Photocatalytic Reduction of Methylene Blue by Surface-Engineered Recombinant Escherichia coli as a Whole-Cell Biocatalyst
Abstract
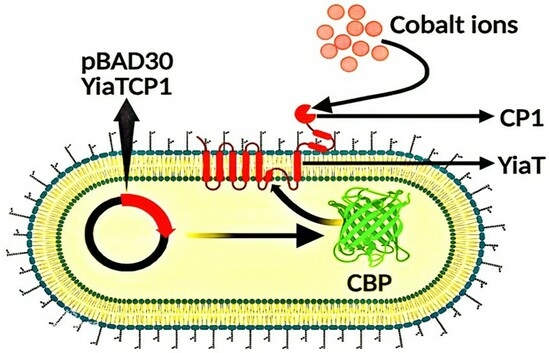
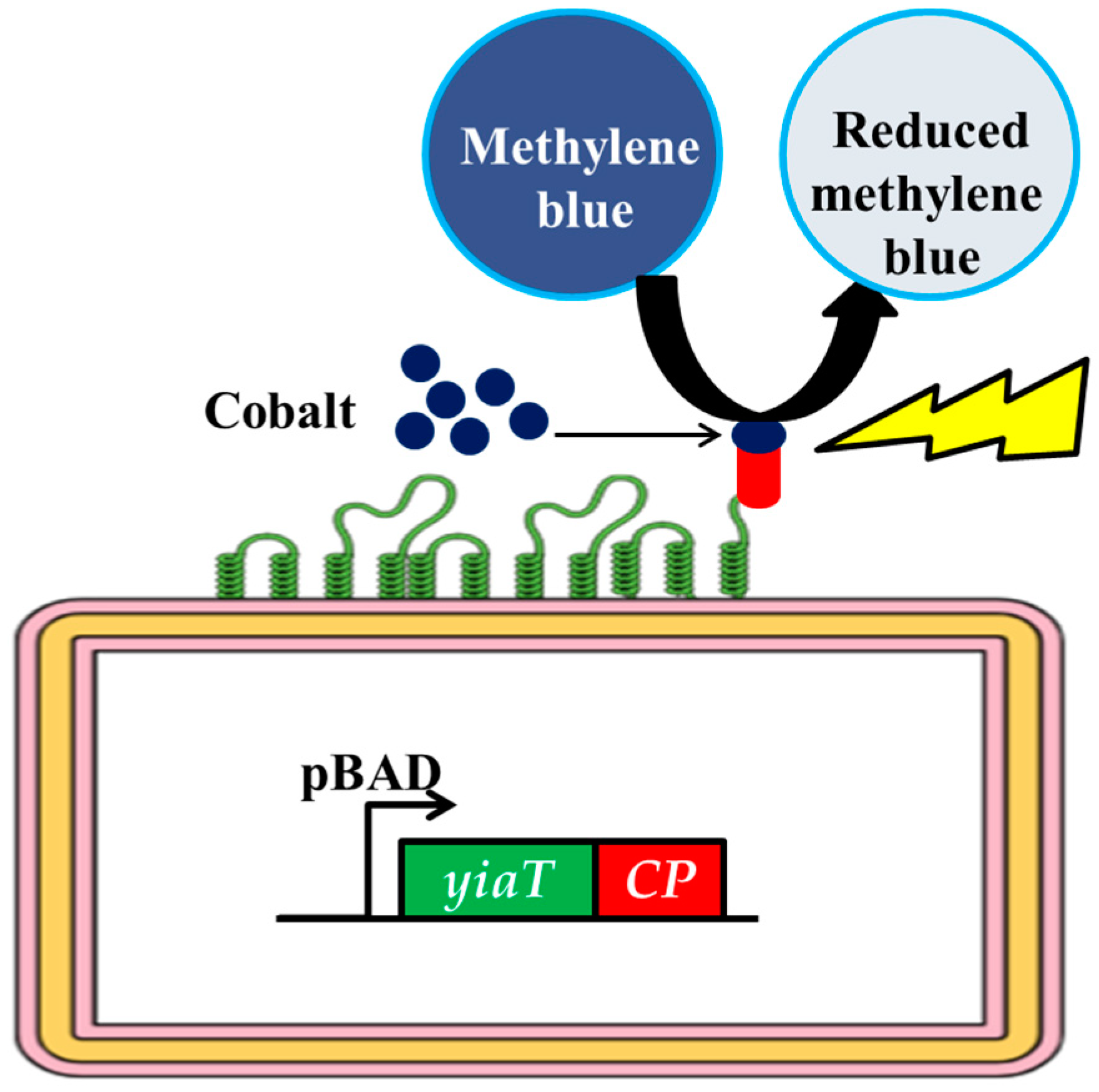
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Recombinant Plasmid Construction
2.3. SDS Page Analysis
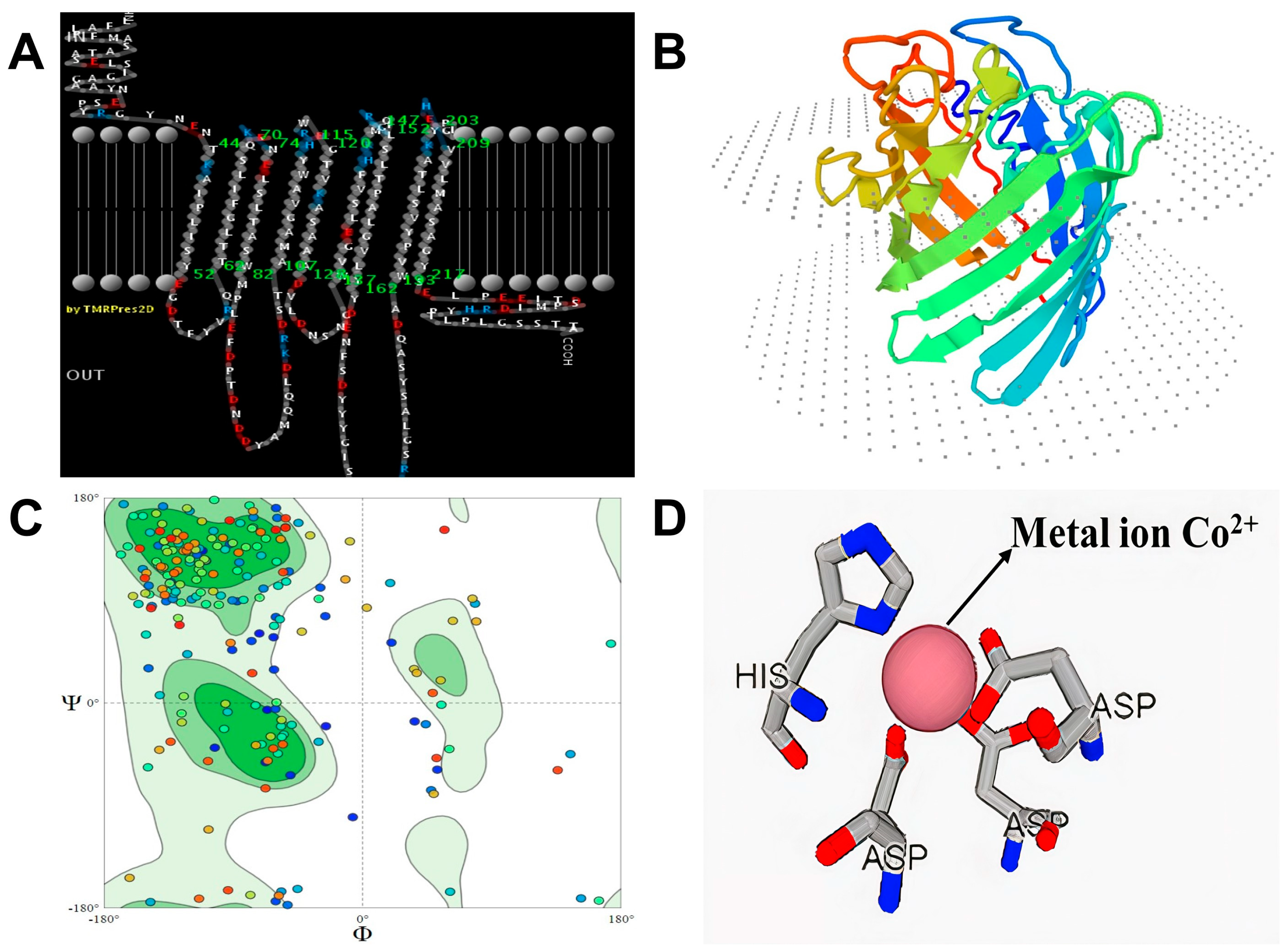
2.4. Molecular Modeling Studies
2.5. Cobalt Recovery Analysis
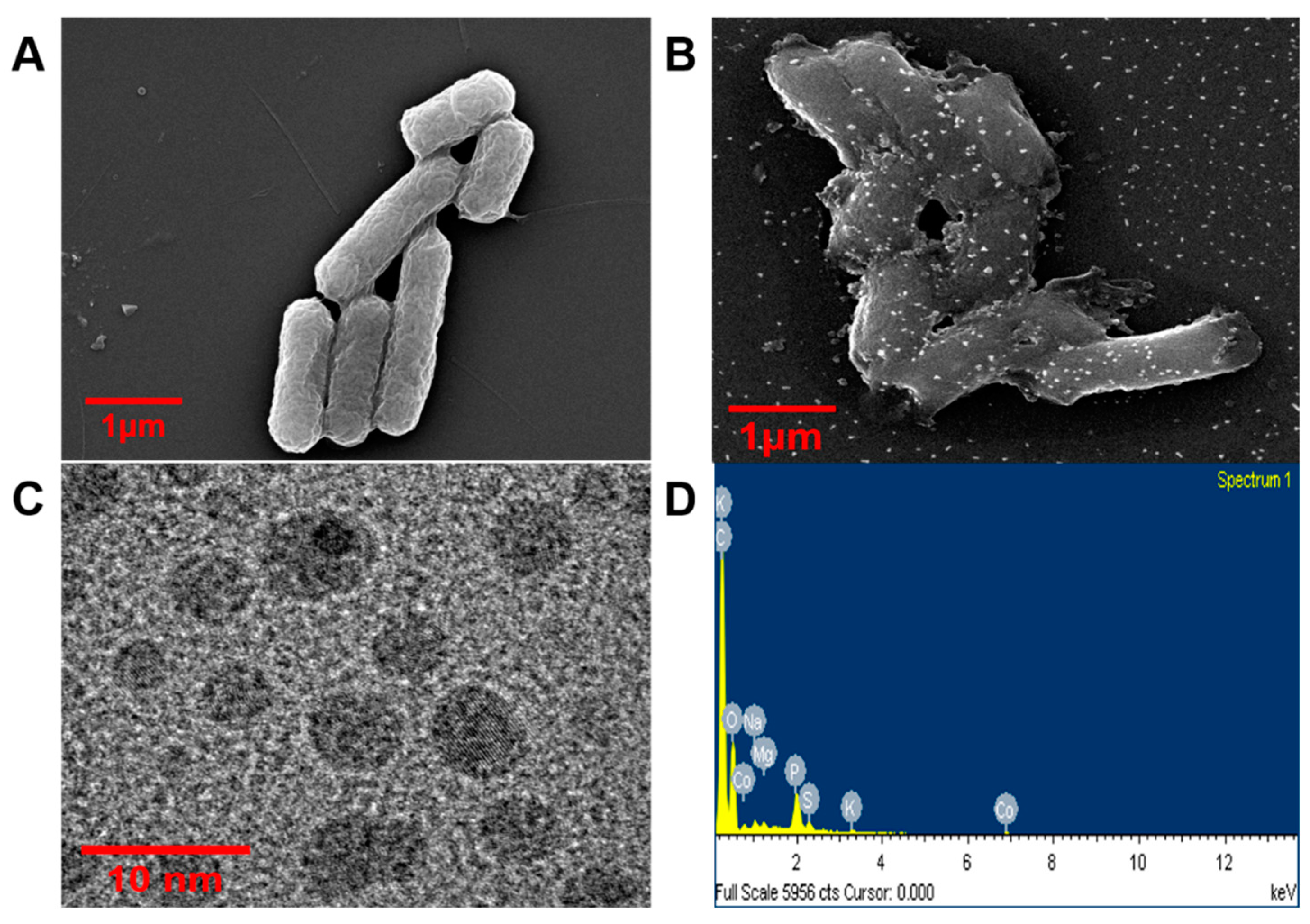
2.6. FE-SEM, TEM, and EDS Analysis
2.7. Photocatalytic Properties of Cobalt Nanoparticle
3. Results and Discussion
3.1. Molecular Modeling and Docking
3.2. Optimization of Expression Conditions
3.3. Cobalt Bio-Adsorption Studies
3.4. Physiochemical Characterization of Bio-Adsorbed Cobalt
FE-SEM, TEM, and EDS Analysis
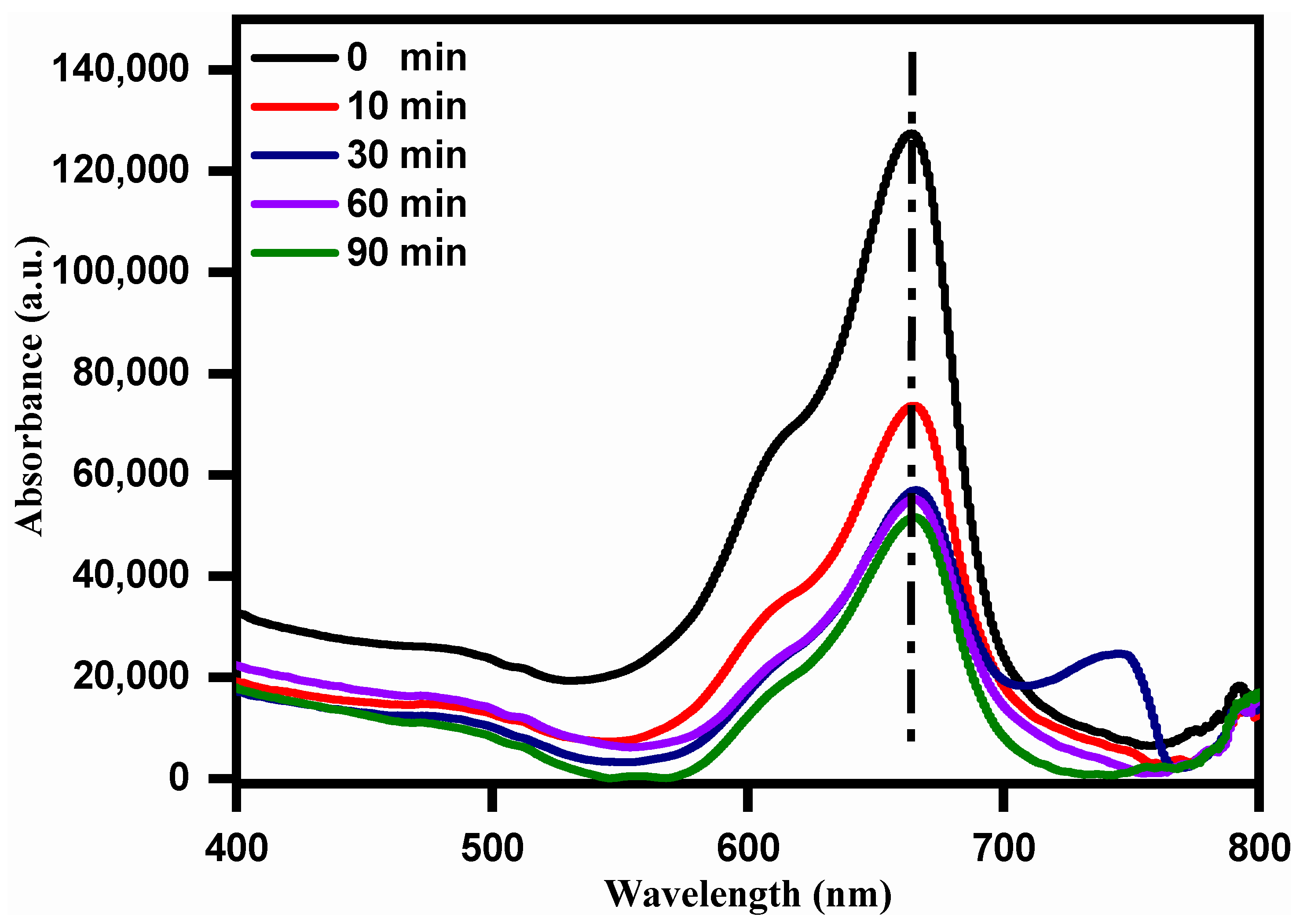
3.5. Photo-Catalytic Degradation Activity with Methylene Blue
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chasapis, C.T.; Spiliopoulou, C.A.; Loutsidou, A.C.; Stefanidou, M.E. Zinc and Human Health: An Update. Arch. Toxicol. 2012, 86, 521–534. [Google Scholar] [CrossRef] [PubMed]
- El-Alfy, M.A.; El-Amier, Y.A.; El-Eraky, T.E. Land Use/Cover and Eco-Toxicity Indices for Identifying Metal Contamination in Sediments of Drains, Manzala Lake, Egypt. Heliyon 2020, 6, e03177. [Google Scholar] [CrossRef] [PubMed]
- Canaza, A.; Pozo, L.; Ferrufino-Guardia, E.; Vargas, V.A.; Canaza, A.; Pozo, L.; Ferrufino-Guardia, E.; Vargas, V.A. Biosorption of Lead (ii) Ions by Dead Bacterial Biomass Isolated from Mine Water. Rev. Boliv. Química 2021, 38, 119–125. [Google Scholar] [CrossRef]
- Ceglia, A.; Meulebroeck, W.; Baert, K.; Wouters, H.; Nys, K.; Thienpont, H.; Terryn, H. Cobalt Absorption Bands for the Differentiation of Historical Na and Ca/K Rich Glass. Surf. Interface Anal. 2011, 44, 219–226. [Google Scholar] [CrossRef]
- Raghu, G.; Balaji, V.; Venkateswaran, G.; Rodrigue, A.; Maruthi Mohan, P. Bioremediation of Trace Cobalt from Simulated Spent Decontamination Solutions of Nuclear Power Reactors Using E. Coli Expressing NiCoT Genes. Appl. Microbiol. Biotechnol. 2008, 81, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Dewulf, J.; Van der Vorst, G.; Denturck, K.; Van Langenhove, H.; Ghyoot, W.; Tytgat, J.; Vandeputte, K. Recycling Rechargeable Lithium Ion Batteries: Critical Analysis of Natural Resource Savings. Resour. Conserv. Recycl. 2010, 54, 229–234. [Google Scholar] [CrossRef]
- Bertuol, D.A.; Toniasso, C.; Jiménez, B.M.; Meili, L.; Dotto, G.L.; Tanabe, E.H.; Aguiar, M.L. Application of Spouted Bed Elutriation in the Recycling of Lithium Ion Batteries. J. Power Sources 2015, 275, 627–632. [Google Scholar] [CrossRef]
- Kang, J.; Senanayake, G.; Sohn, J.; Shin, S.M. Recovery of Cobalt Sulfate from Spent Lithium Ion Batteries by Reductive Leaching and Solvent Extraction with Cyanex 272. Hydrometallurgy 2010, 100, 168–171. [Google Scholar] [CrossRef]
- Li, L.; Dunn, J.B.; Zhang, X.X.; Gaines, L.; Chen, R.J.; Wu, F.; Amine, K. Recovery of Metals from Spent Lithium-Ion Batteries with Organic Acids as Leaching Reagents and Environmental Assessment. J. Power Sources 2013, 233, 180–189. [Google Scholar] [CrossRef]
- United States Environmental Protection Agency. Introduction to Hazardous Waste Identification (40 CFR Parts 261). 2005. Available online: http://www.epa.gov/osw/inforesources/pubs/training/hwid05.pdf (accessed on 30 September 2005).
- Ogunseitan, O.A.; Schoenung, J.M.; Saphores, J.-D.M.; Shapiro, A.A. The Electronics Revolution: From E-Wonderland to E-Wasteland. Science 2009, 326, 670–671. [Google Scholar] [CrossRef]
- Bertuol, D.A.; Machado, C.M.; Silva, M.L.; Calgaro, C.O.; Dotto, G.L.; Tanabe, E.H. Recovery of Cobalt from Spent Lithium-Ion Batteries Using Supercritical Carbon Dioxide Extraction. Waste Manag. 2016, 51, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Oyagbemi, A.A.; Omobowale, T.O.; Awoyomi, O.V.; Ajibade, T.O.; Falayi, O.O.; Ogunpolu, B.S.; Okotie, U.J.; Asenuga, E.R.; Adejumobi, O.A.; Hassan, F.O.; et al. Cobalt Chloride Toxicity Elicited Hypertension and Cardiac Complication via Induction of Oxidative Stress and Upregulation of COX-2/Bax Signaling Pathway. Hum. Exp. Toxicol. 2018, 38, 519–532. [Google Scholar] [CrossRef] [PubMed]
- Venkatraman, V.; Wong, M.K.; Shalita, C.; Parente, B.; Lad, S.P. Cobalt-Induced Toxicity and Spasticity Secondary to Hip Arthroplasty: Case Report and Review of the Literature. Cureus 2020, 12, e12368. [Google Scholar] [CrossRef] [PubMed]
- Krebs, W.; Brombacher, C.; Bosshard, P.P.; Bachofen, R.; Brandl, H. Microbial Recovery of Metals from Solids. FEMS Microbiol. Rev. 1997, 20, 605–617. [Google Scholar] [CrossRef]
- Peres, E.C.; Cunha, J.M.; Dortzbacher, G.F.; Pavan, F.A.; Lima, É.C.; Foletto, E.L.; Dotto, G.L. Treatment of Leachates Containing Cobalt by Adsorption on Spirulina Sp. and Activated Charcoal. J. Environ. Chem. Eng. 2018, 6, 677–685. [Google Scholar] [CrossRef]
- Eksteen, J.J.; Oraby, E.A.; Nguyen, V. Leaching and Ion Exchange Based Recovery of Nickel and Cobalt from a Low Grade, Serpentine-Rich Sulfide Ore Using an Alkaline Glycine Lixiviant System. Min. Eng. 2020, 145, 106073. [Google Scholar] [CrossRef]
- Choo, K.-H.; Kwon, D.-J.; Lee, K.-W.; Choi, S.-J. Selective Removal of Cobalt Species Using Nanofiltration Membranes. Environ. Sci. Technol. 2002, 36, 1330–1336. [Google Scholar] [CrossRef]
- Maloney, M.P.; Moody, G.J.; Thomas, J.D.R. Extraction of Metals from Aqueous Solution with Polyurethane Foam. Analyst 1980, 105, 1087–1097. [Google Scholar] [CrossRef]
- Li, J.; Li, Y.; Cui, K.; Li, H.; Feng, J.; Pu, X.; Xiong, W.; Liu, N.; Yuan, G. Novel MOFs-Based Ion-Imprinted Polymer for Selective Separation of Cobalt Ions from Waste Battery Leaching Solution. Inorganica Chim. Acta 2022, 536, 120922. [Google Scholar] [CrossRef]
- Dotto, G.L.; Cunha, J.M.; Calgaro, C.O.; Tanabe, E.H.; Bertuol, D.A. Surface Modification of Chitin Using Ultrasound-Assisted and Supercritical CO2 Technologies for Cobalt Adsorption. J. Hazard. Mater. 2015, 295, 29–36. [Google Scholar] [CrossRef]
- Freitas, M.B.J.G.; Celante, V.G.; Pietre, M.K. Electrochemical Recovery of Cobalt and Copper from Spent Li-Ion Batteries as Multilayer Deposits. J. Power Sources 2010, 195, 3309–3315. [Google Scholar] [CrossRef]
- Schippers, A.; Hedrich, S.; Vasters, J.; Drobe, M.; Sand, W.; Willscher, S. Biomining: Metal Recovery from Ores with Microorganisms. In Geobiotechnology I: Metal-Related Issues; Schippers, A., Glombitza, F., Sand, W., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 1–47. ISBN 978-3-642-54710-2. [Google Scholar]
- Maruthamuthu, M.k.; Nadarajan, S.P.; Ganesh, I.; Ravikumar, S.; Yun, H.; Yoo, I.; Hong, S.H. Construction of a High Efficiency Copper Adsorption Bacterial System via Peptide Display and Its Application on Copper Dye Polluted Wastewater. Bioprocess Biosyst. Eng. 2015, 38, 2077–2084. [Google Scholar] [CrossRef] [PubMed]
- Pollmann, K.; Kutschke, S.; Matys, S.; Kostudis, S.; Hopfe, S.; Raff, J. Novel Biotechnological Approaches for the Recovery of Metals from Primary and Secondary Resources. Minerals 2016, 6, 54. [Google Scholar] [CrossRef]
- Maruthamuthu, M.k.; Selvamani, V.; Nadarajan, S.P.; Yun, H.; Oh, Y.-K.; Eom, G.T.; Hong, S.H. Manganese and Cobalt Recovery by Surface Display of Metal Binding Peptide on Various Loops of OmpC in Escherichia coli. J. Ind. Microbiol. Biotechnol. 2018, 45, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Ahmmad, B.; Leonard, K.; Islam, M.S.; Kurawaki, J.; Muruganandham, M.; Ohkubo, T.; Kuroda, Y. Green Synthesis of Mesoporous Hematite (α-Fe2O3) Nanoparticles and Their Photocatalytic Activity. Adv. Powder Technol. 2013, 24, 160–167. [Google Scholar] [CrossRef]
- Joy Prabu, H.; Johnson, I. Plant-Mediated Biosynthesis and Characterization of Silver Nanoparticles by Leaf Extracts of Tragia involucrata, Cymbopogon citronella, Solanum verbascifolium and Tylophora ovata. Karbala Int. J. Mod. Sci. 2015, 1, 237–246. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Bagos, P.G.; Liakopoulos, T.D.; Spyropoulos, I.C.; Hamodrakas, S.J. PRED-TMBB: A Web Server for Predicting the Topology of Beta-Barrel Outer Membrane Proteins. Nucleic Acids Res. 2004, 32, W400–W404. [Google Scholar] [CrossRef]
- Zheng, W.; Zhang, C.; Li, Y.; Pearce, R.; Bell, E.W.; Zhang, Y. Folding Non-Homologous Proteins by Coupling Deep-Learning Contact Maps with I-TASSER Assembly Simulations. Cell Rep. Methods 2021, 1, 100014. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Lin, Y.-F.; Cheng, C.-W.; Shih, C.-S.; Hwang, J.-K.; Yu, C.-S.; Lu, C.-H. MIB: Metal Ion-Binding Site Prediction and Docking Server. J. Chem. Inf. Model. 2016, 56, 2287–2291. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.-H.; Lin, Y.-F.; Lin, J.-J.; Yu, C.-S. Prediction of Metal Ion–Binding Sites in Proteins Using the Fragment Transformation Method. PLoS ONE 2012, 7, e39252. [Google Scholar] [CrossRef] [PubMed]
- Soltani, N.; Saion, E.; Hussein, M.Z.; Erfani, M.; Abedini, A.; Bahmanrokh, G.; Navasery, M.; Vaziri, P. Visible Light-Induced Degradation of Methylene Blue in the Presence of Photocatalytic ZnS and CdS Nanoparticles. Int. J. Mol. Sci. 2012, 13, 12242–12258. [Google Scholar] [CrossRef] [PubMed]
- Torres-Martínez, C.L.; Kho, R.; Mian, O.I.; Mehra, R.K. Efficient Photocatalytic Degradation of Environmental Pollutants with Mass-Produced ZnS Nanocrystals. J. Colloid. Interface Sci. 2001, 240, 525–532. [Google Scholar] [CrossRef]
- Pouretedal, H.R.; Norozi, A.; Keshavarz, M.H.; Semnani, A. Nanoparticles of Zinc Sulfide Doped with Manganese, Nickel and Copper as Nanophotocatalyst in the Degradation of Organic Dyes. J. Hazard. Mater. 2009, 162, 674–681. [Google Scholar] [CrossRef]
- Zhao, J.; Yang, X. Photocatalytic Oxidation for Indoor Air Purification: A Literature Review. Build. Environ. 2003, 38, 645–654. [Google Scholar] [CrossRef]
- Das, D.P.; Biswal, N.; Martha, S.; Parida, K.M. Solar-Light Induced Photodegradation of Organic Pollutants over CdS-Pillared Zirconium–Titanium Phosphate (ZTP). J. Mol. Catal. A Chem. 2011, 349, 36–41. [Google Scholar] [CrossRef]



| Strain/Plasmid | Relevant Genotype/Property | Source |
|---|---|---|
| E. coli Strains | ||
| TOP10 | F-mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galE15 galK16 rpsL(Str R) endA1 λ− | Stratagene |
| Plasmids | ||
| pBAD30 | Amp R | NEB a |
| pBADCP1 | pBAD30 containing YiaT-CP1 | This work |
| Name | Sequence (5′ to 3′) |
|---|---|
| C_F | GAGCTCATGTTAATTAATCGCAATATTGTGGCGTTATTTG |
| CP3_R | GGTACCGGTGGTGCTGCTGCCCAGCGGCAGGGTCGGATAATGACGATCAATCATCGGGCTGTCGGTAAT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumaravel, A.; Selvamani, V.; Hong, S.H. Photocatalytic Reduction of Methylene Blue by Surface-Engineered Recombinant Escherichia coli as a Whole-Cell Biocatalyst. Bioengineering 2023, 10, 1389. https://doi.org/10.3390/bioengineering10121389
Kumaravel A, Selvamani V, Hong SH. Photocatalytic Reduction of Methylene Blue by Surface-Engineered Recombinant Escherichia coli as a Whole-Cell Biocatalyst. Bioengineering. 2023; 10(12):1389. https://doi.org/10.3390/bioengineering10121389
Chicago/Turabian StyleKumaravel, Ashokkumar, Vidhya Selvamani, and Soon Ho Hong. 2023. "Photocatalytic Reduction of Methylene Blue by Surface-Engineered Recombinant Escherichia coli as a Whole-Cell Biocatalyst" Bioengineering 10, no. 12: 1389. https://doi.org/10.3390/bioengineering10121389
APA StyleKumaravel, A., Selvamani, V., & Hong, S. H. (2023). Photocatalytic Reduction of Methylene Blue by Surface-Engineered Recombinant Escherichia coli as a Whole-Cell Biocatalyst. Bioengineering, 10(12), 1389. https://doi.org/10.3390/bioengineering10121389