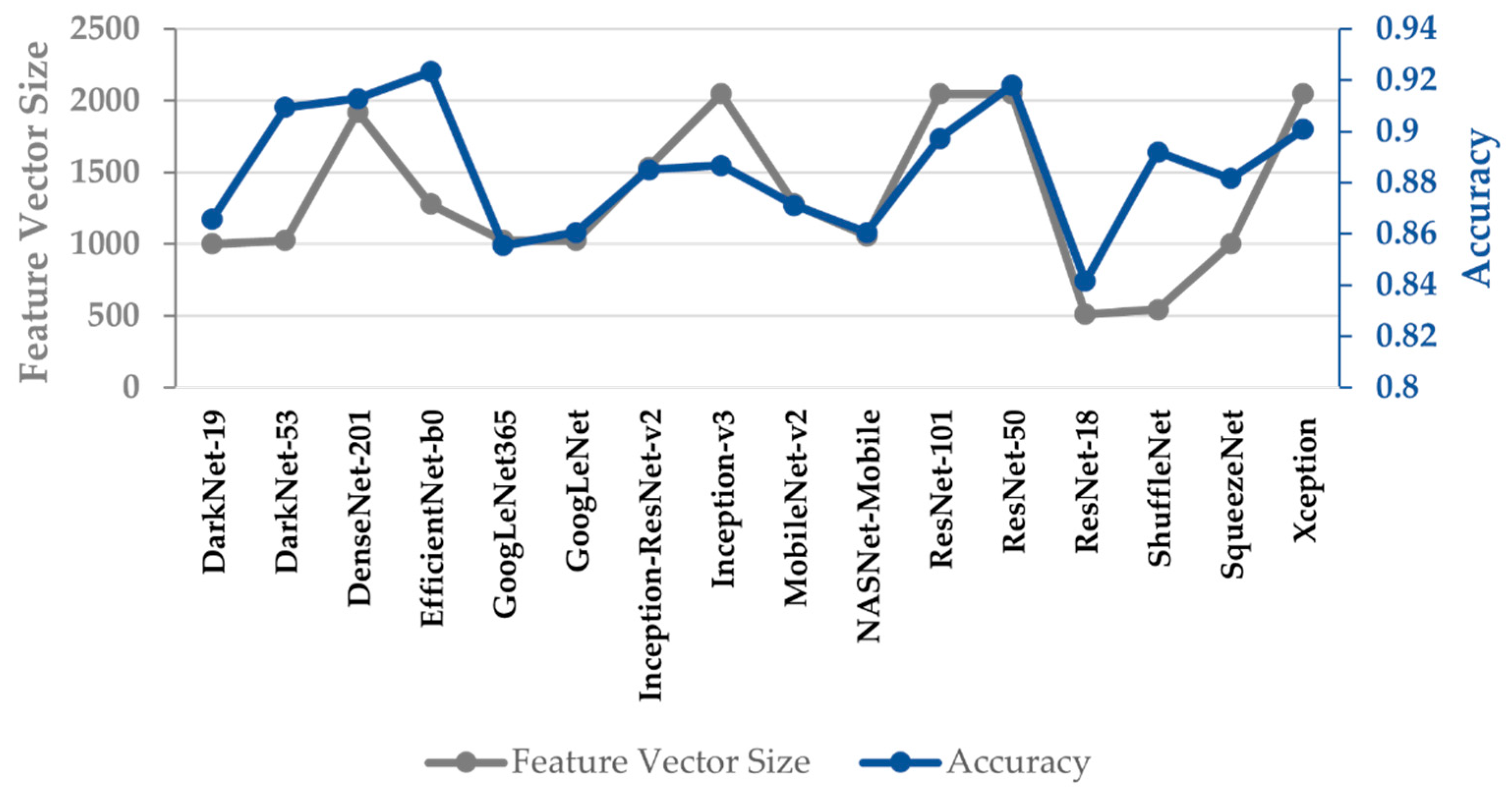
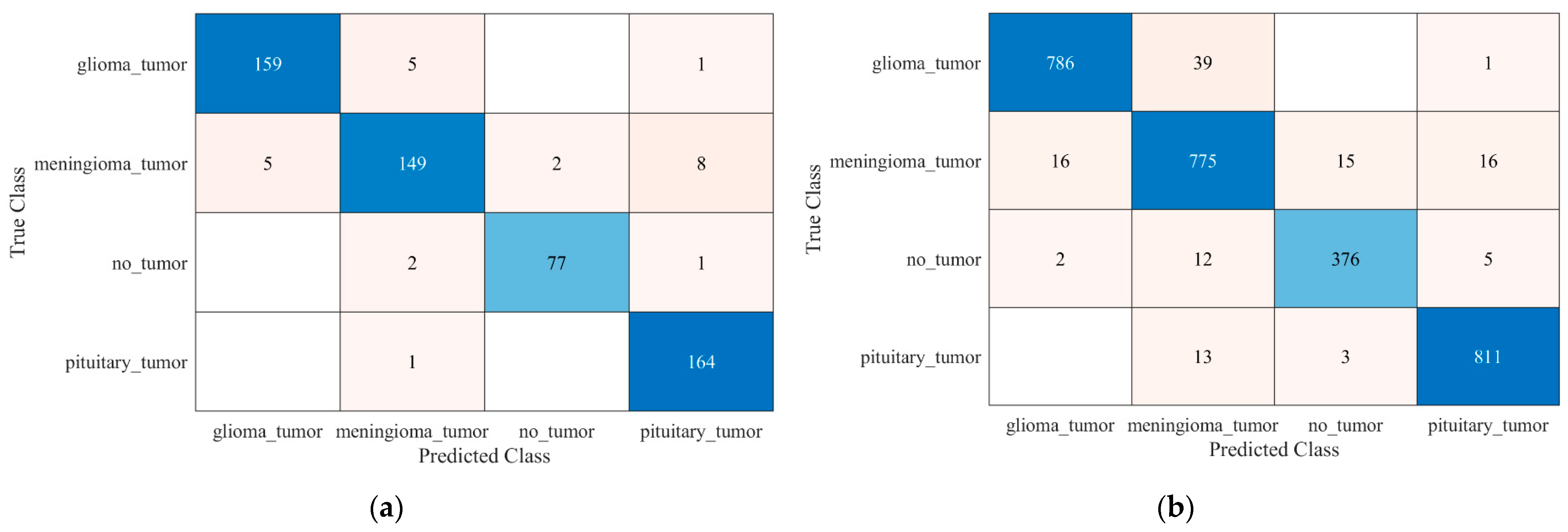
4. Results and Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Haj-Hosseini, N.; Milos, P.; Hildesjö, C.; Hallbeck, M.; Richter, J.; Wårdell, K. Fluorescence spectroscopy and optical coherence tomography for brain tumor detection. In Proceedings of the SPIE Photonics Europe, Biophotonics: Photonic Solutions for Better Health Care, Brussels, Belgium, 3–7 April 2016; pp. 9887–9896. [Google Scholar]
- Ren, W.; Hasanzade Bashkandi, A.; Afshar Jahanshahi, J.; Qasim Mohammad AlHamad, A.; Javaheri, D.; Mohammadi, M. Brain tumor diagnosis using a step-by-step methodology based on courtship learning-based water strider algorithm. Biomed. Signal Process. Control 2023, 83, 104614. [Google Scholar] [CrossRef]
- Brain Tumor Facts. Available online: https://braintumor.org/brain-tumors/about-brain-tumors/brain-tumor-facts/#:~:text=Today%2C%20an%20estimated%20700%2C000%20people,will%20be%20diagnosed%20in%202022 (accessed on 6 March 2023).
- American Cancer Society. Available online: www.cancer.org/cancer.html (accessed on 9 September 2021).
- American Society of Clinical Oncology. Available online: https://www.cancer.net/cancer-types/brain-tumor/diagnosis (accessed on 9 September 2021).
- Wulandari, A.; Sigit, R.; Bachtiar, M.M. Brain tumor segmentation to calculate percentage tumor using MRI. In Proceedings of the 2018 International Electronics Symposium on Knowledge Creation and Intelligent Computing (IES-KCIC), Surabaya, Indonesia, 29–30 October 2018; pp. 292–296. [Google Scholar]
- Xu, Z.; Sheykhahmad, F.R.; Ghadimi, N.; Razmjooy, N. Computer-aided diagnosis of skin cancer based on soft computing techniques. Open Med. 2020, 15, 860–871. [Google Scholar] [CrossRef]
- Kebede, S.R.; Debelee, T.G.; Schwenker, F.; Yohannes, D. Classifier Based Breast Cancer Segmentation. J. Biomim. Biomater. Biomed. Eng. 2020, 47, 41–61. [Google Scholar] [CrossRef]
- Debelee, T.G.; Amirian, M.; Ibenthal, A.; Palm, G.; Schwenker, F. Classification of Mammograms Using Convolutional Neural Network Based Feature Extraction. In Proceedings of the Information and Communication Technology for Development for Africa, Cham, Switzerland, 10–12 December 2018; pp. 89–98. [Google Scholar]
- Gab Allah, A.M.; Sarhan, A.M.; Elshennawy, N.M. Classification of Brain MRI Tumor Images Based on Deep Learning PGGAN Augmentation. Diagnostics 2021, 11, 2343. [Google Scholar] [CrossRef] [PubMed]
- Alanazi, M.F.; Ali, M.U.; Hussain, S.J.; Zafar, A.; Mohatram, M.; Irfan, M.; AlRuwaili, R.; Alruwaili, M.; Ali, N.H.; Albarrak, A.M. Brain Tumor/Mass Classification Framework Using Magnetic-Resonance-Imaging-Based Isolated and Developed Transfer Deep-Learning Model. Sensors 2022, 22, 372. [Google Scholar] [CrossRef]
- Almalki, Y.E.; Ali, M.U.; Kallu, K.D.; Masud, M.; Zafar, A.; Alduraibi, S.K.; Irfan, M.; Basha, M.A.A.; Alshamrani, H.A.; Alduraibi, A.K.; et al. Isolated Convolutional-Neural-Network-Based Deep-Feature Extraction for Brain Tumor Classification Using Shallow Classifier. Diagnostics 2022, 12, 1793. [Google Scholar] [CrossRef]
- Debelee, T.G.; Kebede, S.R.; Schwenker, F.; Shewarega, Z.M. Deep Learning in Selected Cancers’ Image Analysis—A Survey. J. Imaging 2020, 6, 121. [Google Scholar] [CrossRef] [PubMed]
- Pandian, R.; Vedanarayanan, V.; Ravi Kumar, D.N.S.; Rajakumar, R. Detection and classification of lung cancer using CNN and Google net. Meas. Sens. 2022, 24, 100588. [Google Scholar] [CrossRef]
- Allah AM, G.; Sarhan, A.M.; Elshennawy, N.M. Edge U-Net: Brain tumor segmentation using MRI based on deep U-Net model with boundary information. Expert Syst. Appl. 2023, 213, 118833. [Google Scholar] [CrossRef]
- Ma, C.; Luo, G.; Wang, K. Concatenated and Connected Random Forests with Multiscale Patch Driven Active Contour Model for Automated Brain Tumor Segmentation of MR Images. IEEE Trans. Med. Imaging 2018, 37, 1943–1954. [Google Scholar] [CrossRef]
- Almalki, Y.E.; Ali, M.U.; Ahmed, W.; Kallu, K.D.; Zafar, A.; Alduraibi, S.K.; Irfan, M.; Basha, M.A.A.; Alshamrani, H.A.; Alduraibi, A.K. Robust Gaussian and Nonlinear Hybrid Invariant Clustered Features Aided Approach for Speeded Brain Tumor Diagnosis. Life 2022, 12, 1084. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.U.; Kallu, K.D.; Masood, H.; Hussain, S.J.; Ullah, S.; Byun, J.H.; Zafar, A.; Kim, K.S. A Robust Computer-Aided Automated Brain Tumor Diagnosis Approach Using PSO-ReliefF Optimized Gaussian and Non-Linear Feature Space. Life 2022, 12, 2036. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R. SVM classification an approach on detecting abnormality in brain MRI images. Int. J. Eng. Res. Appl. 2013, 3, 1686–1690. [Google Scholar]
- Ayachi, R.; Ben Amor, N. Brain tumor segmentation using support vector machines. In Proceedings of the Symbolic and Quantitative Approaches to Reasoning with Uncertainty: 10th European Conference, ECSQARU 2009, Verona, Italy, 1–3 July 2009; Proceedings 10, pp. 736–747. [Google Scholar]
- Almahfud, M.A.; Setyawan, R.; Sari, C.A.; Setiadi, D.R.I.M.; Rachmawanto, E.H. An Effective MRI Brain Image Segmentation using Joint Clustering (K-Means and Fuzzy C-Means). In Proceedings of the 2018 International Seminar on Research of Information Technology and Intelligent Systems (ISRITI), Yogyakarta, Indonesia, 21–22 November 2018; pp. 11–16. [Google Scholar]
- Abdel-Maksoud, E.; Elmogy, M.; Al-Awadi, R. Brain tumor segmentation based on a hybrid clustering technique. Egypt. Inform. J. 2015, 16, 71–81. [Google Scholar] [CrossRef]
- Kaya, I.E.; Pehlivanlı, A.Ç.; Sekizkardeş, E.G.; Ibrikci, T. PCA based clustering for brain tumor segmentation of T1w MRI images. Comput. Methods Programs Biomed. 2017, 140, 19–28. [Google Scholar] [CrossRef]
- Nazir, M.; Shakil, S.; Khurshid, K. Role of deep learning in brain tumor detection and classification (2015 to 2020): A review. Comput. Med. Imaging Graph. 2021, 91, 101940. [Google Scholar] [CrossRef] [PubMed]
- Pereira, S.; Meier, R.; Alves, V.; Reyes, M.; Silva, C.A. Automatic Brain Tumor Grading from MRI Data Using Convolutional Neural Networks and Quality Assessment; Springer: Cham, Switzerland, 2018; pp. 106–114. [Google Scholar]
- Abiwinanda, N.; Hanif, M.; Hesaputra, S.T.; Handayani, A.; Mengko, T.R. Brain Tumor Classification Using Convolutional Neural Network; Springer: Singapore, 2019; pp. 183–189. [Google Scholar]
- Badža, M.M.; Barjaktarović, M.Č. Classification of Brain Tumors from MRI Images Using a Convolutional Neural Network. Appl. Sci. 2020, 10, 1999. [Google Scholar] [CrossRef]
- Irmak, E. Multi-Classification of Brain Tumor MRI Images Using Deep Convolutional Neural Network with Fully Optimized Framework. Iran. J. Sci. Technol. Trans. Electr. Eng. 2021, 45, 1015–1036. [Google Scholar] [CrossRef]
- Deepak, S.; Ameer, P.M. Brain tumor classification using deep CNN features via transfer learning. Comput. Biol. Med. 2019, 111, 103345. [Google Scholar] [CrossRef]
- Çinar, A.; Yildirim, M. Detection of tumors on brain MRI images using the hybrid convolutional neural network architecture. Med. Hypotheses 2020, 139, 109684. [Google Scholar] [CrossRef]
- Kang, J.; Ullah, Z.; Gwak, J. MRI-Based Brain Tumor Classification Using Ensemble of Deep Features and Machine Learning Classifiers. Sensors 2021, 21, 2222. [Google Scholar] [CrossRef] [PubMed]
- Dokeroglu, T.; Deniz, A.; Kiziloz, H.E. A comprehensive survey on recent metaheuristics for feature selection. Neurocomputing 2022, 494, 269–296. [Google Scholar] [CrossRef]
- Chakrabarty, N.; Kanchan, S. Brain Tumor Classification (MRI). Available online: https://www.kaggle.com/datasets/sartajbhuvaji/brain-tumor-classification-mri?select=Training (accessed on 17 March 2022).
- Rosebrock, A. Finding extreme points in contours with Open CV. Available online: https://www.pyimagesearch.com/2016/04/11/finding-extreme-points-in-contours-with-opencv/ (accessed on 9 September 2021).
- Dash, M.; Liu, H. Feature selection for classification. Intell. Data Anal. 1997, 1, 131–156. [Google Scholar] [CrossRef]
- Guyon, I.; Elisseeff, A. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- Kohavi, R.; John, G.H. Wrappers for feature subset selection. Artif. Intell. 1997, 97, 273–324. [Google Scholar] [CrossRef]
- Abdel-Basset, M.; Abdel-Fatah, L.; Sangaiah, A.K. Metaheuristic algorithms: A comprehensive review. Comput. Intell. Multimed. Big Data Cloud Eng. Appl. 2018, 185–231. [Google Scholar] [CrossRef]
- Yang, X.-S. Chapter 1—Introduction to Algorithms. In Nature-Inspired Optimization Algorithms, 2nd ed.; Yang, X.-S., Ed.; Academic Press: Cambridge, MA, USA, 2021; pp. 1–22. [Google Scholar]
- Liu, W.; Wang, J. A Brief Survey on Nature-Inspired Metaheuristics for Feature Selection in Classification in this Decade. In Proceedings of the 2019 IEEE 16th International Conference on Networking, Sensing and Control (ICNSC), Banff, AB, Canada, 9–11 May 2019; pp. 424–429. [Google Scholar]
- Agrawal, P.; Abutarboush, H.F.; Ganesh, T.; Mohamed, A.W. Metaheuristic Algorithms on Feature Selection: A Survey of One Decade of Research (2009–2019). IEEE Access 2021, 9, 26766–26791. [Google Scholar] [CrossRef]
- Faramarzi, A.; Heidarinejad, M.; Mirjalili, S.; Gandomi, A.H. Marine Predators Algorithm: A nature-inspired metaheuristic. Expert Syst. Appl. 2020, 152, 113377. [Google Scholar] [CrossRef]
- Rai, R.; Dhal, K.G.; Das, A.; Ray, S. An Inclusive Survey on Marine Predators Algorithm: Variants and Applications. Arch. Comput. Methods Eng. 2023. [Google Scholar] [CrossRef]
- Ewees, A.A.; Ismail, F.H.; Ghoniem, R.M.; Gaheen, M.A. Enhanced Marine Predators Algorithm for Solving Global Optimization and Feature Selection Problems. Mathematics 2022, 10, 4154. [Google Scholar] [CrossRef]
- Zhao, W.; Wang, L.; Zhang, Z. Atom search optimization and its application to solve a hydrogeologic parameter estimation problem. Knowl.-Based Syst. 2019, 163, 283–304. [Google Scholar] [CrossRef]
- Kamel, S.; Hamour, H.; Ahmed, M.H.; Nasrat, L. Atom Search optimization Algorithm for Optimal Radial Distribution System Reconfiguration. In Proceedings of the 2019 International Conference on Computer, Control, Electrical, and Electronics Engineering (ICCCEEE), Khartoum, Sudan, 21–23 September 2019; pp. 1–5. [Google Scholar]
- Bairathi, D.; Gopalani, D. A novel swarm intelligence based optimization method: Harris' hawk optimization. In Proceedings of the Intelligent Systems Design and Applications: 18th International Conference on Intelligent Systems Design and Applications (ISDA 2018), Vellore, India, 6–8 December 2018; Volume 2, pp. 832–842. [Google Scholar]
- Heidari, A.A.; Mirjalili, S.; Faris, H.; Aljarah, I.; Mafarja, M.; Chen, H. Harris hawks optimization: Algorithm and applications. Future Gener. Comput. Syst. 2019, 97, 849–872. [Google Scholar] [CrossRef]
- Arora, S.; Singh, S. Butterfly optimization algorithm: A novel approach for global optimization. Soft Comput. 2019, 23, 715–734. [Google Scholar] [CrossRef]
- Zhou, H.; Cheng, H.-Y.; Wei, Z.-L.; Zhao, X.; Tang, A.-D.; Xie, L. A Hybrid Butterfly Optimization Algorithm for Numerical Optimization Problems. Comput. Intell. Neurosci. 2021, 2021, 7981670. [Google Scholar] [CrossRef]
- Mirjalili, S.; Lewis, A. The whale optimization algorithm. Adv. Eng. Softw. 2016, 95, 51–67. [Google Scholar] [CrossRef]
- Mirjalili, S.; Mirjalili, S.M.; Lewis, A. Grey wolf optimizer. Adv. Eng. Softw. 2014, 69, 46–61. [Google Scholar] [CrossRef]
- Emary, E.; Zawbaa, H.M.; Hassanien, A.E. Binary grey wolf optimization approaches for feature selection. Neurocomputing 2016, 172, 371–381. [Google Scholar] [CrossRef]
- Tu, Q.; Chen, X.; Liu, X. Multi-strategy ensemble grey wolf optimizer and its application to feature selection. Appl. Soft Comput. 2019, 76, 16–30. [Google Scholar] [CrossRef]
- Yang, X.-S. A new metaheuristic bat-inspired algorithm. In Nature Inspired Cooperative Strategies for Optimization (NICSO 2010); Springer: Berlin/Heidelberg, Germany, 2010; pp. 65–74. [Google Scholar]
- Yang, X.-S. Chapter 11—Bat Algorithms. In Nature-Inspired Optimization Algorithms, 2nd ed.; Yang, X.-S., Ed.; Academic Press: Cambridge, MA, USA, 2021; pp. 157–173. [Google Scholar]
- Yang, X.-S.; He, X. Bat algorithm: Literature review and applications. Int. J. Bio-Inspired Comput. 2013, 5, 141–149. [Google Scholar] [CrossRef]
- Yang, X.-S. Firefly algorithm, stochastic test functions and design optimisation. Int. J. Bio-Inspired Comput. 2010, 2, 78–84. [Google Scholar] [CrossRef]
- Fister, I.; Fister Jr, I.; Yang, X.-S.; Brest, J. A comprehensive review of firefly algorithms. Swarm Evol. Comput. 2013, 13, 34–46. [Google Scholar] [CrossRef]
- Cristianini, N.; Shawe-Taylor, J. An Introduction to Support Vector Machines and Other Kernel-Based Learning Methods; Cambridge University Press: Cambridge, UK, 2000. [Google Scholar]
- Smola, A.J.; Schölkopf, B. A tutorial on support vector regression. Stat. Comput. 2004, 14, 199–222. [Google Scholar] [CrossRef]
- Jun, C. Brain Tumor Dataset. 2017. Available online: https://figshare.com/articles/dataset/brain_tumor_dataset/1512427 (accessed on 9 September 2021).
- Cheng, J.; Huang, W.; Cao, S.; Yang, R.; Yang, W.; Yun, Z.; Wang, Z.; Feng, Q. Enhanced Performance of Brain Tumor Classification via Tumor Region Augmentation and Partition. PLoS ONE 2015, 10, e0140381. [Google Scholar] [CrossRef]
- Rehman, A.; Naz, S.; Razzak, M.I.; Akram, F.; Imran, M. A Deep Learning-Based Framework for Automatic Brain Tumors Classification Using Transfer Learning. Circuits Syst. Signal Process. 2020, 39, 757–775. [Google Scholar] [CrossRef]



| Glioma Tumor | Meningioma Tumor | No Tumor | Pituitary Tumor | |
|---|---|---|---|---|
| Brain MRI images | 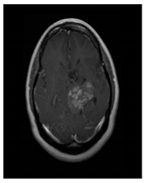 | 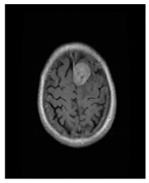 | 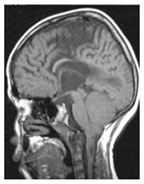 | 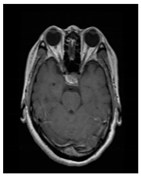 |
| No. of images per class | 826 | 822 | 395 | 827 |
| MPA | ASOA | HHOA | BOA | WOA | GWOA | BA | FA |
|---|---|---|---|---|---|---|---|
| Number of iterations = 50 | Number of iterations = 50 | Number of iterations = 50 | Number of iterations = 50 | Number of iterations = 50 | Number of iterations = 50 | Number of iterations = 50 | Number of iterations = 50 |
| Population size = 10 | Population size = 10 | Population size = 10 | Population size = 10 | Population size = 10 | Population size = 10 | Population size = 10 | Population size = 10 |
| Fish aggregating devices effect = 0.2 | Depth weight = 50 | Levy component = 1.5 | Modular modality = 0.01 | Constant = 1 | Maximum frequency = 2 | Absorption coefficient = 1 | |
| Constant = 0.5 | Multiplier weight = 0.2 | Switch probability = 0.8 | Minimum frequency = 0 | Constant = 1 | |||
| Levy component = 1.5 | Constant = 0.9 | Light amplitude =1 | |||||
| Maximum loudness = 2 | Control alpha = 0.97 | ||||||
| Maximum pulse rate = 1 |
| Network | MPA | ASOA | HHOA | BOA | WOA | GWOA | BA | FA | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accuracy | Feature Vector Size | Accuracy | Feature Vector Size | Accuracy | Feature Vector Size | Accuracy | Feature Vector Size | Accuracy | Feature Vector Size | Accuracy | Feature Vector Size | Accuracy | Feature Vector Size | Accuracy | Feature Vector Size | |
| DarkNet-19 | 0.906 | 267 | 0.895 | 516 | 0.894 | 172 | 0.880 | 413 | 0.895 | 304 | 0.909 | 303 | 0.894 | 494 | 0.892 | 447 |
| DarkNet-53 | 0.930 | 732 | 0.922 | 504 | 0.923 | 575 | 0.913 | 481 | 0.918 | 712 | 0.934 | 362 | 0.913 | 491 | 0.920 | 521 |
| DenseNet-201 | 0.930 | 939 | 0.916 | 979 | 0.922 | 1270 | 0.894 | 912 | 0.909 | 970 | 0.946 | 651 | 0.908 | 964 | 0.918 | 950 |
| EfficientNet-b0 | 0.944 | 821 | 0.949 | 641 | 0.939 | 862 | 0.925 | 551 | 0.944 | 654 | 0.939 | 451 | 0.934 | 669 | 0.936 | 621 |
| GoogLeNet365 | 0.894 | 388 | 0.901 | 499 | 0.885 | 487 | 0.878 | 472 | 0.882 | 542 | 0.899 | 401 | 0.887 | 522 | 0.887 | 479 |
| GoogLeNet | 0.889 | 667 | 0.873 | 531 | 0.883 | 501 | 0.852 | 521 | 0.873 | 690 | 0.895 | 338 | 0.864 | 518 | 0.871 | 493 |
| Inception-ResNet-v2 | 0.915 | 533 | 0.909 | 753 | 0.908 | 739 | 0.909 | 802 | 0.902 | 782 | 0.925 | 488 | 0.904 | 754 | 0.908 | 763 |
| Inception-v3 | 0.904 | 878 | 0.911 | 997 | 0.908 | 1023 | 0.895 | 764 | 0.895 | 1174 | 0.923 | 768 | 0.901 | 990 | 0.895 | 966 |
| MobileNet-v2 | 0.913 | 812 | 0.902 | 647 | 0.890 | 770 | 0.883 | 623 | 0.887 | 738 | 0.894 | 512 | 0.892 | 659 | 0.897 | 633 |
| NASNet-Mobile | 0.885 | 575 | 0.869 | 505 | 0.871 | 714 | 0.864 | 461 | 0.873 | 854 | 0.887 | 364 | 0.866 | 497 | 0.871 | 549 |
| ResNet-101 | 0.925 | 1279 | 0.927 | 1057 | 0.915 | 1231 | 0.923 | 927 | 0.915 | 1536 | 0.927 | 826 | 0.913 | 985 | 0.911 | 1005 |
| ResNet-50 | 0.934 | 1228 | 0.937 | 1013 | 0.939 | 1254 | 0.916 | 877 | 0.932 | 1068 | 0.939 | 692 | 0.927 | 1017 | 0.934 | 1016 |
| ResNet-18 | 0.878 | 274 | 0.876 | 242 | 0.875 | 315 | 0.854 | 214 | 0.864 | 428 | 0.887 | 210 | 0.880 | 242 | 0.873 | 233 |
| ShuffleNet | 0.916 | 217 | 0.904 | 274 | 0.911 | 374 | 0.887 | 271 | 0.895 | 321 | 0.918 | 218 | 0.894 | 286 | 0.901 | 256 |
| SqueezeNet | 0.904 | 519 | 0.913 | 499 | 0.902 | 619 | 0.885 | 516 | 0.894 | 852 | 0.901 | 385 | 0.889 | 485 | 0.890 | 483 |
| Xception | 0.920 | 809 | 0.929 | 1035 | 0.916 | 1149 | 0.908 | 889 | 0.911 | 978 | 0.930 | 775 | 0.916 | 1002 | 0.915 | 1009 |
| Validation | Class | TPR (%) | FNR (%) | PPV (%) | FDR (%) | Accuracy (%) |
|---|---|---|---|---|---|---|
| 0.2-holdout | glioma_tumor | 96.4 | 3.6 | 97.0 | 3.0 | 95.6 |
| meningioma_tumor | 90.9 | 9.2 | 94.9 | 5.1 | ||
| no_tumor | 96.3 | 3.8 | 97.5 | 2.5 | ||
| pituitary_tumor | 99.4 | 0.6 | 94.3 | 5.7 | ||
| Five-fold cross-validation | glioma_tumor | 95.2 | 4.8 | 97.8 | 2.2 | 95.7 |
| meningioma_tumor | 94.3 | 5.7 | 92.4 | 7.6 | ||
| no_tumor | 95.2 | 4.8 | 95.4 | 4.6 | ||
| pituitary_tumor | 98.1 | 1.9 | 97.4 | 2.6 |
| Validation | Class | TPR (%) | FNR (%) | PPV (%) | FDR (%) | Accuracy (%) |
|---|---|---|---|---|---|---|
| 0.2-holdout | glioma_tumor | 95.8 | 4.2 | 98.9 | 1.1 | 96.7 |
| meningioma_tumor | 95.7 | 4.3 | 91.2 | 8.8 | ||
| pituitary_tumor | 98.9 | 1.1 | 97.9 | 2.1 | ||
| Five-fold cross-validation | glioma_tumor | 96.9 | 3.1 | 97.7 | 2.3 | 96.6 |
| meningioma_tumor | 93.1 | 6.9 | 92.6 | 7.4 | ||
| pituitary_tumor | 98.7 | 1.3 | 98.0 | 2.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, M.U.; Hussain, S.J.; Zafar, A.; Bhutta, M.R.; Lee, S.W. WBM-DLNets: Wrapper-Based Metaheuristic Deep Learning Networks Feature Optimization for Enhancing Brain Tumor Detection. Bioengineering 2023, 10, 475. https://doi.org/10.3390/bioengineering10040475
Ali MU, Hussain SJ, Zafar A, Bhutta MR, Lee SW. WBM-DLNets: Wrapper-Based Metaheuristic Deep Learning Networks Feature Optimization for Enhancing Brain Tumor Detection. Bioengineering. 2023; 10(4):475. https://doi.org/10.3390/bioengineering10040475
Chicago/Turabian StyleAli, Muhammad Umair, Shaik Javeed Hussain, Amad Zafar, Muhammad Raheel Bhutta, and Seung Won Lee. 2023. "WBM-DLNets: Wrapper-Based Metaheuristic Deep Learning Networks Feature Optimization for Enhancing Brain Tumor Detection" Bioengineering 10, no. 4: 475. https://doi.org/10.3390/bioengineering10040475
APA StyleAli, M. U., Hussain, S. J., Zafar, A., Bhutta, M. R., & Lee, S. W. (2023). WBM-DLNets: Wrapper-Based Metaheuristic Deep Learning Networks Feature Optimization for Enhancing Brain Tumor Detection. Bioengineering, 10(4), 475. https://doi.org/10.3390/bioengineering10040475







