Advanced Vaccine Design Strategies against SARS-CoV-2 and Emerging Variants
Abstract
1. Introduction
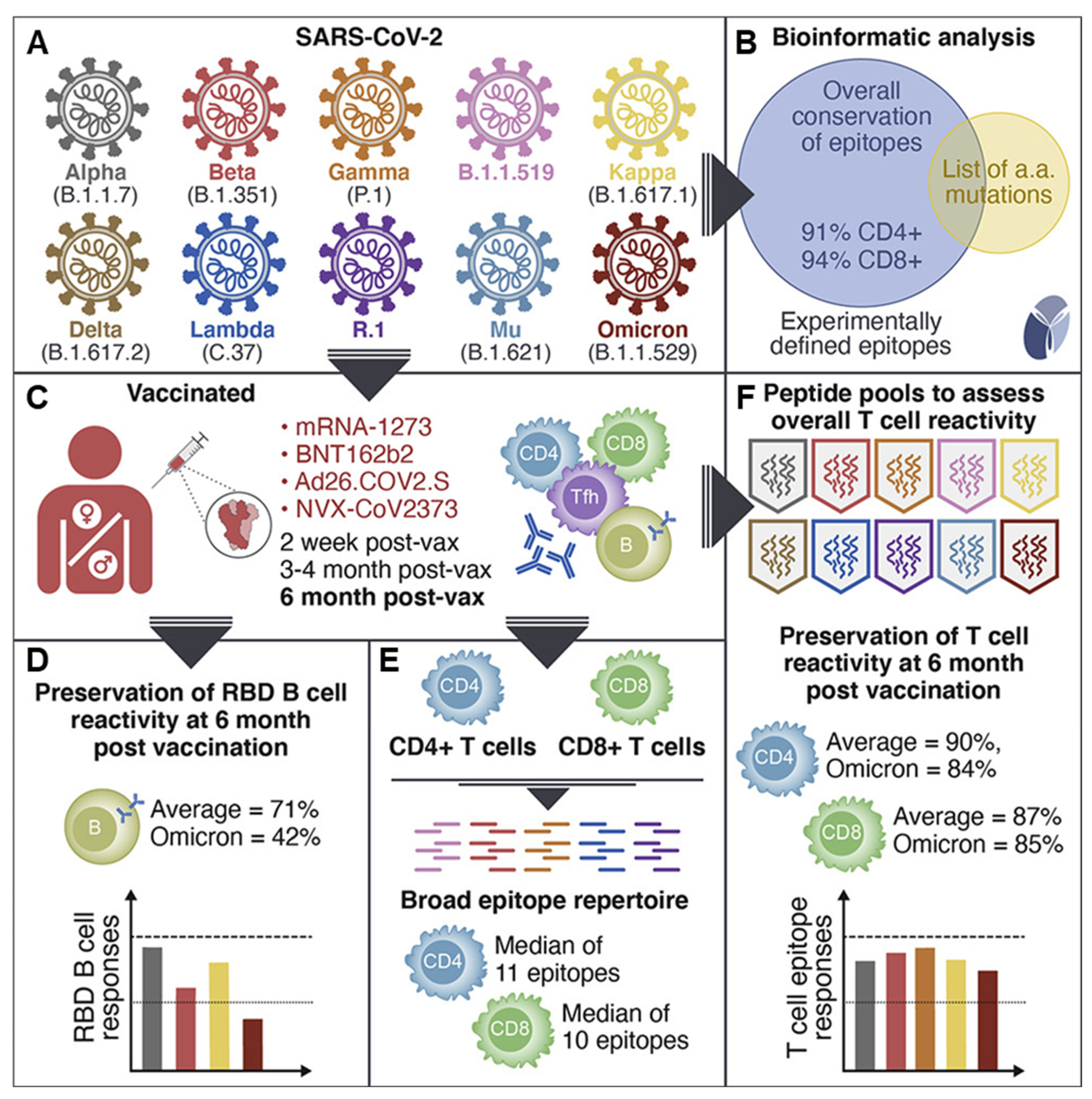
2. Overview of SARS-CoV-2 and Its Variants
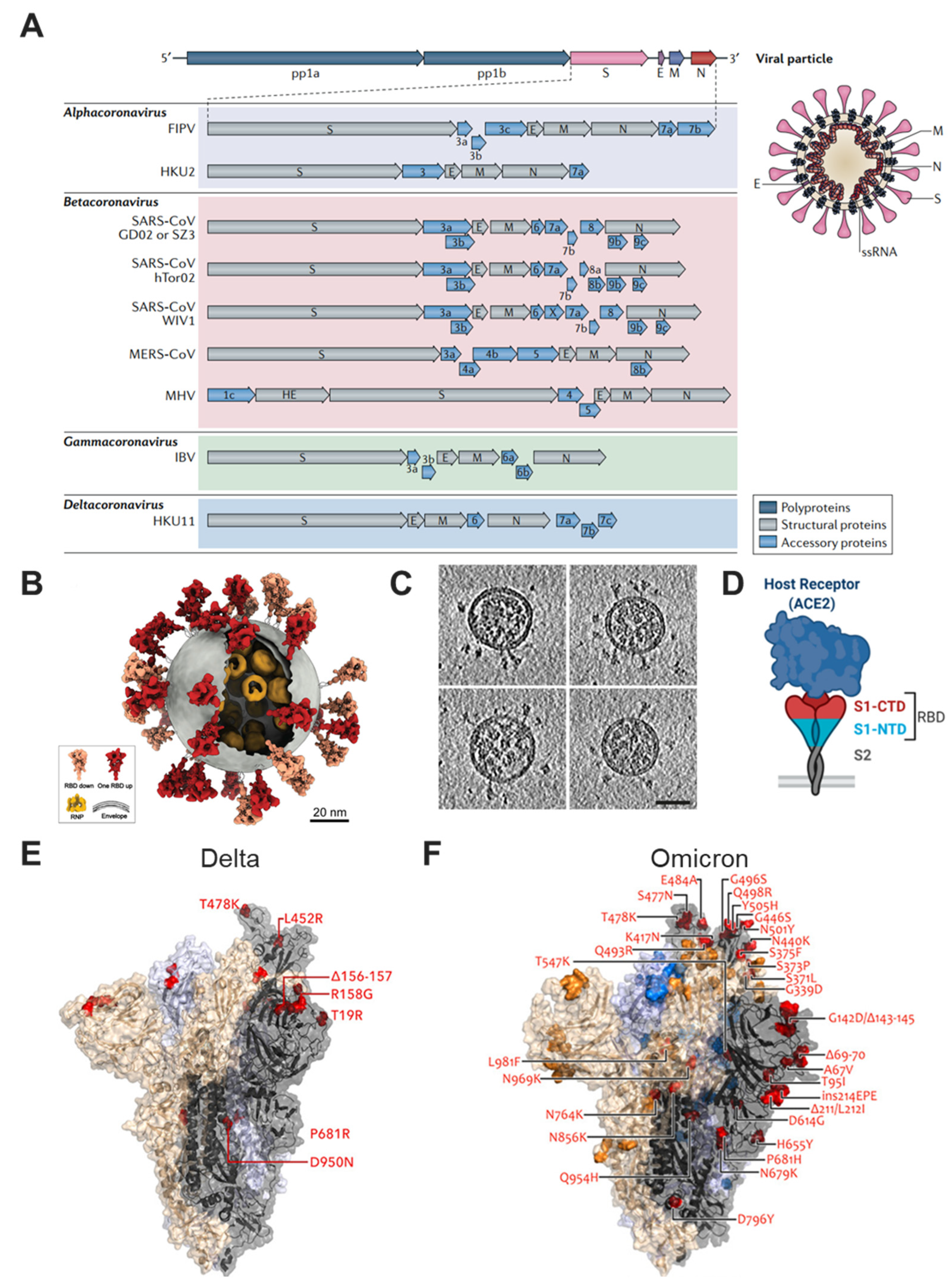
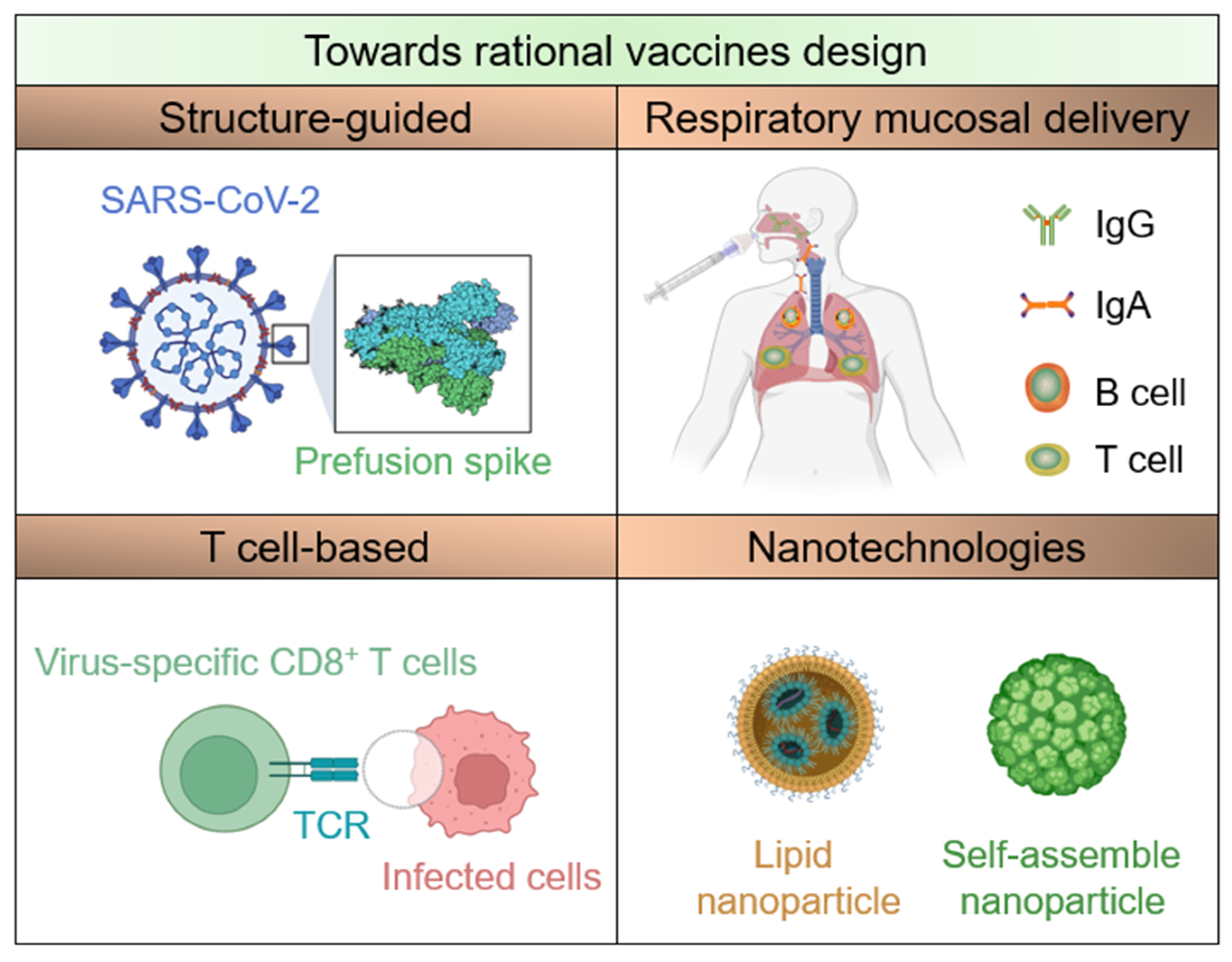
3. Structure-Guided Vaccine Design

4. T-Cell-Based Vaccines
5. Respiratory Mucosal Delivery
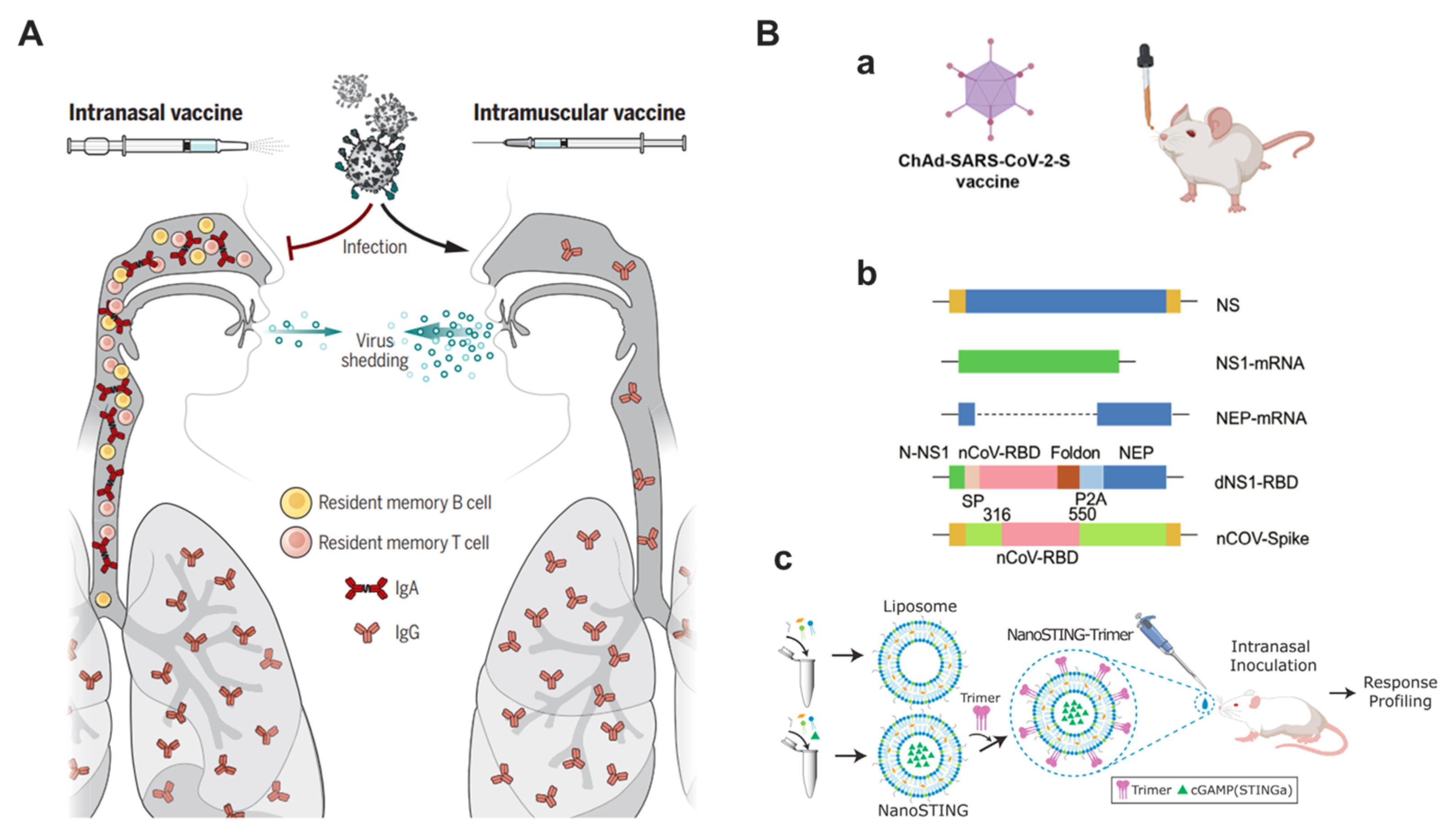
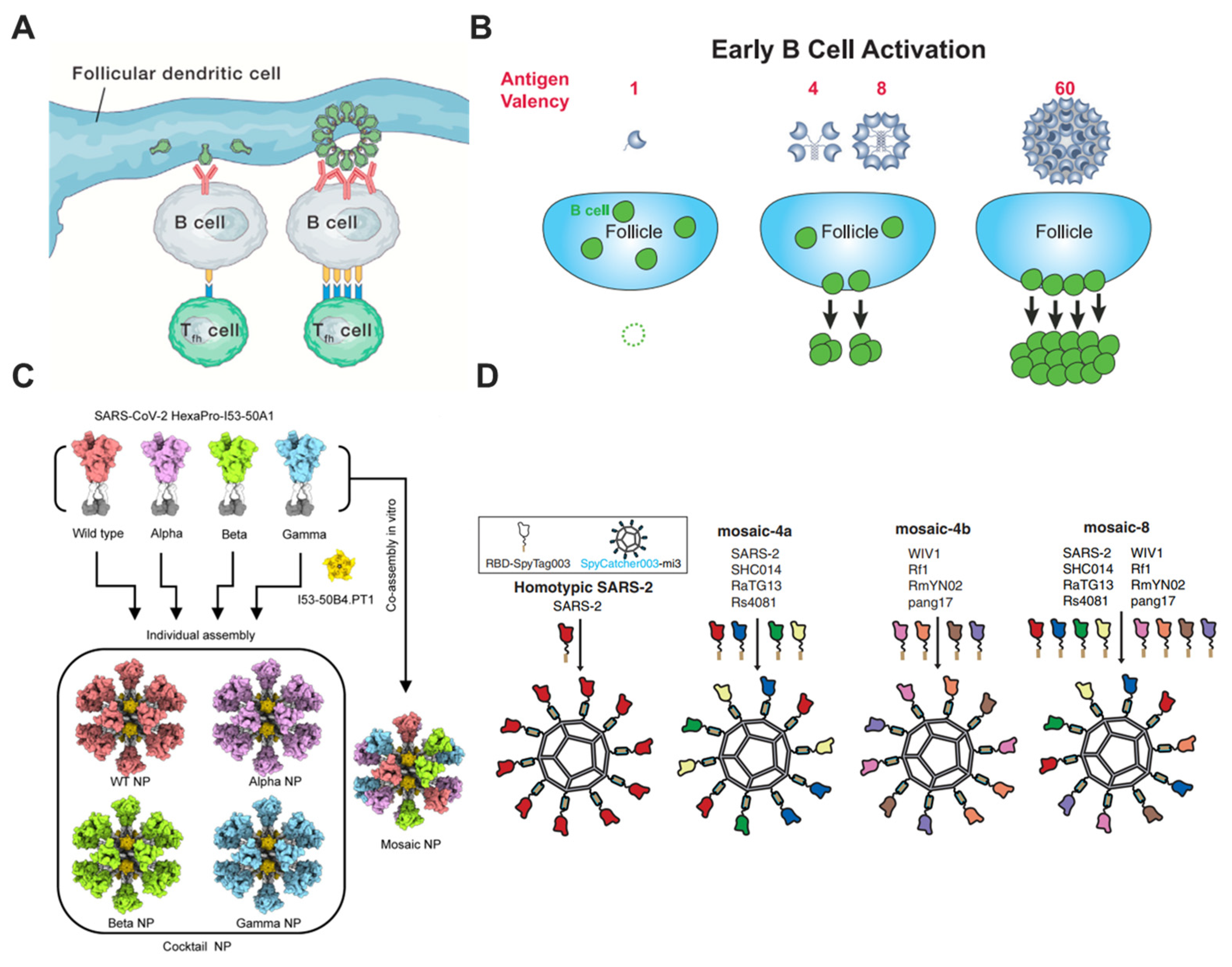
6. Nanotechnologies
7. Challenges and Future Perspective
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, P.; Wang, X. COVID-19: A new challenge for human beings. Cell. Mol. Immunol. 2020, 17, 555–557. [Google Scholar] [CrossRef] [PubMed]
- De Gregorio, E.; Rappuoli, R. From empiricism to rational design: A personal perspective of the evolution of vaccine development. Nat. Rev. Immunol. 2014, 14, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Lee, L.A.; Liu, Y.; Scherpbier, R.W.; Wen, N.; Zhang, G.; Zhu, X.; Ning, G.; Wang, F.; Li, Y.; et al. Vaccine-preventable disease control in the People’s Republic of China: 1949–2016. Vaccine 2018, 36, 8131–8137. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Wang, Y.; Cao, L.; Wang, Y.; Zhao, Q.; Tang, S.; Gong, W.; Guo, L.; Liu, Z.; Wen, Z.; et al. Impact of immunization programs on 11 childhood vaccine-preventable diseases in China: 1950–2018. Innovation 2021, 2, 100113. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, Y.; Huang, B.; Deng, W.; Quan, Y.; Wang, W.; Xu, W.; Zhao, Y.; Li, N.; Zhang, J.; et al. Development of an Inactivated Vaccine Candidate, BBIBP-CorV, with Potent Protection against SARS-CoV-2. Cell 2020, 182, 713–721.e719. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Bao, L.; Mao, H.; Wang, L.; Xu, K.; Yang, M.; Li, Y.; Zhu, L.; Wang, N.; Lv, Z.; et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science 2020, 369, 77–81. [Google Scholar] [CrossRef]
- Zhu, M. SARS Immunity and Vaccination. Cell Mol. Immunol. 2004, 1, 193–198. [Google Scholar]
- Lu, L.; Liu, Q.; Du, L.; Jiang, S. Middle East respiratory syndrome coronavirus (MERS-CoV): Challenges in identifying its source and controlling its spread. Microbes Infect. 2013, 15, 625–629. [Google Scholar] [CrossRef]
- He, W.; Zhang, W.; Yan, H.; Xu, H.; Xie, Y.; Wu, Q.; Wang, C.; Dong, G. Distribution and evolution of H1N1 influenza A viruses with adamantanes-resistant mutations worldwide from 1918 to 2019. J. Med. Virol. 2021, 93, 3473–3483. [Google Scholar] [CrossRef]
- Al Kaabi, N.; Zhang, Y.; Xia, S.; Yang, Y.; Al Qahtani, M.M.; Abdulrazzaq, N.; Al Nusair, M.; Hassany, M.; Jawad, J.S.; Abdalla, J.; et al. Effect of 2 Inactivated SARS-CoV-2 Vaccines on Symptomatic COVID-19 Infection in Adults: A Randomized Clinical Trial. JAMA 2021, 326, 35–45. [Google Scholar] [CrossRef]
- Trimpert, J.; Dietert, K.; Firsching, T.C.; Ebert, N.; Thi Nhu Thao, T.; Vladimirova, D.; Kaufer, S.; Labroussaa, F.; Abdelgawad, A.; Conradie, A.; et al. Development of safe and highly protective live-attenuated SARS-CoV-2 vaccine candidates by genome recoding. Cell Rep. 2021, 36, 109493. [Google Scholar] [CrossRef]
- Sanchez-Felipe, L.; Vercruysse, T.; Sharma, S.; Ma, J.; Lemmens, V.; Van Looveren, D.; Arkalagud Javarappa, M.P.; Boudewijns, R.; Malengier-Devlies, B.; Liesenborghs, L.; et al. A single-dose live-attenuated YF17D-vectored SARS-CoV-2 vaccine candidate. Nature 2021, 590, 320–325. [Google Scholar] [CrossRef]
- Wu, Y.; Huang, X.; Yuan, L.; Wang, S.; Zhang, Y.; Xiong, H.; Chen, R.; Ma, J.; Qi, R.; Nie, M.; et al. A recombinant spike protein subunit vaccine confers protective immunity against SARS-CoV-2 infection and transmission in hamsters. Sci. Transl. Med. 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Yisimayi, A.; Bai, Y.; Huang, W.; Li, X.; Zhang, Z.; Yuan, T.; An, R.; Wang, J.; Xiao, T.; et al. Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines. Cell Res. 2021, 31, 732–741. [Google Scholar] [CrossRef] [PubMed]
- Bravo, L.; Smolenov, I.; Han, H.H.; Li, P.; Hosain, R.; Rockhold, F.; Clemens, S.A.C.; Roa, C.; Borja-Tabora, C.; Quinsaat, A.; et al. Efficacy of the adjuvanted subunit protein COVID-19 vaccine, SCB-2019: A phase 2 and 3 multicentre, double-blind, randomised, placebo-controlled trial. Lancet 2022, 399, 461–472. [Google Scholar] [CrossRef]
- Zhu, F.-C.; Li, Y.-H.; Guan, X.-H.; Hou, L.-H.; Wang, W.-J.; Li, J.-X.; Wu, S.-P.; Wang, B.-S.; Wang, Z.; Wang, L.; et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: A dose-escalation, open-label, non-randomised, first-in-human trial. Lancet 2020, 395, 1845–1854. [Google Scholar] [CrossRef]
- Wu, S.; Huang, J.; Zhang, Z.; Wu, J.; Zhang, J.; Hu, H.; Zhu, T.; Zhang, J.; Luo, L.; Fan, P.; et al. Safety, tolerability, and immunogenicity of an aerosolised adenovirus type-5 vector-based COVID-19 vaccine (Ad5-nCoV) in adults: Preliminary report of an open-label and randomised phase 1 clinical trial. Lancet Infect. Dis. 2021, 21, 1654–1664. [Google Scholar] [CrossRef]
- Zhu, F.-C.; Guan, X.-H.; Li, Y.-H.; Huang, J.-Y.; Jiang, T.; Hou, L.-H.; Li, J.-X.; Yang, B.-F.; Wang, L.; Wang, W.-J.; et al. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: A randomised, double-blind, placebo-controlled, phase 2 trial. Lancet 2020, 396, 479–488. [Google Scholar] [CrossRef]
- Yu, J.; Tostanoski, L.H.; Peter, L.; Mercado, N.B.; McMahan, K.; Mahrokhian, S.H.; Nkolola, J.P.; Liu, J.; Li, Z.; Chandrashekar, A.; et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 2020, 369, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Perez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef] [PubMed]
- COVID-19 Vaccine Tracker and Landscape. Available online: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed on 13 December 2022).
- Hirabara, S.M.; Serdan, T.D.A.; Gorjao, R.; Masi, L.N.; Pithon-Curi, T.C.; Covas, D.T.; Curi, R.; Durigon, E.L. SARS-COV-2 Variants: Differences and Potential of Immune Evasion. Front. Cell Infect. Microbiol. 2021, 11, 781429. [Google Scholar] [CrossRef] [PubMed]
- Tian, D.; Sun, Y.; Xu, H.; Ye, Q. The emergence and epidemic characteristics of the highly mutated SARS-CoV-2 Omicron variant. J. Med. Virol. 2022, 94, 2376–2383. [Google Scholar] [CrossRef] [PubMed]
- Andrews, N.; Stowe, J.; Kirsebom, F.; Toffa, S.; Rickeard, T.; Gallagher, E.; Gower, C.; Kall, M.; Groves, N.; O’Connell, A.M.; et al. COVID-19 Vaccine Effectiveness against the Omicron (B.1.1.529) Variant. N. Engl. J. Med. 2022, 386, 1532–1546. [Google Scholar] [CrossRef] [PubMed]
- McKeigue, P.M.; McAllister, D.A.; Hutchinson, S.J.; Robertson, C.; Stockton, D.; Colhoun, H.M. Vaccine efficacy against severe COVID-19 in relation to delta variant (B.1.617.2) and time since second dose in patients in Scotland (REACT-SCOT): A case-control study. Lancet Respir. Med. 2022, 10, 566–572. [Google Scholar] [CrossRef]
- Qu, L.; Yi, Z.; Shen, Y.; Lin, L.; Chen, F.; Xu, Y.; Wu, Z.; Tang, H.; Zhang, X.; Tian, F.; et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell 2022, 185, 1728–1744.e1716. [Google Scholar] [CrossRef]
- Liu, Z.; Xu, W.; Xia, S.; Gu, C.; Wang, X.; Wang, Q.; Zhou, J.; Wu, Y.; Cai, X.; Qu, D.; et al. RBD-Fc-based COVID-19 vaccine candidate induces highly potent SARS-CoV-2 neutralizing antibody response. Signal Transduct. Target Ther. 2020, 5, 282. [Google Scholar] [CrossRef]
- Su, D.; Li, X.; He, C.; Huang, X.; Chen, M.; Wang, Q.; Qin, W.; Liang, Y.; Xu, R.; Wu, J.; et al. Broad neutralization against SARS-CoV-2 variants induced by a modified B.1.351 protein-based COVID-19 vaccine candidate. bioRxiv 2021. [Google Scholar] [CrossRef]
- Xu, K.; Gao, P.; Liu, S.; Lu, S.; Lei, W.; Zheng, T.; Liu, X.; Xie, Y.; Zhao, Z.; Guo, S.; et al. Protective prototype-Beta and Delta-Omicron chimeric RBD-dimer vaccines against SARS-CoV-2. Cell 2022, 185, 2265–2278.e2214. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, Z.; He, J.; Liao, Y.; Li, Y.; Pei, R.; Fang, X.; Zeng, P.; Fan, R.; Ou, Z.; et al. Safety and immunogenicity of a recombinant interferon-armed RBD dimer vaccine (V-01) for COVID-19 in healthy adults: A randomized, double-blind, placebo-controlled, Phase I trial. Emerg. Microbes Infect. 2021, 10, 1589–1597. [Google Scholar] [CrossRef]
- Zhu, F.; Zhuang, C.; Chu, K.; Zhang, L.; Zhao, H.; Huang, S.; Su, Y.; Lin, H.; Yang, C.; Jiang, H.; et al. Safety and immunogenicity of a live-attenuated influenza virus vector-based intranasal SARS-CoV-2 vaccine in adults: Randomised, double-blind, placebo-controlled, phase 1 and 2 trials. Lancet Respir. Med. 2022, 10, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, P.; Yuan, L.; Zhang, L.; Zhang, L.; Zhao, H.; Chen, C.; Wang, X.; Han, J.; Chen, Y.; et al. A live attenuated virus-based intranasal COVID-19 vaccine provides rapid, prolonged, and broad protection against SARS-CoV-2. Sci. Bull 2022, 67, 1372–1387. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Miranda, M.C.; Schafer, A.; Pham, M.N.; Greaney, A.; Arunachalam, P.S.; Navarro, M.J.; Tortorici, M.A.; Rogers, K.; O’Connor, M.A.; et al. Elicitation of broadly protective sarbecovirus immunity by receptor-binding domain nanoparticle vaccines. Cell 2021, 184, 5432–5447.e5416. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.A.; Gnanapragasam, P.N.P.; Lee, Y.E.; Hoffman, P.R.; Ou, S.; Kakutani, L.M.; Keeffe, J.R.; Wu, H.-J.; Howarth, M.; West, A.P.; et al. Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science 2021, 371, 735–741. [Google Scholar] [CrossRef] [PubMed]
- Crank, M.C.; Ruckwardt, T.J.; Chen, M.; Morabito, K.M.; Phung, E.; Costner, P.J.; Holman, L.A.; Hickman, S.P.; Berkowitz, N.M.; Gordon, I.J.; et al. A proof of concept for structure-based vaccine design targeting RSV in humans. Science 2019, 365, 505–509. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.L.; Goldsmith, J.A.; Schaub, J.M.; DiVenere, A.M.; Kuo, H.-C.; Javanmardi, K.; Le, K.C.; Wrapp, D.; Lee, A.G.; Liu, Y.; et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 2020, 369, 1501–1505. [Google Scholar] [CrossRef]
- Nathan, A.; Rossin, E.J.; Kaseke, C.; Park, R.J.; Khatri, A.; Koundakjian, D.; Urbach, J.M.; Singh, N.K.; Bashirova, A.; Tano-Menka, R.; et al. Structure-guided T cell vaccine design for SARS-CoV-2 variants and sarbecoviruses. Cell 2021, 184, 4401–4413.e4410. [Google Scholar] [CrossRef]
- Jarjour, N.N.; Masopust, D.; Jameson, S.C. T Cell Memory: Understanding COVID-19. Immunity 2021, 54, 14–18. [Google Scholar] [CrossRef]
- Niessl, J.; Sekine, T.; Buggert, M. T cell immunity to SARS-CoV-2. Semin. Immunol. 2021, 55, 101505. [Google Scholar] [CrossRef]
- Lapuente, D.; Fuchs, J.; Willar, J.; Vieira Antao, A.; Eberlein, V.; Uhlig, N.; Issmail, L.; Schmidt, A.; Oltmanns, F.; Peter, A.S.; et al. Protective mucosal immunity against SARS-CoV-2 after heterologous systemic prime-mucosal boost immunization. Nat. Commun. 2021, 12, 6871. [Google Scholar] [CrossRef]
- An, X.; Martinez-Paniagua, M.; Rezvan, A.; Sefat, S.R.; Fathi, M.; Singh, S.; Biswas, S.; Pourpak, M.; Yee, C.; Liu, X.; et al. Single-dose intranasal vaccination elicits systemic and mucosal immunity against SARS-CoV-2. iScience 2021, 24, 103037. [Google Scholar] [CrossRef] [PubMed]
- Afkhami, S.; D’Agostino, M.R.; Zhang, A.; Stacey, H.D.; Marzok, A.; Kang, A.; Singh, R.; Bavananthasivam, J.; Ye, G.; Luo, X.; et al. Respiratory mucosal delivery of next-generation COVID-19 vaccine provides robust protection against both ancestral and variant strains of SARS-CoV-2. Cell 2022, 185, 896–915.e819. [Google Scholar] [CrossRef] [PubMed]
- Shin, M.D.; Shukla, S.; Chung, Y.H.; Beiss, V.; Chan, S.K.; Ortega-Rivera, O.A.; Wirth, D.M.; Chen, A.; Sack, M.; Pokorski, J.K.; et al. COVID-19 vaccine development and a potential nanomaterial path forward. Nat. Nanotechnol. 2020, 15, 646–655. [Google Scholar] [CrossRef] [PubMed]
- Vu, M.N.; Kelly, H.G.; Kent, S.J.; Wheatley, A.K. Current and future nanoparticle vaccines for COVID-19. EBioMedicine 2021, 74, 103699. [Google Scholar] [CrossRef]
- Cui, J.; Li, F.; Shi, Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Troyano-Hernaez, P.; Reinosa, R.; Holguin, A. Evolution of SARS-CoV-2 Envelope, Membrane, Nucleocapsid, and Spike Structural Proteins from the Beginning of the Pandemic to September 2020: A Global and Regional Approach by Epidemiological Week. Viruses 2021, 13, 243. [Google Scholar] [CrossRef]
- Arya, R.; Kumari, S.; Pandey, B.; Mistry, H.; Bihani, S.C.; Das, A.; Prashar, V.; Gupta, G.D.; Panicker, L.; Kumar, M. Structural insights into SARS-CoV-2 proteins. J. Mol. Biol. 2021, 433, 166725. [Google Scholar] [CrossRef]
- Yao, H.; Song, Y.; Chen, Y.; Wu, N.; Xu, J.; Sun, C.; Zhang, J.; Weng, T.; Zhang, Z.; Wu, Z.; et al. Molecular Architecture of the SARS-CoV-2 Virus. Cell 2020, 183, 730–738.e713. [Google Scholar] [CrossRef]
- Ke, Z.; Oton, J.; Qu, K.; Cortese, M.; Zila, V.; McKeane, L.; Nakane, T.; Zivanov, J.; Neufeldt, C.J.; Cerikan, B.; et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 2020, 588, 498–502. [Google Scholar] [CrossRef]
- Atzrodt, C.L.; Maknojia, I.; McCarthy, R.D.P.; Oldfield, T.M.; Po, J.; Ta, K.T.L.; Stepp, H.E.; Clements, T.P. A Guide to COVID-19: A global pandemic caused by the novel coronavirus SARS-CoV-2. FEBS J. 2020, 287, 3633–3650. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Batra, H.; Yadav, P.; Chand, S. COVID-19 Vaccines Currently under Preclinical and Clinical Studies, and Associated Antiviral Immune Response. Vaccines 2020, 8, 649. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e286. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Yang, Y.; Zhang, X. Neutralizing antibodies for the prevention and treatment of COVID-19. Cell Mol. Immunol. 2021, 18, 2293–2306. [Google Scholar] [CrossRef]
- Piccoli, L.; Park, Y.J.; Tortorici, M.A.; Czudnochowski, N.; Walls, A.C.; Beltramello, M.; Silacci-Fregni, C.; Pinto, D.; Rosen, L.E.; Bowen, J.E.; et al. Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell 2020, 183, 1024–1042.e1021. [Google Scholar] [CrossRef]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501.e1415. [Google Scholar] [CrossRef]
- Dai, L.; Gao, G.F. Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 2021, 21, 73–82. [Google Scholar] [CrossRef]
- Hou, Y.J.; Chiba, S.; Halfmann, P.; Ehre, C.; Kuroda, M.; Dinnon, K.H., III; Leist, S.R.; Schäfer, A.; Nakajima, N.; Takahashi, K.; et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 2020, 370, 1464–1468. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, Q.; Wei, P.; Chen, Z.; Aviszus, K.; Yang, J.; Downing, W.; Jiang, C.; Liang, B.; Reynoso, L.; et al. The basis of a more contagious 501Y.V1 variant of SARS-CoV-2. Cell Res. 2021, 31, 720–722. [Google Scholar] [CrossRef]
- Motozono, C.; Toyoda, M.; Zahradnik, J.; Saito, A.; Nasser, H.; Tan, T.S.; Ngare, I.; Kimura, I.; Uriu, K.; Kosugi, Y.; et al. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe 2021, 29, 1124–1136.e1111. [Google Scholar] [CrossRef] [PubMed]
- Weisblum, Y.; Schmidt, F.; Zhang, F.; DaSilva, J.; Poston, D.; Lorenzi, J.C.; Muecksch, F.; Rutkowska, M.; Hoffmann, H.H.; Michailidis, E.; et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife 2020, 9, e61312. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Wang, J.; Jian, F.; Xiao, T.; Song, W.; Yisimayi, A.; Huang, W.; Li, Q.; Wang, P.; An, R.; et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 2022, 602, 657–663. [Google Scholar] [CrossRef]
- Kumar, S.; Thambiraja, T.S.; Karuppanan, K.; Subramaniam, G. Omicron and Delta variant of SARS-CoV-2: A comparative computational study of spike protein. J. Med. Virol. 2022, 94, 1641–1649. [Google Scholar] [CrossRef]
- Li, X. Omicron: Call for updated vaccines. J. Med. Virol. 2022, 94, 1261–1263. [Google Scholar] [CrossRef]
- Anderson, E.J.; Rouphael, N.G.; Widge, A.T.; Jackson, L.A.; Roberts, P.C.; Makhene, M.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; Pruijssers, A.J.; et al. Safety and Immunogenicity of SARS-CoV-2 mRNA-1273 Vaccine in Older Adults. N. Engl. J. Med. 2020, 383, 2427–2438. [Google Scholar] [CrossRef] [PubMed]
- Lien, C.E.; Kuo, T.Y.; Lin, Y.J.; Lian, W.C.; Lin, M.Y.; Liu, L.T.; Cheng, J.; Chou, Y.C.; Chen, C. Evaluating the Neutralizing Ability of a CpG-Adjuvanted S-2P Subunit Vaccine Against Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Variants of Concern. Clin. Infect. Dis. 2022, 74, 1899–1905. [Google Scholar] [CrossRef]
- Park, H.S.; Matsuoka, Y.; Luongo, C.; Yang, L.; Santos, C.; Liu, X.; Ahlers, L.R.H.; Moore, I.N.; Afroz, S.; Johnson, R.F.; et al. Intranasal immunization with avian paramyxovirus type 3 expressing SARS-CoV-2 spike protein protects hamsters against SARS-CoV-2. NPJ Vaccines 2022, 7, 72. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.G.; Su, D.; Song, T.Z.; Zeng, Y.; Huang, W.; Wu, J.; Xu, R.; Luo, P.; Yang, X.; Zhang, X.; et al. S-Trimer, a COVID-19 subunit vaccine candidate, induces protective immunity in nonhuman primates. Nat. Commun. 2021, 12, 1346. [Google Scholar] [CrossRef]
- Sun, S.; Cai, Y.; Song, T.Z.; Pu, Y.; Cheng, L.; Xu, H.; Sun, J.; Meng, C.; Lin, Y.; Huang, H.; et al. Interferon-armed RBD dimer enhances the immunogenicity of RBD for sterilizing immunity against SARS-CoV-2. Cell Res. 2021, 31, 1011–1023. [Google Scholar] [CrossRef] [PubMed]
- Heitmann, J.S.; Bilich, T.; Tandler, C.; Nelde, A.; Maringer, Y.; Marconato, M.; Reusch, J.; Jager, S.; Denk, M.; Richter, M.; et al. A COVID-19 peptide vaccine for the induction of SARS-CoV-2 T cell immunity. Nature 2022, 601, 617–622. [Google Scholar] [CrossRef] [PubMed]
- García-Arriaza, J.; Garaigorta, U.; Pérez, P.; Lázaro-Frías, A.; Zamora, C.; Gastaminza, P.; Del Fresno, C.; Casasnovas, J.M.; Sorzano, C.Ó.S.; Sancho, D.; et al. COVID-19 vaccine candidates based on modified vaccinia virus Ankara expressing the SARS-CoV-2 spike induce robust T- and B-cell immune responses and full efficacy in mice. J. Virol. 2021, 95, e02260-20. [Google Scholar] [CrossRef] [PubMed]
- Routhu, N.K.; Cheedarla, N.; Gangadhara, S.; Bollimpelli, V.S.; Boddapati, A.K.; Shiferaw, A.; Rahman, S.A.; Sahoo, A.; Edara, V.V.; Lai, L.; et al. A modified vaccinia Ankara vector-based vaccine protects macaques from SARS-CoV-2 infection, immune pathology, and dysfunction in the lungs. Immunity 2021, 54, 542–556 e549. [Google Scholar] [CrossRef]
- Cao, H.; Mai, J.; Zhou, Z.; Li, Z.; Duan, R.; Watt, J.; Chen, Z.; Bandara, R.A.; Li, M.; Ahn, S.K.; et al. Intranasal HD-Ad vaccine protects the upper and lower respiratory tracts of hACE2 mice against SARS-CoV-2. Cell Biosci. 2021, 11, 202. [Google Scholar] [CrossRef] [PubMed]
- Royal, J.M.; Simpson, C.A.; McCormick, A.A.; Phillips, A.; Hume, S.; Morton, J.; Shepherd, J.; Oh, Y.; Swope, K.; DeBeauchamp, J.L.; et al. Development of a SARS-CoV-2 Vaccine Candidate Using Plant-Based Manufacturing and a Tobacco Mosaic Virus-like Nano-Particle. Vaccines 2021, 9, 1347. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.F.; Sun, C.; Sun, J.; Xie, C.; Zhuang, Z.; Xu, H.Q.; Liu, Z.; Liu, Y.H.; Peng, S.; Yuan, R.Y.; et al. Quadrivalent mosaic HexaPro-bearing nanoparticle vaccine protects against infection of SARS-CoV-2 variants. Nat. Commun. 2022, 13, 2674. [Google Scholar] [CrossRef]
- Ma, X.; Zou, F.; Yu, F.; Li, R.; Yuan, Y.; Zhang, Y.; Zhang, X.; Deng, J.; Chen, T.; Song, Z.; et al. Nanoparticle Vaccines Based on the Receptor Binding Domain (RBD) and Heptad Repeat (HR) of SARS-CoV-2 Elicit Robust Protective Immune Responses. Immunity 2020, 53, 1315–1330 e1319. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Lin, X.; Wang, Y.; Abraham, C.; Sou, C.; Ngo, T.; Zhang, Y.; Wilson, I.A.; Zhu, J. Single-component, self-assembling, protein nanoparticles presenting the receptor binding domain and stabilized spike as SARS-CoV-2 vaccine candidates. Sci. Adv. 1591. [Google Scholar] [CrossRef] [PubMed]
- Graham, B.S.; Gilman, M.S.A.; McLellan, J.S. Structure-Based Vaccine Antigen Design. Annu. Rev. Med. 2019, 70, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Mangala Prasad, V.; Leaman, D.P.; Lovendahl, K.N.; Croft, J.T.; Benhaim, M.A.; Hodge, E.A.; Zwick, M.B.; Lee, K.K. Cryo-ET of Env on intact HIV virions reveals structural variation and positioning on the Gag lattice. Cell 2022, 185, 641–653 e617. [Google Scholar] [CrossRef]
- Derking, R.; Sanders, R.W. Structure-guided envelope trimer design in HIV-1 vaccine development: A narrative review. J. Int. AIDS Soc. 2021, 24 (Suppl. 7), e25797. [Google Scholar] [CrossRef]
- Skehel, J.J.; Wiley, D.C. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu. Rev. Biochem. 2000, 69, 531–569. [Google Scholar] [CrossRef] [PubMed]
- Stevens, J.; Corper, A.L.; Basler, C.F.; Taubenberger, J.K.; Palese, P.; Wilson, I.A. Structure of the Uncleaved Human H1 Hemagglutinin from the Extinct 1918 Influenza Virus. Science 2004, 303, 1866–1870. [Google Scholar] [CrossRef] [PubMed]
- Bangaru, S.; Ozorowski, G.; Turner, H.L.; Antanasijevic, A.; Huang, D.; Wang, X.; Torres, J.L.; Diedrich, J.K.; Tian, J.-H.; Portnoff, A.D.; et al. Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate. Science 2020, 370, 1089–1094. [Google Scholar] [CrossRef] [PubMed]
- McMillan, C.L.D.; Amarilla, A.A.; Modhiran, N.; Choo, J.J.Y.; Azuar, A.; Honeyman, K.E.; Khromykh, A.A.; Young, P.R.; Watterson, D.; Muller, D.A. Skin-patch delivered subunit vaccine induces broadly neutralising antibodies against SARS-CoV-2 variants of concern. Vaccine 2022, 40, 4929–4932. [Google Scholar] [CrossRef] [PubMed]
- Pallesen, J.; Wang, N.; Corbett, K.S.; Wrapp, D.; Kirchdoerfer, R.N.; Turner, H.L.; Cottrell, C.A.; Becker, M.M.; Wang, L.; Shi, W.; et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA 2017, 114, E7348–E7357. [Google Scholar] [CrossRef] [PubMed]
- Arnold, C. Stalking new vaccines: Methods that target the stems of viral proteins could put universal vaccines within reach. Nat. Med. 2017, 23, 1248–1250. [Google Scholar] [CrossRef] [PubMed]
- Rey, F.A.; Lok, S.M. Common Features of Enveloped Viruses and Implications for Immunogen Design for Next-Generation Vaccines. Cell 2018, 172, 1319–1334. [Google Scholar] [CrossRef] [PubMed]
- Sadoff, J.; Gray, G.; Vandebosch, A.; Cardenas, V.; Shukarev, G.; Grinsztejn, B.; Goepfert, P.A.; Truyers, C.; Fennema, H.; Spiessens, B.; et al. Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against COVID-19. N. Engl. J. Med. 2021, 384, 2187–2201. [Google Scholar] [CrossRef]
- Sadoff, J.; Le Gars, M.; Shukarev, G.; Heerwegh, D.; Truyers, C.; de Groot, A.M.; Stoop, J.; Tete, S.; Van Damme, W.; Leroux-Roels, I.; et al. Interim Results of a Phase 1-2a Trial of Ad26.COV2.S COVID-19 Vaccine. N. Engl. J. Med. 2021, 384, 1824–1835. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, S.M.; Liu, W.D.; Huang, Y.S.; Lin, Y.J.; Hsieh, E.F.; Lian, W.C.; Chen, C.; Janssen, R.; Shih, S.R.; Huang, C.G.; et al. Safety and immunogenicity of a Recombinant Stabilized Prefusion SARS-CoV-2 Spike Protein Vaccine (MVC-COV1901) Adjuvanted with CpG 1018 and Aluminum Hydroxide in healthy adults: A Phase 1, dose-escalation study. EClinicalMedicine 2021, 38, 100989. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.P.; Do, Q.; Phan, L.T.; Dinh, D.V.; Khong, H.; Hoang, L.V.; Nguyen, T.V.; Pham, H.N.; Chu, M.V.; Nguyen, T.T.; et al. Safety and immunogenicity of Nanocovax, a SARS-CoV-2 recombinant spike protein vaccine: Interim results of a double-blind, randomised controlled phase 1 and 2 trial. Lancet Reg. Health West Pac. 2022, 24, 100474. [Google Scholar] [CrossRef] [PubMed]
- Heath, P.T.; Galiza, E.P.; Baxter, D.N.; Boffito, M.; Browne, D.; Burns, F.; Chadwick, D.R.; Clark, R.; Cosgrove, C.; Galloway, J.; et al. Safety and Efficacy of NVX-CoV2373 COVID-19 Vaccine. N. Engl. J. Med. 2021, 385, 1172–1183. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Chamblee, M.; Zhang, Y.; Ye, C.; Dravid, P.; Park, J.G.; Mahesh, K.C.; Trivedi, S.; Murthy, S.; Sharma, H.; et al. SARS-CoV-2 prefusion spike protein stabilized by six rather than two prolines is more potent for inducing antibodies that neutralize viral variants of concern. Proc. Natl. Acad. Sci. USA 2022, 119, e2110105119. [Google Scholar] [CrossRef]
- Pitisuttithum, P.; Luvira, V.; Lawpoolsri, S.; Muangnoicharoen, S.; Kamolratanakul, S.; Sivakorn, C.; Narakorn, P.; Surichan, S.; Prangpratanporn, S.; Puksuriwong, S.; et al. Safety and immunogenicity of an inactivated recombinant Newcastle disease virus vaccine expressing SARS-CoV-2 spike: Interim results of a randomised, placebo-controlled, phase 1 trial. EClinicalMedicine 2022, 45, 101323. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Su, D.; Sun, Y.; Huang, X.; Liang, Y.; Fang, L.; Ma, Y.; Li, W.; Liang, P.; Zheng, S. Cryo-EM structure of S-Trimer, a subunit vaccine candidate for COVID-19. J. Virol. 2021, 95, e00194-21. [Google Scholar] [CrossRef] [PubMed]
- Premkumar, L.; Segovia-Chumbez, B.; Jadi, R.; Martinez, D.R.; Raut, R.; Markmann, A.; Cornaby, C.; Bartelt, L.; Weiss, S.; Park, Y.; et al. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol 2020, 5, eabc8413. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Zheng, T.; Xu, K.; Han, Y.; Xu, L.; Huang, E.; An, Y.; Cheng, Y.; Li, S.; Liu, M.; et al. A Universal Design of Betacoronavirus Vaccines against COVID-19, MERS, and SARS. Cell 2020, 182, 722–733.e711. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Gao, L.; Tao, L.; Hadinegoro, S.R.; Erkin, M.; Ying, Z.; He, P.; Girsang, R.T.; Vergara, H.; Akram, J.; et al. Efficacy and Safety of the RBD-Dimer-Based COVID-19 Vaccine ZF2001 in Adults. N. Engl. J. Med. 2022, 386, 2097–2111. [Google Scholar] [CrossRef] [PubMed]
- Zang, J.; Zhu, Y.; Zhou, Y.; Gu, C.; Yi, Y.; Wang, S.; Xu, S.; Hu, G.; Du, S.; Yin, Y.; et al. Yeast-produced RBD-based recombinant protein vaccines elicit broadly neutralizing antibodies and durable protective immunity against SARS-CoV-2 infection. Cell Discov. 2021, 7, 71. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Zhu, L.; Ni, Z.; Meng, H.; You, L. Duration of serum neutralizing antibodies for SARS-CoV-2: Lessons from SARS-CoV infection. J. Microbiol. Immunol. Infect. 2020, 53, 821–822. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Beltran, W.F.; Lam, E.C.; St Denis, K.; Nitido, A.D.; Garcia, Z.H.; Hauser, B.M.; Feldman, J.; Pavlovic, M.N.; Gregory, D.J.; Poznansky, M.C.; et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 2021, 184, 2372–2383 e2379. [Google Scholar] [CrossRef]
- Schmidt, A.; Lapuente, D. T Cell Immunity against Influenza: The Long Way from Animal Models Towards a Real-Life Universal Flu Vaccine. Viruses 2021, 13, 199. [Google Scholar] [CrossRef] [PubMed]
- Matyushenko, V.; Kotomina, T.; Kudryavtsev, I.; Mezhenskaya, D.; Prokopenko, P.; Matushkina, A.; Sivak, K.; Muzhikyan, A.; Rudenko, L.; Isakova-Sivak, I. Conserved T-cell epitopes of respiratory syncytial virus (RSV) delivered by recombinant live attenuated influenza vaccine viruses efficiently induce RSV-specific lung-localized memory T cells and augment influenza-specific resident memory T-cell responses. Antiviral Res. 2020, 182, 104864. [Google Scholar] [CrossRef] [PubMed]
- Son, Y.M.; Cheon, I.S.; Wu, Y.; Li, C.; Wang, Z.; Gao, X.; Chen, Y.; Takahashi, Y.; Fu, Y.X.; Dent, A.L.; et al. Tissue-resident CD4(+) T helper cells assist the development of protective respiratory B and CD8(+) T cell memory responses. Sci. Immunol. 2021, 6, eabb6852. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Kolesnikov, A.; Yin, R.; Guest, J.D.; Gowthaman, R.; Shmelev, A.; Serdyuk, Y.; Dianov, D.V.; Efimov, G.A.; Pierce, B.G.; et al. Structural assessment of HLA-A2-restricted SARS-CoV-2 spike epitopes recognized by public and private T-cell receptors. Nat. Commun. 2022, 13, 19. [Google Scholar] [CrossRef]
- Tarke, A.; Coelho, C.H.; Zhang, Z.; Dan, J.M.; Yu, E.D.; Methot, N.; Bloom, N.I.; Goodwin, B.; Phillips, E.; Mallal, S.; et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell 2022, 185, 847–859.e811. [Google Scholar] [CrossRef] [PubMed]
- Poran, A.; Harjanto, D.; Malloy, M.; Arieta, C.M.; Rothenberg, D.A.; Lenkala, D.; van Buuren, M.M.; Addona, T.A.; Rooney, M.S.; Srinivasan, L.; et al. Sequence-based prediction of SARS-CoV-2 vaccine targets using a mass spectrometry-based bioinformatics predictor identifies immunogenic T cell epitopes. Genome Med. 2020, 12, 70. [Google Scholar] [CrossRef] [PubMed]
- Gao, A.; Chen, Z.; Amitai, A.; Doelger, J.; Mallajosyula, V.; Sundquist, E.; Pereyra Segal, F.; Carrington, M.; Davis, M.M.; Streeck, H.; et al. Learning from HIV-1 to predict the immunogenicity of T cell epitopes in SARS-CoV-2. iScience 2021, 24, 102311. [Google Scholar] [CrossRef] [PubMed]
- Guerrera, G.; Picozza, M.; D’Orso, S.; Placido, R.; Pirronello, M.; Verdiani, A.; Termine, A.; Fabrizio, C.; Giannessi, F.; Sambucci, M.; et al. BNT162b2 vaccination induces durable SARS-CoV-2-specific T cells with a stem cell memory phenotype. Sci. Immunol. 2021, 6, eabl5344. [Google Scholar] [CrossRef]
- Sahin, U.; Muik, A.; Derhovanessian, E.; Vogler, I.; Kranz, L.M.; Vormehr, M.; Baum, A.; Pascal, K.; Quandt, J.; Maurus, D.; et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature 2020, 586, 594–599. [Google Scholar] [CrossRef] [PubMed]
- GeurtsvanKessel, C.H.; Geers, D.; Schmitz, K.S.; Mykytyn, A.Z.; Lamers, M.M.; Bogers, S.; Scherbeijn, S.; Gommers, L.; Sablerolles, R.S.G.; Nieuwkoop, N.N.; et al. Divergent SARS-CoV-2 Omicron-reactive T and B cell responses in COVID-19 vaccine recipients. Sci. Immunol. 2022, 7, eabo2202. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, X.; Jin, J.; Ma, Z.; Liu, Y.; Zhang, X.; Su, B. T-cell responses to SARS-CoV-2 Omicron spike epitopes with mutations after the third booster dose of an inactivated vaccine. J. Med. Virol. 2022, 94, 3998–4004. [Google Scholar] [CrossRef] [PubMed]
- Alu, A.; Chen, L.; Lei, H.; Wei, Y.; Tian, X.; Wei, X. Intranasal COVID-19 vaccines: From bench to bed. EBioMedicine 2022, 76, 103841. [Google Scholar] [CrossRef]
- Lavelle, E.C.; Ward, R.W. Mucosal vaccines—fortifying the frontiers. Nat. Rev. Immunol. 2022, 22, 236–250. [Google Scholar] [CrossRef] [PubMed]
- Langel, S.N.; Johnson, S.; Martinez, C.I.; Tedjakusuma, S.N.; Peinovich, N.; Dora, E.G.; Kuehl, P.J.; Irshad, H.; Barrett, E.G.; Werts, A.D.; et al. Adenovirus type 5 SARS-CoV-2 vaccines delivered orally or intranasally reduced disease severity and transmission in a hamster model. Sci. Transl. Med. 2022, 14, eabn6868. [Google Scholar] [CrossRef] [PubMed]
- Lund, F.E.; Randall, T.D. Scent of a vaccine. Science 2021, 373, 397–399. [Google Scholar] [CrossRef]
- Chavda, V.P.; Vora, L.K.; Pandya, A.K.; Patravale, V.B. Intranasal vaccines for SARS-CoV-2: From challenges to potential in COVID-19 management. Drug. Discov. Today 2021, 26, 2619–2636. [Google Scholar] [CrossRef]
- Hassan, A.O.; Kafai, N.M.; Dmitriev, I.P.; Fox, J.M.; Smith, B.K.; Harvey, I.B.; Chen, R.E.; Winkler, E.S.; Wessel, A.W.; Case, J.B.; et al. A Single-Dose Intranasal ChAd Vaccine Protects Upper and Lower Respiratory Tracts against SARS-CoV-2. Cell 2020, 183, 169–184.e113. [Google Scholar] [CrossRef]
- Ye, Z.W.; Ong, C.P.; Tang, K.; Fan, Y.; Luo, C.; Zhou, R.; Luo, P.; Cheng, Y.; Gray, V.S.; Wang, P.; et al. Intranasal administration of a single dose of a candidate live attenuated vaccine derived from an NSP16-deficient SARS-CoV-2 strain confers sterilizing immunity in animals. Cell Mol. Immunol. 2022, 19, 588–601. [Google Scholar] [CrossRef]
- Li, J.-X.; Wu, S.-P.; Guo, X.-L.; Tang, R.; Huang, B.-Y.; Chen, X.-Q.; Chen, Y.; Hou, L.-H.; Liu, J.-X.; Zhong, J.; et al. Safety and immunogenicity of heterologous boost immunisation with an orally administered aerosolised Ad5-nCoV after two-dose priming with an inactivated SARS-CoV-2 vaccine in Chinese adults: A randomised, open-label, single-centre trial. Lancet Respir. Med. 2022, 10, 739–748. [Google Scholar] [CrossRef]
- Rappuoli, R.; Serruto, D. Self-Assembling Nanoparticles Usher in a New Era of Vaccine Design. Cell 2019, 176, 1245–1247. [Google Scholar] [CrossRef]
- Mohsen, M.O.; Zha, L.; Cabral-Miranda, G.; Bachmann, M.F. Major findings and recent advances in virus-like particle (VLP)-based vaccines. Semin. Immunol. 2017, 34, 123–132. [Google Scholar] [CrossRef]
- Cai, X.; Zheng, W.; Pan, S.; Zhang, S.; Xie, Y.; Guo, H.; Wang, G.; Li, Z.; Luo, M. A virus-like particle of the hepatitis B virus preS antigen elicits robust neutralizing antibodies and T cell responses in mice. Antiviral Res. 2018, 149, 48–57. [Google Scholar] [CrossRef]
- Wang, D.; Liu, X.; Wei, M.; Qian, C.; Song, S.; Chen, J.; Wang, Z.; Xu, Q.; Yang, Y.; He, M.; et al. Rational design of a multi-valent human papillomavirus vaccine by capsomere-hybrid co-assembly of virus-like particles. Nat. Commun. 2020, 11, 2841. [Google Scholar] [CrossRef]
- Kato, Y.; Abbott, R.K.; Freeman, B.L.; Haupt, S.; Groschel, B.; Silva, M.; Menis, S.; Irvine, D.J.; Schief, W.R.; Crotty, S. Multifaceted Effects of Antigen Valency on B Cell Response Composition and Differentiation In Vivo. Immunity 2020, 53, 548–563 e548. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, B.; Tolia, N.H. Protein-based antigen presentation platforms for nanoparticle vaccines. NPJ Vaccines 2021, 6, 70. [Google Scholar] [CrossRef] [PubMed]
- Frey, S.; Castro, A.; Arsiwala, A.; Kane, R.S. Bionanotechnology for vaccine design. Curr. Opin. Biotechnol. 2018, 52, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Ueda, G.; Antanasijevic, A.; Fallas, J.A.; Sheffler, W.; Copps, J.; Ellis, D.; Hutchinson, G.B.; Moyer, A.; Yasmeen, A.; Tsybovsky, Y.; et al. Tailored design of protein nanoparticle scaffolds for multivalent presentation of viral glycoprotein antigens. eLife 2020, 9, e57659. [Google Scholar] [CrossRef] [PubMed]
- Subbarao, K.; Geng, Q.; Tai, W.; Baxter, V.K.; Shi, J.; Wan, Y.; Zhang, X.; Montgomery, S.A.; Taft-Benz, S.A.; Anderson, E.J.; et al. Novel virus-like nanoparticle vaccine effectively protects animal model from SARS-CoV-2 infection. PLoS Pathog. 2021, 17, e1009897. [Google Scholar] [CrossRef]
- Keech, C.; Albert, G.; Cho, I.; Robertson, A.; Reed, P.; Neal, S.; Plested, J.S.; Zhu, M.; Cloney-Clark, S.; Zhou, H.; et al. Phase 1-2 Trial of a SARS-CoV-2 Recombinant Spike Protein Nanoparticle Vaccine. N. Engl. J. Med. 2020, 383, 2320–2332. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.A.; van Doremalen, N.; Greaney, A.J.; Andersen, H.; Sharma, A.; Starr, T.N.; Keeffe, J.R.; Fan, C.; Schulz, J.E.; Gnanapragasam, P.N.; et al. Mosaic RBD nanoparticles protect against challenge by diverse sarbecoviruses in animal models. Science 2022, 377, eabq0839. [Google Scholar] [CrossRef]
- Bergwerk, M.; Gonen, T.; Lustig, Y.; Amit, S.; Lipsitch, M.; Cohen, C.; Mandelboim, M.; Levin, E.G.; Rubin, C.; Indenbaum, V.; et al. COVID-19 Breakthrough Infections in Vaccinated Health Care Workers. N. Engl. J. Med. 2021, 385, 1474–1484. [Google Scholar] [CrossRef]
- Hacisuleyman, E.; Hale, C.; Saito, Y.; Blachere, N.E.; Bergh, M.; Conlon, E.G.; Schaefer-Babajew, D.J.; DaSilva, J.; Muecksch, F.; Gaebler, C.; et al. Vaccine Breakthrough Infections with SARS-CoV-2 Variants. N. Engl. J. Med. 2021, 384, 2212–2218. [Google Scholar] [CrossRef]
- Chia, W.N.; Zhu, F.; Ong, S.W.X.; Young, B.E.; Fong, S.-W.; Le Bert, N.; Tan, C.W.; Tiu, C.; Zhang, J.; Tan, S.Y.; et al. Dynamics of SARS-CoV-2 neutralising antibody responses and duration of immunity: A longitudinal study. Lancet Microbe 2021, 2, e240–e249. [Google Scholar] [CrossRef]
- Milne, G.; Hames, T.; Scotton, C.; Gent, N.; Johnsen, A.; Anderson, R.M.; Ward, T. Does infection with or vaccination against SARS-CoV-2 lead to lasting immunity? Lancet Respir. Med. 2021, 9, 1450–1466. [Google Scholar] [CrossRef]
- He, Q.; Mao, Q.; An, C.; Zhang, J.; Gao, F.; Bian, L.; Li, C.; Liang, Z.; Xu, M.; Wang, J. Heterologous prime-boost: Breaking the protective immune response bottleneck of COVID-19 vaccine candidates. Emerg. Microbes Infect. 2021, 10, 629–637. [Google Scholar] [CrossRef]
- Sapkota, B.; Saud, B.; Shrestha, R.; Al-Fahad, D.; Sah, R.; Shrestha, S.; Rodriguez-Morales, A.J. Heterologous prime-boost strategies for COVID-19 vaccines. J. Travel Med. 2022, 29, taab191. [Google Scholar] [CrossRef]
- Westrop, S.J.; Whitaker, H.J.; Powell, A.A.; Power, L.; Whillock, C.; Campbell, H.; Simmons, R.; Warrener, L.; Ramsay, M.E.; Ladhani, S.N.; et al. Real-world data on immune responses following heterologous prime-boost COVID-19 vaccination schedule with Pfizer and AstraZeneca vaccines in England. J. Infect. 2022, 84, 692–700. [Google Scholar] [CrossRef] [PubMed]
- Collier, D.A.; Ferreira, I.A.T.M.; Kotagiri, P.; Datir, R.P.; Lim, E.Y.; Touizer, E.; Meng, B.; Abdullahi, A.; Baker, S.; Dougan, G.; et al. Age-related immune response heterogeneity to SARS-CoV-2 vaccine BNT162b2. Nature 2021, 596, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Patone, M.; Mei, X.W.; Handunnetthi, L.; Dixon, S.; Zaccardi, F.; Shankar-Hari, M.; Watkinson, P.; Khunti, K.; Harnden, A.; Coupland, C.A.C.; et al. Risk of Myocarditis After Sequential Doses of COVID-19 Vaccine and SARS-CoV-2 Infection by Age and Sex. Circulation 2022, 146, 743–754. [Google Scholar] [CrossRef] [PubMed]
| Approaches | Principles | Examples |
|---|---|---|
| Structure-guided vaccine design | The metastable prefusion conformation is the functional state for spike proteins, stabilizing the prefusion form could preserve neutralization-sensitive epitopes. | S-2P [37,68,69], S-6P [70], S-trimer [15,71], RBD dimer [30,31,72], etc. |
| T-cell-based vaccines | Circulating antibodies may be short-lived, or of low magnitude and/or potency, T cells have an important role for COVID-19 outcome and maintenance of SARS-CoV-2 immunity. | Peptide vaccine [73], MVA-S [74,75], etc. |
| Respiratory mucosal delivery | To elicit mucosal IgA and T-cell mediated immune responses in the respiratory tract. | CA4-dNS1-nCoV-RBD [32,33], Ad-vectored [43,76], intranasal subunit vaccine [42], etc. |
| Nanotechnologies | Multivalent display of antigen enhances B-cell responses and can provide longer-lasting immunity than monovalent antigens. | Mosaic nanoparticles [35,77,78], Ferritin-based nanoparticles [79,80], etc. |
| Approaches | Advantages | Limits |
|---|---|---|
| Structure-guided vaccine design | The immunogenicity and yield of target antigens are improved | The high-resolution structures are required |
| T-cell-based vaccines | Stronger CD8+ T-cell responses are provided | T-cell epitopes need to be identified |
| Respiratory mucosal delivery | Mucosal immunity in the respiratory tract can be elicited rapidly | Serological antibody titers are relatively lower than intramuscular injection |
| Nanotechnologies | Multivalent antigens can be displayed in one particle | The nanotoxicity needs further clinical investigation |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Xia, Y.; Liu, X.; Liu, G. Advanced Vaccine Design Strategies against SARS-CoV-2 and Emerging Variants. Bioengineering 2023, 10, 148. https://doi.org/10.3390/bioengineering10020148
Zhang J, Xia Y, Liu X, Liu G. Advanced Vaccine Design Strategies against SARS-CoV-2 and Emerging Variants. Bioengineering. 2023; 10(2):148. https://doi.org/10.3390/bioengineering10020148
Chicago/Turabian StyleZhang, Jianzhong, Yutian Xia, Xuan Liu, and Gang Liu. 2023. "Advanced Vaccine Design Strategies against SARS-CoV-2 and Emerging Variants" Bioengineering 10, no. 2: 148. https://doi.org/10.3390/bioengineering10020148
APA StyleZhang, J., Xia, Y., Liu, X., & Liu, G. (2023). Advanced Vaccine Design Strategies against SARS-CoV-2 and Emerging Variants. Bioengineering, 10(2), 148. https://doi.org/10.3390/bioengineering10020148






