High Precision Cervical Precancerous Lesion Classification Method Based on ConvNeXt
Abstract
:1. Introduction
2. Related Works
2.1. Cervical Cancer Diagnosis
2.2. Deep Learning Models for Cervical Cytology Analysis
3. Analysis of the DCCL Dataset
3.1. Dataset Overview
3.2. Dataset Processing
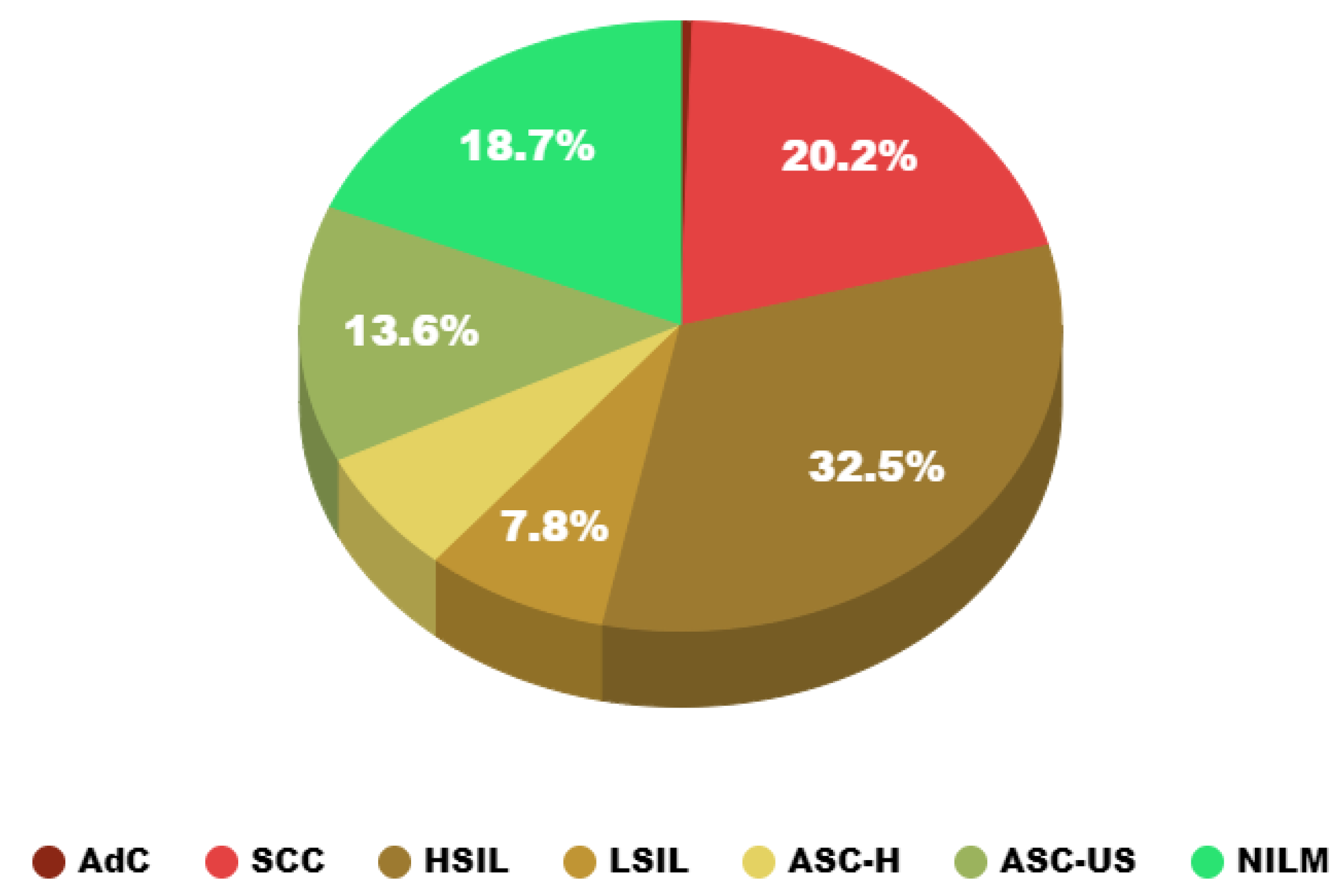
3.3. Data Characteristic Analysis
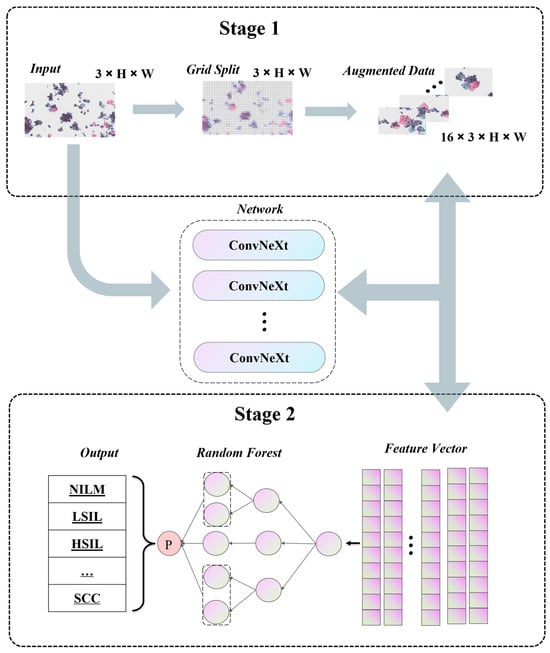
4. Methodology
4.1. Pipeline
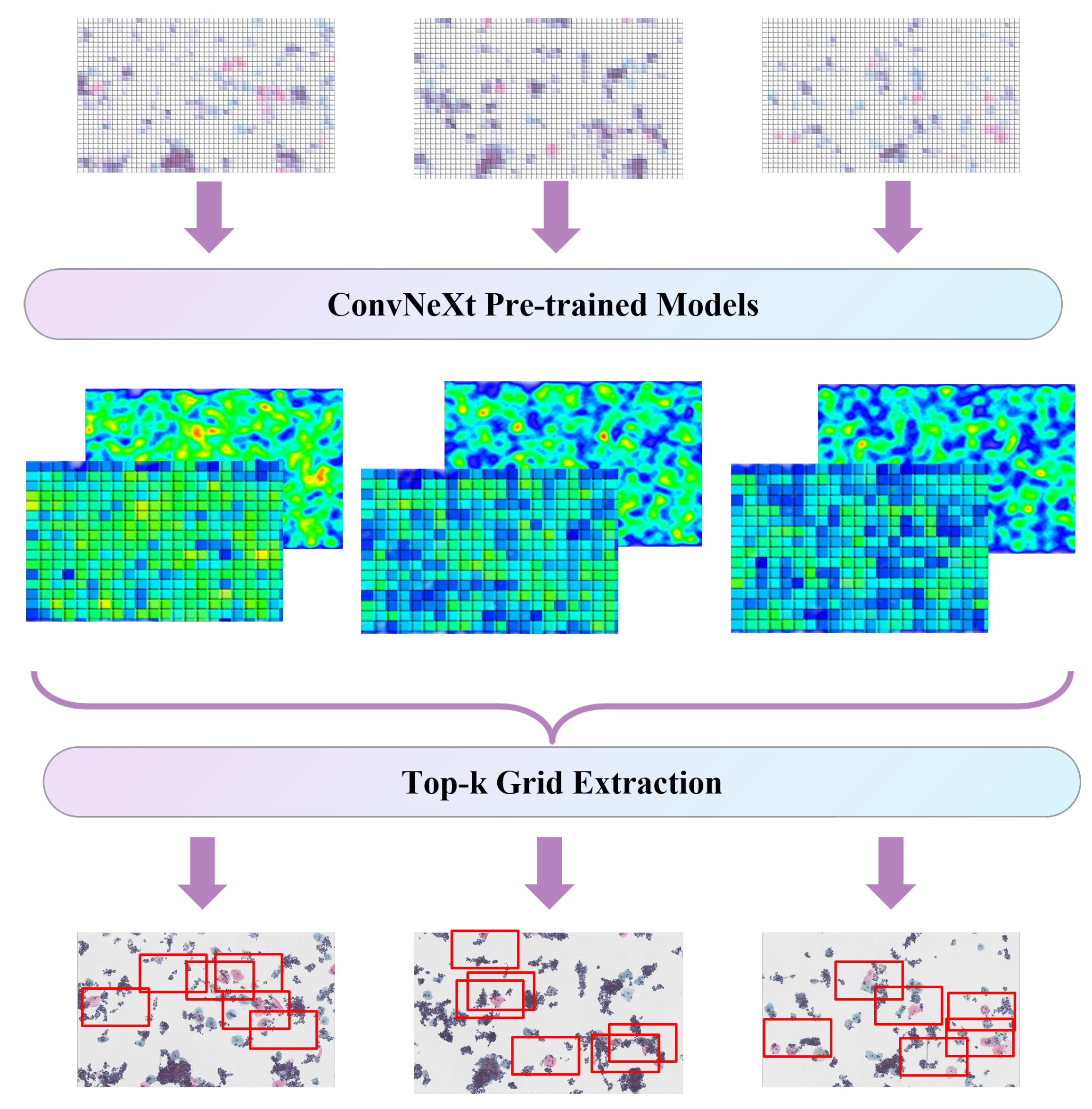
4.2. Self-Supervised Data Augmentation
4.3. Ensemble Learning Strategy
5. Experiments
5.1. Experiment Setup
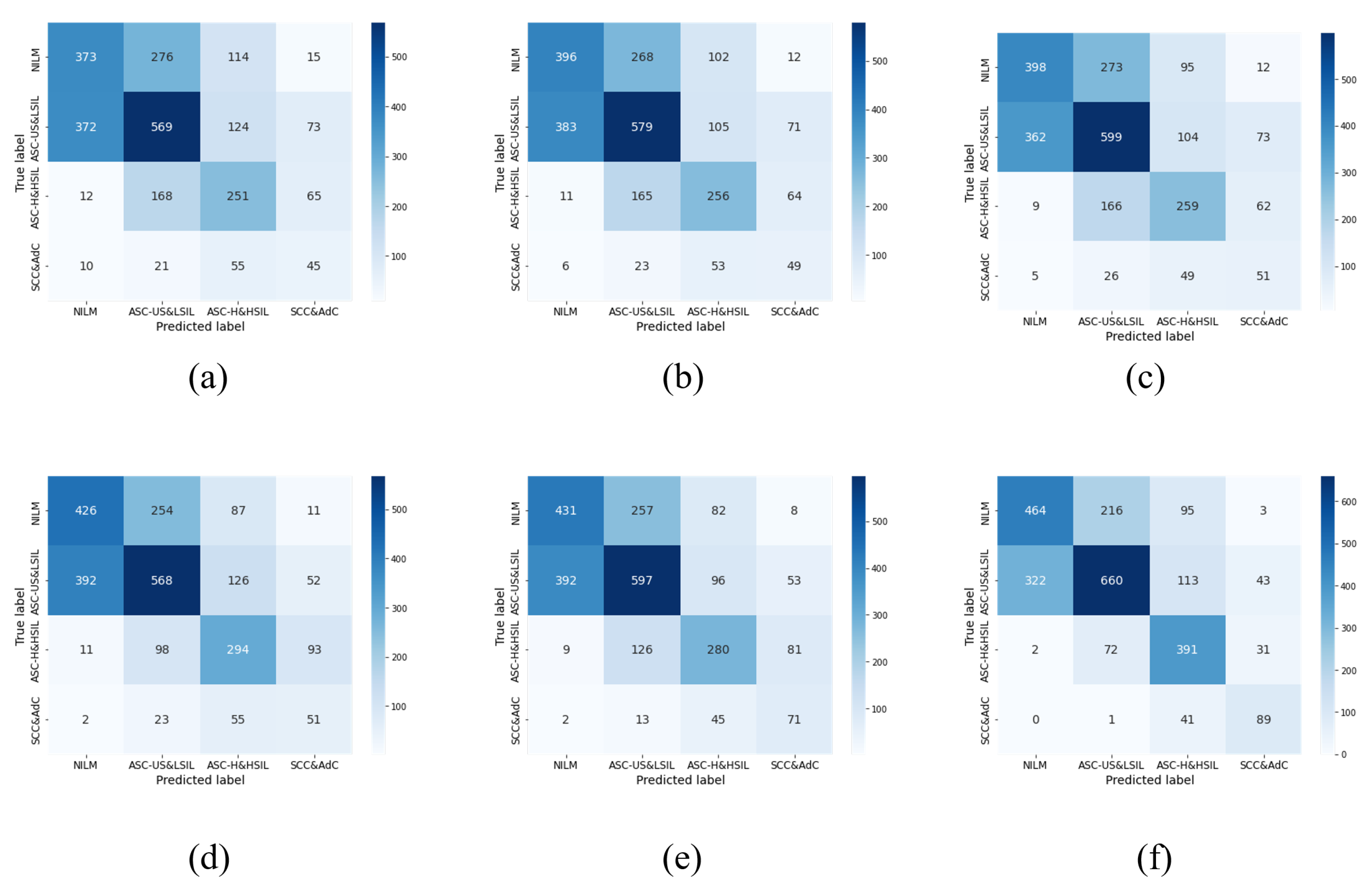
5.2. Fine-Tuning Policy Verification
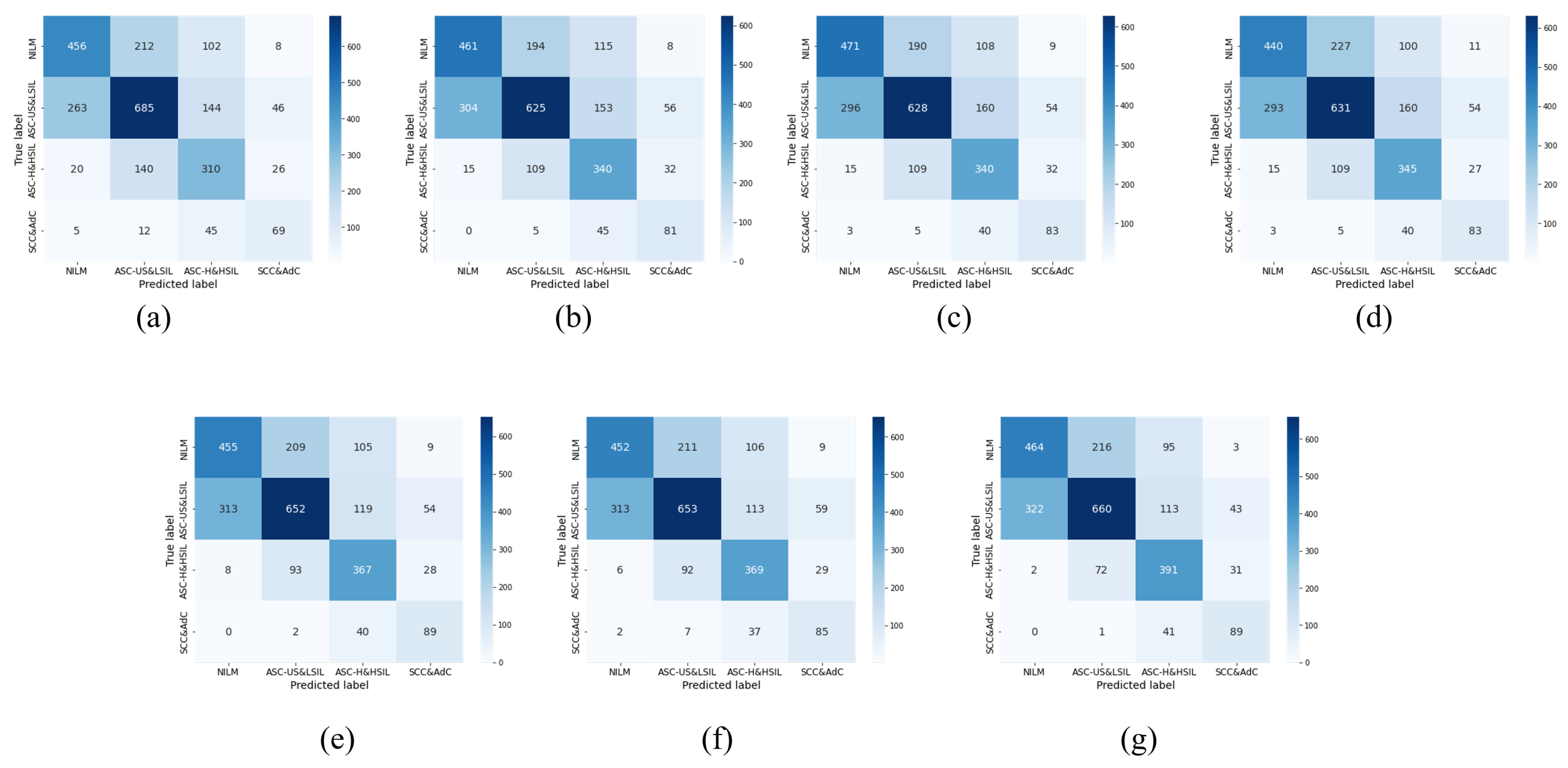
5.3. Comparison with Advanced Methods
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Wuerthner, B.A.; Avila-Wallace, M. Cervical cancer: Screening, management, and prevention. Nurse Pract. 2016, 41, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Ducatman, B.S. Cytology: Diagnostic Principles and Clinical Correlates; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Zhao, X.; Cui, Y.; Jiang, S.; Meng, Y.; Liu, A.; Wei, L.; Liu, T.; Han, H.; Liu, X.; Liu, F.; et al. Comparative study of HR HPV E6/E7 mRNA and HR-HPV DNA in cervical cancer screening. Zhonghua Yi Xue Za Zhi 2014, 94, 3432–3435. [Google Scholar] [PubMed]
- Shen, Y.; Xia, J.; Li, H.; Xu, Y.; Xu, S. Human papillomavirus infection rate, distribution characteristics, and risk of age in pre-and postmenopausal women. BMC Women’s Health 2021, 21, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Latsuzbaia, A.; Van Keer, S.; Broeck, D.V.; Weyers, S.; Donders, G.; De Sutter, P.; Tjalma, W.; Doyen, J.; Vorsters, A.; Arbyn, M. Clinical accuracy of Alinity m HR HPV assay on self-versus clinician-taken samples using the VALHUDES protocol. J. Mol. Diagn. 2023, 25, 957–966. [Google Scholar] [CrossRef]
- Vink, F.; Lissenberg-Witte, B.I.; Meijer, C.; Berkhof, J.; van Kemenade, F.; Siebers, A.; Steenbergen, R.; Bleeker, M.; Heideman, D. FAM19A4/miR124-2 methylation analysis as a triage test for HPV-positive women: Cross-sectional and longitudinal data from a Dutch screening cohort. Clin. Microbiol. Infect. 2021, 27, 125.e1–125.e6. [Google Scholar] [CrossRef]
- Liu, S.; Yuan, Z.; Qiao, X.; Liu, Q.; Song, K.; Kong, B.; Su, X. Light scattering pattern specific convolutional network static cytometry for label-free classification of cervical cells. Cytom. Part A 2021, 99, 610–621. [Google Scholar] [CrossRef]
- Bhatla, N.; Singhal, S.; Saraiya, U.; Srivastava, S.; Bhalerao, S.; Shamsunder, S.; Chavan, N.; Basu, P.; Purandare, C.; on behalf of FOGSI Expert Group. Screening and management of preinvasive lesions of the cervix: Good clinical practice recommendations from the Federation of Obstetrics and Gynaecologic Societies of India (FOGSI). J. Obstet. Gynaecol. Res. 2020, 46, 201–214. [Google Scholar] [CrossRef]
- Liu, Y.; Liao, J.; Yi, X.; Pan, Z.; Pan, J.; Sun, C.; Zhou, H.; Meng, Y. Diagnostic value of colposcopy in patients with cytology-negative and HR-HPV-positive cervical lesions. Arch. Gynecol. Obstet. 2022, 306, 1161–1169. [Google Scholar] [CrossRef] [PubMed]
- Papanicolaou, G. A new procedure for staining vaginal smears. Science 1942, 95, 438–439. [Google Scholar] [CrossRef]
- Silva-López, M.S.; Ilizaliturri Hernández, C.A.; Navarro Contreras, H.R.; Rodríguez Vázquez, Á.G.; Ortiz-Dosal, A.; Kolosovas-Machuca, E.S. Raman spectroscopy of individual cervical exfoliated cells in premalignant and malignant lesions. Appl. Sci. 2022, 12, 2419. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, D.; Wang, L.; Li, Y.; Chen, X.; Luo, R.; Che, S.; Liang, H.; Li, Y.; Liu, S.; et al. DCCL: A benchmark for cervical cytology analysis. In Proceedings of the Machine Learning in Medical Imaging: 10th International Workshop, MLMI 2019, Held in Conjunction with MICCAI 2019, Shenzhen, China, 13 October 2019; Proceedings 10. Springer: Cham, Switzerland, 2019; pp. 63–72. [Google Scholar]
- Cohen, P.A.; Jhingran, A.; Oaknin, A.; Denny, L. Cervical cancer. Lancet 2019, 393, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Miyoshi, A.; Ueda, Y.; Tanaka, Y.; Nakae, R.; Morimoto, A.; Shiomi, M.; Enomoto, T.; Sekine, M.; Sasagawa, T.; et al. An artificial intelligence-assisted diagnostic system improves the accuracy of image diagnosis of uterine cervical lesions. Mol. Clin. Oncol. 2022, 16, 1–6. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the dimensionality of data with neural networks. Science 2006, 313, 504–507. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Sun, J.; Tárnok, A.; Su, X. Deep learning-based single-cell optical image studies. Cytom. Part A 2020, 97, 226–240. [Google Scholar] [CrossRef]
- George, K.; Sankaran, P. Computer assisted recognition of breast cancer in biopsy images via fusion of nucleus-guided deep convolutional features. Comput. Methods Programs Biomed. 2020, 194, 105531. [Google Scholar] [CrossRef]
- Anwar, S.M.; Majid, M.; Qayyum, A.; Awais, M.; Alnowami, M.; Khan, M.K. Medical image analysis using convolutional neural networks: A review. J. Med. Syst. 2018, 42, 226. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.K.; Chen, M.; Malcolm, G.P.; Hempler, N.; Dholakia, K.; Powis, S.J. Label-free optical hemogram of granulocytes enhanced by artificial neural networks. Opt. Express 2019, 27, 13706–13720. [Google Scholar] [CrossRef] [PubMed]
- Mahbod, A.; Schaefer, G.; Wang, C.; Dorffner, G.; Ecker, R.; Ellinger, I. Transfer learning using a multi-scale and multi-network ensemble for skin lesion classification. Comput. Methods Programs Biomed. 2020, 193, 105475. [Google Scholar] [CrossRef] [PubMed]
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell 2018, 172, 1122–1131. [Google Scholar] [CrossRef] [PubMed]
- Coudray, N.; Ocampo, P.S.; Sakellaropoulos, T.; Narula, N.; Snuderl, M.; Fenyö, D.; Moreira, A.L.; Razavian, N.; Tsirigos, A. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat. Med. 2018, 24, 1559–1567. [Google Scholar] [CrossRef]
- Pan, C.; Schoppe, O.; Parra-Damas, A.; Cai, R.; Todorov, M.I.; Gondi, G.; von Neubeck, B.; Böğürcü-Seidel, N.; Seidel, S.; Sleiman, K.; et al. Deep learning reveals cancer metastasis and therapeutic antibody targeting in the entire body. Cell 2019, 179, 1661–1676. [Google Scholar] [CrossRef]
- Song, Y.; Zhang, L.; Chen, S.; Ni, D.; Li, B.; Zhou, Y.; Lei, B.; Wang, T. A deep learning based framework for accurate segmentation of cervical cytoplasm and nuclei. In Proceedings of the 2014 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Chicago, IL, USA, 26–30 August 2014; IEEE: Piscataway, NJ, USA, 2014; pp. 2903–2906. [Google Scholar]
- Zhang, L.; Kong, H.; Ting Chin, C.; Liu, S.; Fan, X.; Wang, T.; Chen, S. Automation-assisted cervical cancer screening in manual liquid-based cytology with hematoxylin and eosin staining. Cytom. Part A 2014, 85, 214–230. [Google Scholar] [CrossRef]
- Pramanik, R.; Biswas, M.; Sen, S.; de Souza Júnior, L.A.; Papa, J.P.; Sarkar, R. A fuzzy distance-based ensemble of deep models for cervical cancer detection. Comput. Methods Programs Biomed. 2022, 219, 106776. [Google Scholar] [CrossRef]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 2818–2826. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Jantzen, J.; Norup, J.; Dounias, G.; Bjerregaard, B. Pap-smear benchmark data for pattern classification. In Nature Inspired Smart Information Systems (NiSIS 2005); NiSIS: Albufeira, Portugal, 2005; pp. 1–9. [Google Scholar]
- Plissiti, M.E.; Dimitrakopoulos, P.; Sfikas, G.; Nikou, C.; Krikoni, O.; Charchanti, A. SIPAKMED: A new dataset for feature and image based classification of normal and pathological cervical cells in Pap smear images. In Proceedings of the 2018 25th IEEE International Conference on Image Processing (ICIP), Athens, Greece, 7–10 October 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 3144–3148. [Google Scholar]
- Liu, Z.; Mao, H.; Wu, C.Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A convnet for the 2020s. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 11976–11986. [Google Scholar]
- Lu, J.; Song, E.; Ghoneim, A.; Alrashoud, M. Machine learning for assisting cervical cancer diagnosis: An ensemble approach. Future Gener. Comput. Syst. 2020, 106, 199–205. [Google Scholar] [CrossRef]
- Chandran, V.; Sumithra, M.; Karthick, A.; George, T.; Deivakani, M.; Elakkiya, B.; Subramaniam, U.; Manoharan, S. Diagnosis of cervical cancer based on ensemble deep learning network using colposcopy images. Biomed Res. Int. 2021, 2021, 5584004. [Google Scholar] [CrossRef] [PubMed]
- Adweb, K.M.A.; Cavus, N.; Sekeroglu, B. Cervical cancer diagnosis using very deep networks over different activation functions. IEEE Access 2021, 9, 46612–46625. [Google Scholar] [CrossRef]
- Xu, T.; Liu, P.; Li, P.; Wang, X.; Xue, H.; Guo, J.; Dong, B.; Sun, P. RACNet: Risk assessment Net of cervical lesions in colposcopic images. Connect. Sci. 2022, 34, 2139–2157. [Google Scholar] [CrossRef]
- Soni, V.D.; Soni, A.N. Cervical cancer diagnosis using convolution neural network with conditional random field. In Proceedings of the 2021 Third International Conference on Inventive Research in Computing Applications (ICIRCA), Coimbatore, India, 2–4 September 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 1749–1754. [Google Scholar]
- Fang, M.; Lei, X.; Liao, B.; Wu, F.X. A Deep Neural Network for Cervical Cell Classification Based on Cytology Images. IEEE Access 2022, 10, 130968–130980. [Google Scholar] [CrossRef]
- Mohammed, B.A.; Senan, E.M.; Al-Mekhlafi, Z.G.; Alazmi, M.; Alayba, A.M.; Alanazi, A.A.; Alreshidi, A.; Alshahrani, M. Hybrid Techniques for Diagnosis with WSIs for Early Detection of Cervical Cancer Based on Fusion Features. Appl. Sci. 2022, 12, 8836. [Google Scholar] [CrossRef]
- Kavitha, R.; Jothi, D.K.; Saravanan, K.; Swain, M.P.; Gonzáles, J.L.A.; Bhardwaj, R.J.; Adomako, E. Ant colony optimization-enabled CNN deep learning technique for accurate detection of cervical cancer. Biomed Res. Int. 2023, 2023, 1742891. [Google Scholar] [CrossRef] [PubMed]
- Attallah, O. CerCan· Net: Cervical Cancer Classification Model via Multi-layer Feature Ensembles of Lightweight CNNs and Transfer Learning. Expert Syst. Appl. 2023, 229, 120624. [Google Scholar] [CrossRef]
- Zaki, N.; Qin, W.; Krishnan, A. Graph-based methods for cervical cancer segmentation: Advancements, limitations, and future directions. AI Open 2023, 4, 42–55. [Google Scholar] [CrossRef]
- Bnouni, N.; Amor, H.B.; Rekik, I.; Rhim, M.S.; Solaiman, B.; Amara, N.E.B. Boosting CNN learning by ensemble image preprocessing methods for cervical cancer segmentation. In Proceedings of the 2021 18th International Multi-Conference on Systems, Signals & Devices (SSD), Monastir, Tunisia, 22–25 March 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 264–269. [Google Scholar]
- Sellamuthu Palanisamy, V.; Athiappan, R.K.; Nagalingam, T. Pap smear based cervical cancer detection using residual neural networks deep learning architecture. Concurr. Comput. Pract. Exp. 2022, 34, e6608. [Google Scholar] [CrossRef]
- de Lima, C.R.; Khan, S.G.; Shah, S.H.; Ferri, L. Mask region-based CNNs for cervical cancer progression diagnosis on pap smear examinations. Heliyon 2023, 9, e21388. [Google Scholar] [CrossRef]
- Wita, D.S. Image Segmentation of Normal Pap Smear Thinprep using U-Net with Mobilenetv2 Encoder. J. Med. Inform. Technol. 2023, 1, 31–35. [Google Scholar] [CrossRef]
- Taha, B.; Dias, J.; Werghi, N. Classification of cervical-cancer using pap-smear images: A convolutional neural network approach. In Proceedings of the Medical Image Understanding and Analysis: 21st Annual Conference, MIUA 2017, Edinburgh, UK, 11–13 July 2017; Proceedings 21. Springer: Cham, Switzerland, 2017; pp. 261–272. [Google Scholar]
- Ghoneim, A.; Muhammad, G.; Hossain, M.S. Cervical cancer classification using convolutional neural networks and extreme learning machines. Future Gener. Comput. Syst. 2020, 102, 643–649. [Google Scholar] [CrossRef]
- Lin, H.; Hu, Y.; Chen, S.; Yao, J.; Zhang, L. Fine-grained classification of cervical cells using morphological and appearance based convolutional neural networks. IEEE Access 2019, 7, 71541–71549. [Google Scholar] [CrossRef]
- Fekri-Ershad, S.; Alsaffar, M.F. Developing a Tuned Three-Layer Perceptron Fed with Trained Deep Convolutional Neural Networks for Cervical Cancer Diagnosis. Diagnostics 2023, 13, 686. [Google Scholar] [CrossRef] [PubMed]
- Kalbhor, M.; Shinde, S.; Joshi, H.; Wajire, P. Pap smear-based cervical cancer detection using hybrid deep learning and performance evaluation. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 2023, 11, 1615–1624. [Google Scholar] [CrossRef]
- Kundu, R.; Chattopadhyay, S. Deep features selection through genetic algorithm for cervical pre-cancerous cell classification. Multimed. Tools Appl. 2023, 82, 13431–13452. [Google Scholar] [CrossRef]
- Tucker, J. CERVISCAN: An image analysis system for experiments in automatic cervical smear prescreening. Comput. Biomed. Res. 1976, 9, 93–107. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, R.; Matsubara, T.; Uehara, K. Data augmentation using random image cropping and patching for deep CNNs. IEEE Trans. Circuits Syst. Video Technol. 2019, 30, 2917–2931. [Google Scholar] [CrossRef]
- Yun, S.; Han, D.; Oh, S.J.; Chun, S.; Choe, J.; Yoo, Y. Cutmix: Regularization strategy to train strong classifiers with localizable features. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 6023–6032. [Google Scholar]
- Cubuk, E.D.; Zoph, B.; Mane, D.; Vasudevan, V.; Le, Q.V. Autoaugment: Learning augmentation policies from data. arXiv 2018, arXiv:1805.09501. [Google Scholar]
- Cubuk, E.D.; Zoph, B.; Shlens, J.; Le, Q.V. Randaugment: Practical automated data augmentation with a reduced search space. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, Seattle, WA, USA, 13–19 June 2020; pp. 702–703. [Google Scholar]
- Selvaraju, R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Grad-Cam, B. Visual explanations from deep networks via gradient-based localization. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
- Bao, H.; Dong, L.; Piao, S.; Wei, F. Beit: Bert pre-training of image transformers. arXiv 2021, arXiv:2106.08254. [Google Scholar]




| Cell Type | Train | Val | Test | Total |
|---|---|---|---|---|
| NILM | 2588 | 1540 | 2292 | 6420 |
| ASC-US | 2471 | 838 | 1378 | 4687 |
| ASC-H | 1147 | 543 | 591 | 2281 |
| LSIL | 1739 | 346 | 595 | 2680 |
| HSIL | 5890 | 1807 | 3482 | 11,179 |
| SCC | 3006 | 1225 | 2731 | 6962 |
| AdC | 122 | 20 | 31 | 173 |
| Total | 16,963 | 6319 | 11,100 | 34,382 |
| Cell Type | Train | Val | Test | Total |
|---|---|---|---|---|
| NILM | 1046 | 494 | 778 | 2318 |
| ASC-US&LSIL | 2108 | 731 | 1138 | 3977 |
| ASC-H&HSIL | 992 | 401 | 496 | 1889 |
| SCC&AdC | 243 | 61 | 131 | 435 |
| Total | 4389 | 1687 | 2543 | 8619 |
| Dataset | Patients | Labelled Patches | Labelled Cells | Lesion Cell Types | Classification Annotations | Detection Annotations | Open Source |
|---|---|---|---|---|---|---|---|
| CerviSCAN [60] | 82 | 900 | 12,043 | 3 | ✓ | × | ✓ |
| Herlev [37] | - | - | 917 | 3 | ✓ | × | ✓ |
| HEMLBC [30] | 200 | - | 2370 | 4 | ✓ | ✓ | × |
| DCCL [15] | 1167 | 14,432 | 34,392 | 6 | ✓ | ✓ | ✓ |
| Method | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) |
|---|---|---|---|---|
| Raw ConvNeXt | 59.77 | 56.12 | 58.49 | 57.09 |
| +CutMix [62] | 59.26 | 55.98 | 61.14 | 57.83 |
| +Autoaug [63] | 59.85 | 56.62 | 61.91 | 58.53 |
| +Randaug [64] | 58.95 | 56.11 | 61.23 | 58.02 |
| +SDA | 61.46 | 58.61 | 64.43 | 60.80 |
| +ELS | 61.30 | 58.01 | 63.69 | 60.15 |
| Our Method | 63.08 | 60.78 | 66.10 | 62.82 |
| Method | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) |
|---|---|---|---|---|
| ResNet [35] | 48.68 | 43.13 | 45.72 | 44.08 |
| Inception [32] | 50.33 | 45.08 | 47.70 | 46.04 |
| DenseNet [36] | 51.39 | 46.14 | 48.74 | 47.09 |
| Swin [67] | 52.65 | 47.11 | 50.72 | 48.32 |
| Beit [68] | 54.23 | 50.20 | 54.63 | 51.71 |
| Our Method | 63.08 | 60.78 | 66.10 | 62.82 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, J.; Zhang, T.; Gong, Z.; Huang, X. High Precision Cervical Precancerous Lesion Classification Method Based on ConvNeXt. Bioengineering 2023, 10, 1424. https://doi.org/10.3390/bioengineering10121424
Tang J, Zhang T, Gong Z, Huang X. High Precision Cervical Precancerous Lesion Classification Method Based on ConvNeXt. Bioengineering. 2023; 10(12):1424. https://doi.org/10.3390/bioengineering10121424
Chicago/Turabian StyleTang, Jing, Ting Zhang, Zeyu Gong, and Xianjun Huang. 2023. "High Precision Cervical Precancerous Lesion Classification Method Based on ConvNeXt" Bioengineering 10, no. 12: 1424. https://doi.org/10.3390/bioengineering10121424
APA StyleTang, J., Zhang, T., Gong, Z., & Huang, X. (2023). High Precision Cervical Precancerous Lesion Classification Method Based on ConvNeXt. Bioengineering, 10(12), 1424. https://doi.org/10.3390/bioengineering10121424