Genomic Insights on the Carbon-Negative Workhorse: Systematical Comparative Genomic Analysis on 56 Synechococcus Strains
Abstract
1. Introduction
2. Materials and Methods
2.1. Synechococcus Strains and Database Accession Numbers
2.2. Construction of Phylogenetic Trees
2.3. Pan-Core Genomes Analysis
2.4. Analysis of Genes Exclusively Related to Temperature and Salinity
3. Results
3.1. General Features of 56 Synechococcus Strains
3.2. Phylogenetic and Comparative Genome Analyses
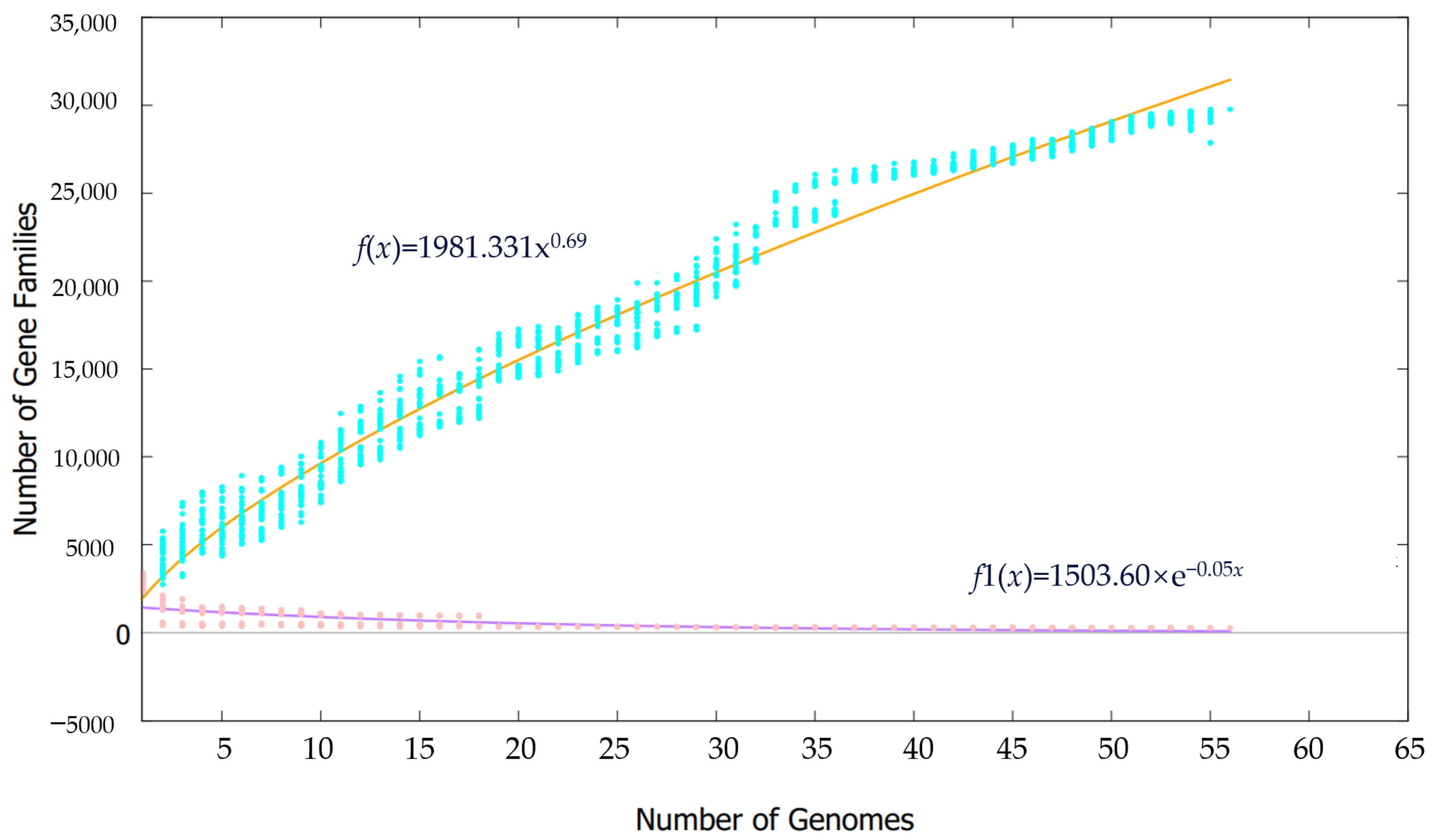
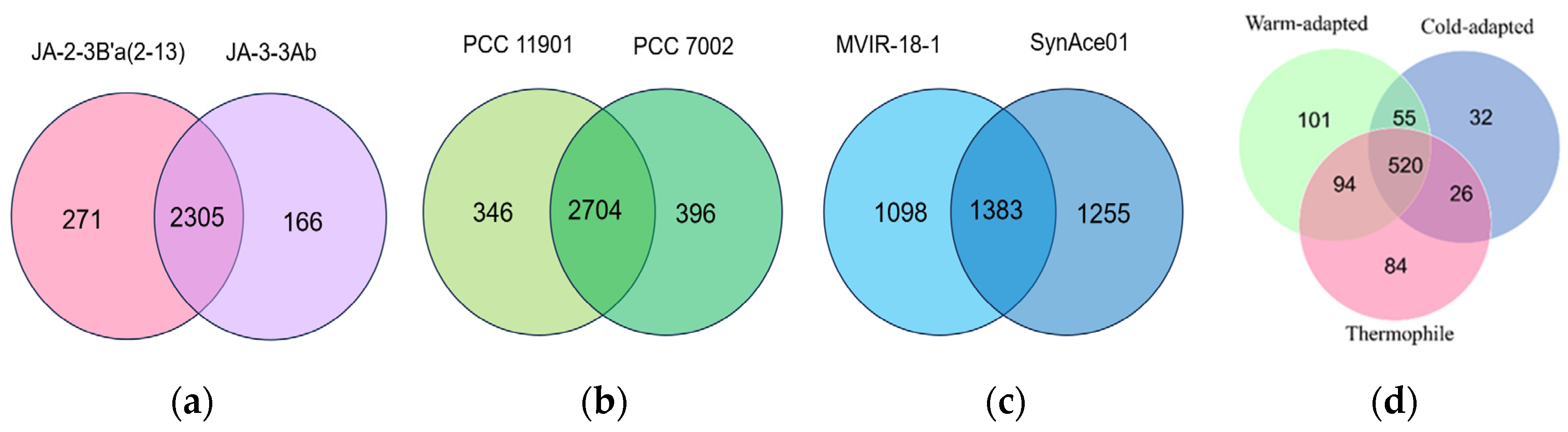
3.3. Core and Pan Genomic Analyses of the 56 Synechococcus Strains
3.4. Temperature Adaptation Mechanism of Synechococcus and Comparative Genomics
3.4.1. Growth Temperature and Structural Characteristics of the Synechococcus Genome
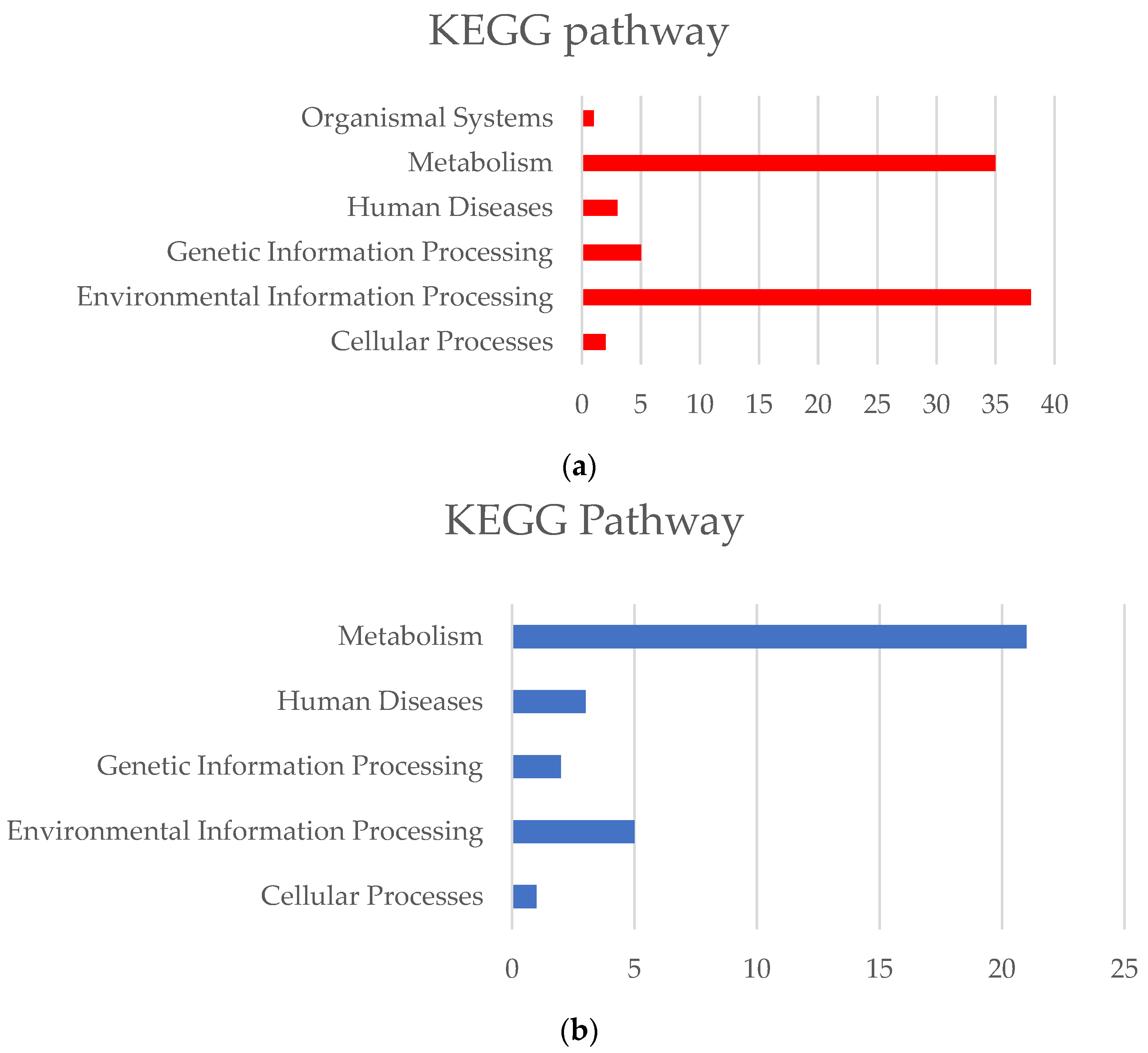
3.4.2. Temperature Adaptation Mechanisms
3.4.3. Thermal Adaptation Strategies
3.4.4. Cold Adaptation Strategies
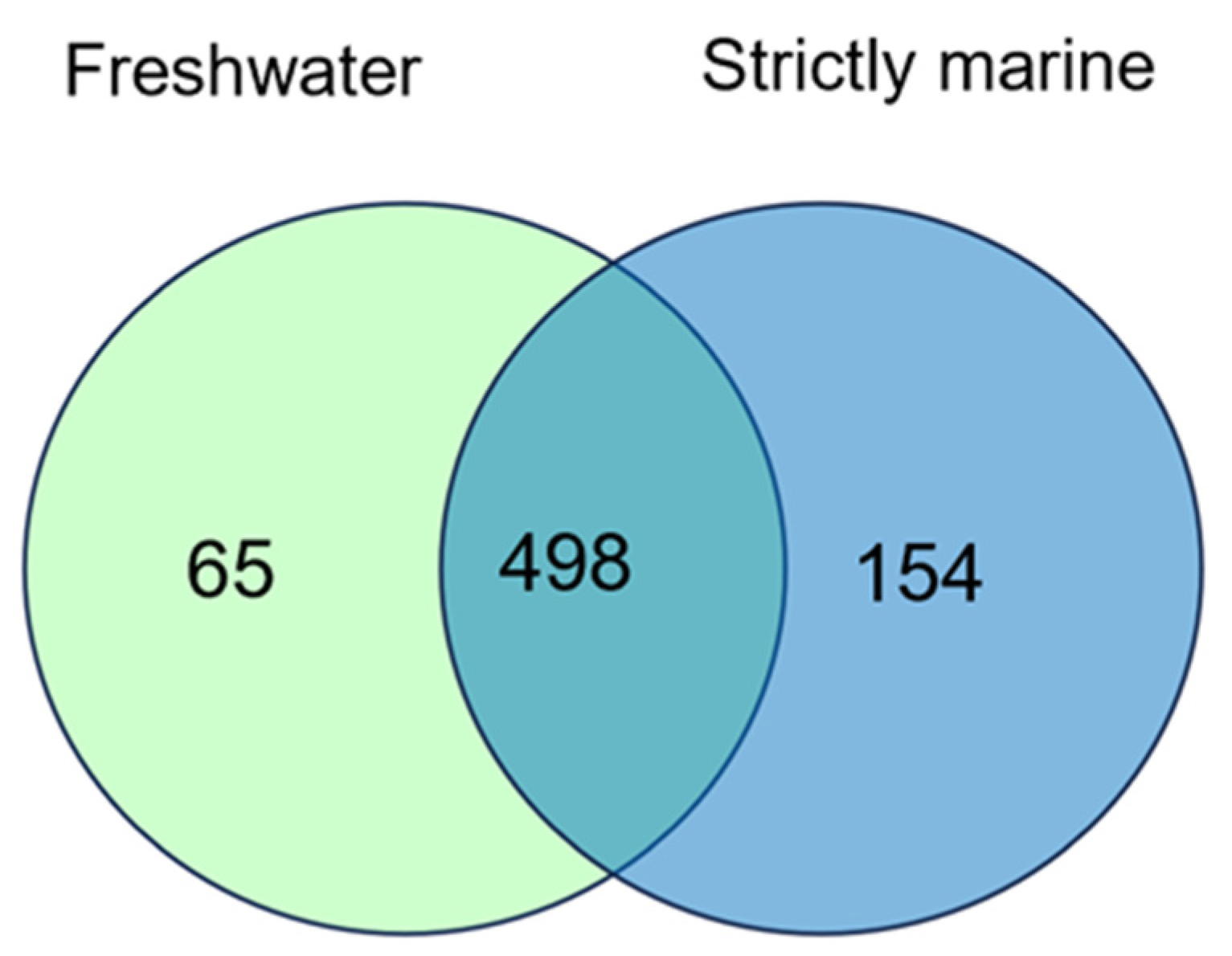
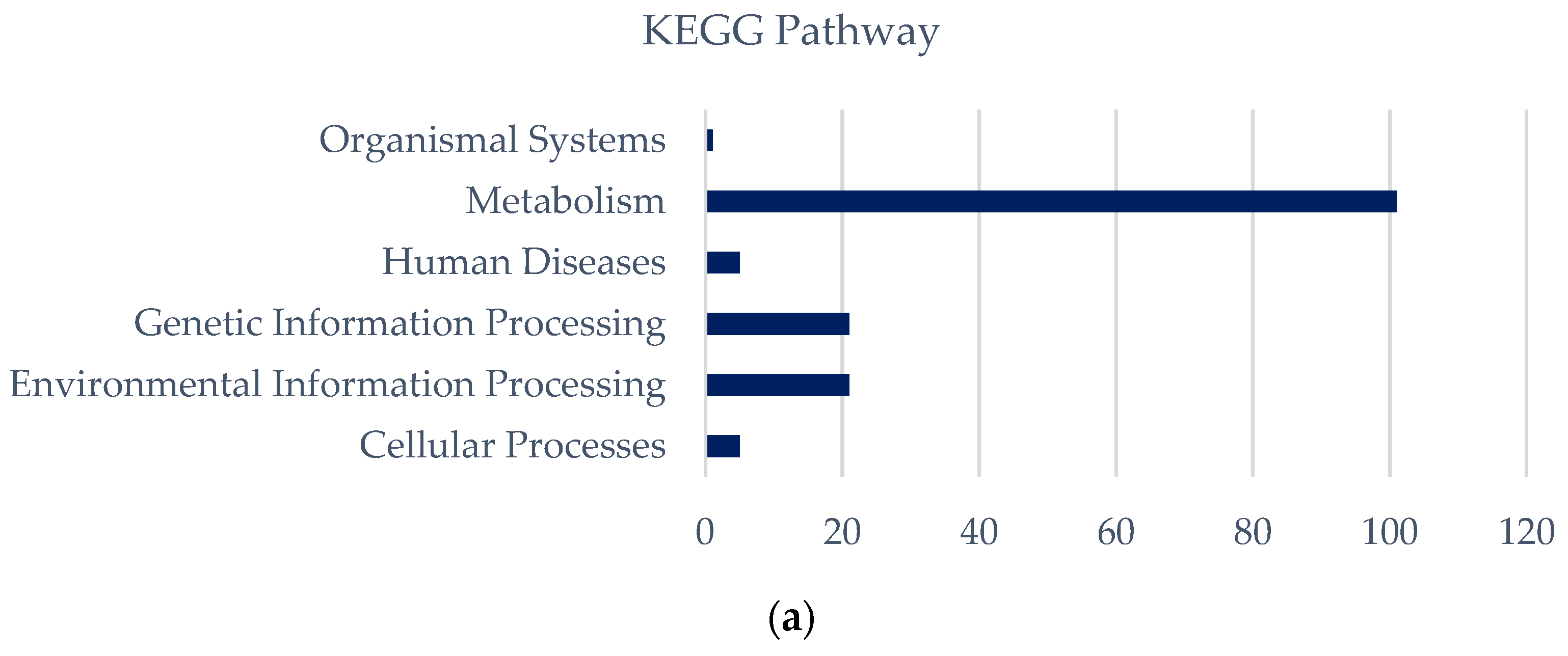
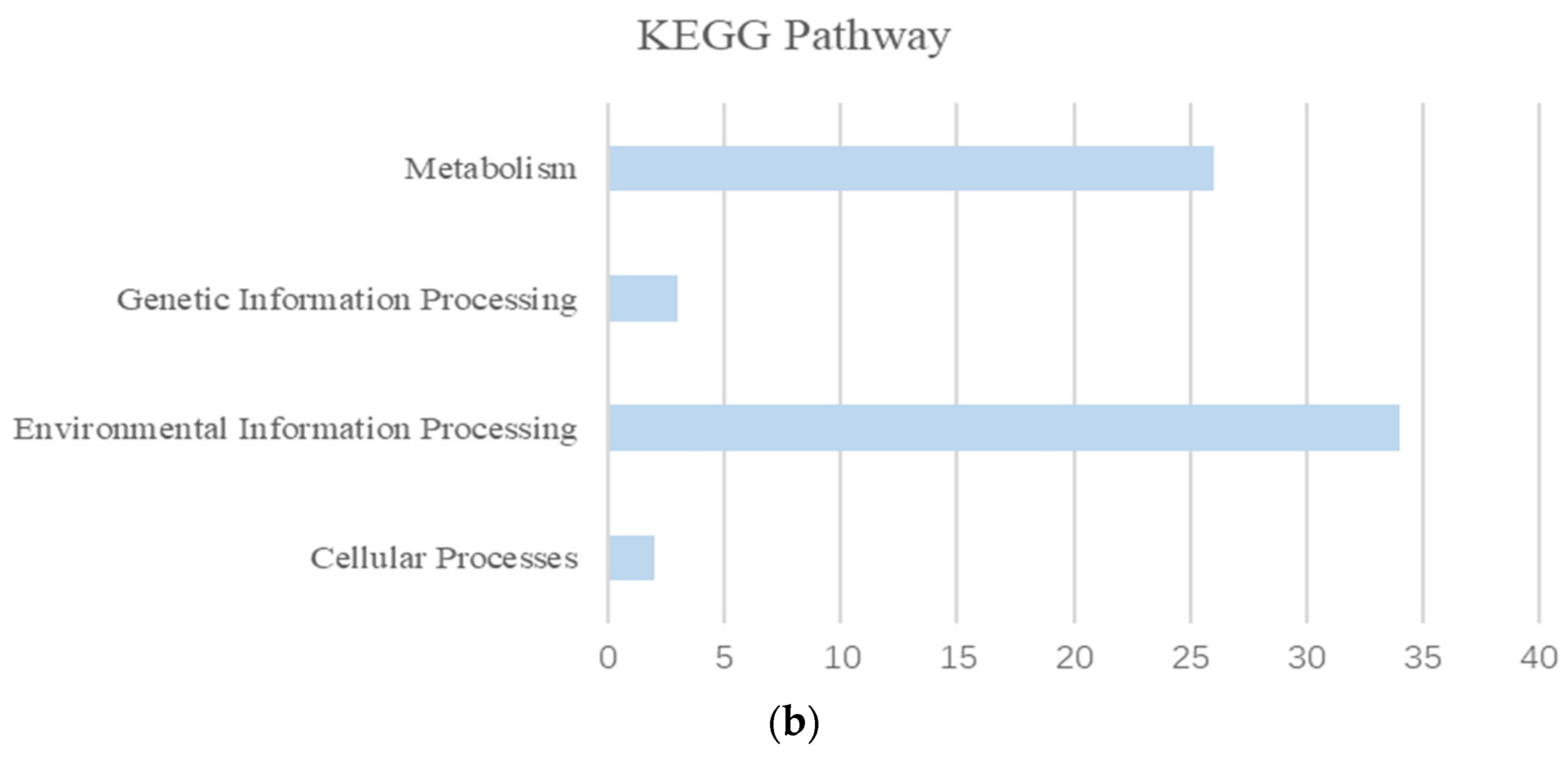
3.5. Salt Tolerance and Comparative Genomics in Synechococcus
3.5.1. Genome Features of Synechococcus with Different Salinity Growth Conditions
3.5.2. Salinity Adaptation Mechanisms
3.5.3. High-Salt Adaptation Strategies
- Synthesis of compatible solutes
- 2.
- Antioxidant defense
- 3.
- Ion transport and distribution
3.5.4. Freshwater Adaptation Strategies
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huang, S.; Wilhelm, S.W. Novel lineages of Prochlorococcus and Synechococcus in the global oceans. ISME J. 2012, 6, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.; Tao, F. Direct carbon capture for the production of high-performance biodegradable plastics by cyanobacterial cell factories. Green Chem. 2022, 24, 4470–4483. [Google Scholar] [CrossRef]
- Salazar, V.W.; Tschoeke, D.A. A new genomic taxonomy system for the Synechococcus collective. Environ. Microbiol. 2020, 22, 4557–4570. [Google Scholar] [CrossRef] [PubMed]
- Deviram, G.; Mathimani, T. Applications of microalgal and cyanobacterial biomass on a way to safe, cleaner and a sustainable environment. J. Clean. Prod. 2020, 253, 119770. [Google Scholar] [CrossRef]
- Komárek, J.; Johansen, J.R. Phylogeny and taxonomy of Synechococcus-like cyanobacteria. Fottea 2020, 20, 171–191. [Google Scholar] [CrossRef]
- Johnson, P.W.; Sieburth, J.M. Chroococcoid cyanobacteria in the sea: A ubiquitous and diverse phototrophic biomass1. Limnol. Oceanogr. 1979, 24, 928–935. [Google Scholar] [CrossRef]
- Stanier, R.Y.; Cohen-Bazire, G. Phototrophic prokaryotes: The cyanobacteria. Annu. Rev. Microbiol. 1977, 31, 225–274. [Google Scholar] [CrossRef] [PubMed]
- Yadav, I.; Rautela, A. Approaches in the photosynthetic production of sustainable fuels by cyanobacteria using tools of synthetic biology. World J. Microbiol. Biotechnol. 2021, 37, 201. [Google Scholar] [CrossRef]
- Luan, G.; Lu, X. Tailoring cyanobacterial cell factory for improved industrial properties. Biotechnol. Adv. 2018, 36, 430–442. [Google Scholar] [CrossRef]
- Roh, H.; Lee, J.S. Improved CO-derived polyhydroxybutyrate (PHB) production by engineering fast-growing cyanobacterium Synechococcus elongatus UTEX 2973 for potential utilization of flue gas. Bioresour. Technol. 2021, 327, 124789. [Google Scholar] [CrossRef]
- Tan, C.; Xu, P. Carbon-negative synthetic biology: Challenges and emerging trends of cyanobacterial technology. Trends Biotechnol. 2022, 40, 1488–1502. [Google Scholar] [CrossRef]
- Sini, P.; Dang, T.B.C. Cyanobacteria, cyanotoxins, and neurodegenerative diseases: Dangerous liaisons. Int. J. Mol. Sci. 2021, 22, 8726. [Google Scholar] [CrossRef]
- Palenik, B.; Brahamsha, B. The genome of a motile marine Synechococcus. Nature 2003, 424, 1037–1042. [Google Scholar] [CrossRef]
- Sun, Z.; Blanchard, J.L. Strong genome-wide selection early in the evolution of Prochlorococcus resulted in a reduced genome through the loss of a large number of small effect genes. PLoS ONE 2014, 9, e88837. [Google Scholar] [CrossRef]
- Swan, B.K.; Tupper, B. Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. Proc. Natl. Acad. Sci. USA 2013, 110, 11463–11468. [Google Scholar] [CrossRef]
- Cui, H.; Wang, Y. Genome-wide analysis of putative peroxiredoxin in unicellular and filamentous cyanobacteria. BMC Evol. Biol. 2012, 12, 220. [Google Scholar] [CrossRef] [PubMed]
- Grébert, T.; Garczarek, L. Diversity and evolution of pigment types in marine Synechococcus cyanobacteria. Genome Biol. Evol. 2022, 14, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Herdman, H. Form-genus XIII. Synechococcus. In Bergey’s Manual of Systematic Bacteriology; Springer: Berlin/Heidelberg, Germany, 2001; Volume 1, pp. 508–512. [Google Scholar]
- Tettelin, H.; Riley, D. Comparative genomics: The bacterial pan-genome. Curr. Opin. Microbiol. 2008, 11, 472–477. [Google Scholar] [CrossRef]
- Dzaman-Serafin, S.; Telatyńska-Mieszek, B. Heat shock proteins and their characteristics. Pol. Merkur. Lek. 2005, 19, 215–219. [Google Scholar]
- Jung, S.J.; Jung, Y. Proper insertion and topogenesis of membrane proteins in the ER depend on Sec63. Biochim. Biophys. Acta Gen. Subj. 2019, 1863, 1371–1380. [Google Scholar] [CrossRef]
- Thoms, S. Cdc48 can distinguish between native and non-native proteins in the absence of cofactors. FEBS Lett. 2002, 520, 107–110. [Google Scholar] [CrossRef]
- Miao, F.; Bouziane, M. 3-Methyladenine-DNA glycosylase (MPG potein) interacts with human RAD23 proteins. J. Biol. Chem. 2000, 275, 28433–28438. [Google Scholar] [CrossRef]
- Chen, C.; Su, L. Enhanced the catalytic efficiency and thermostability of maltooligosyltrehalose synthase from Arthrobacter ramosus by directed evolution. Biochem. Eng. J. 2020, 162, 107724. [Google Scholar] [CrossRef]
- Laudenbach, D.E.; Grossman, A.R. Characterization and mutagenesis of sulfur-regulated genes in a cyanobacterium: Evidence for function in sulfate transport. J. Bacteriol. 1991, 173, 2739–2750. [Google Scholar] [CrossRef] [PubMed]
- Horn, C.; Jenewein, S. Biochemical and structural analysis of the Bacillus subtilis ABC transporter OpuA and its isolated subunits. J. Mol. Microbiol. Biotechnol. 2005, 10, 76–91. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Shi, W.W. Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC. Biochem. J. 2011, 436, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Zhai, H. A lycopene β-cyclase gene, IbLCYB2, enhances carotenoid contents and abiotic stress tolerance in transgenic sweetpotato. Plant Sci. 2018, 272, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Moreno, J.C.; Pizarro, L. Levels of lycopene β-cyclase 1 modulate carotenoid gene expression and accumulation in Daucus carota. PLoS ONE 2013, 8, e58144. [Google Scholar] [CrossRef]
- Dauvillée, D.; Kinderf, I.S. Role of the Escherichia coli glgX gene in glycogen metabolism. J. Bacteriol. 2005, 187, 1465–1473. [Google Scholar] [CrossRef]
- LeBlanc, J.; Labrie, A. Glycogen and nonspecific adaptation to cold. J. Appl. Physiol. 1981, 51, 1428–1432. [Google Scholar] [CrossRef]
- Williams, C.P.; Stanley, W.A. Peroxin 5: A cycling receptor for protein translocation into peroxisomes. Int. J. Biochem. Cell B 2010, 42, 1771–1774. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, A.A.; Vicente, A.M. Association of the cold shock DEAD-box RNA helicase RhlE to the RNA degradosome in Caulobacter crescentus. J. Bacteriol. 2017, 199, 10–1128. [Google Scholar] [CrossRef]
- Jennings, M.J.; Hathazi, D. Intracellular lipid accumulation and mitochondrial dysfunction accompanies endoplasmic reticulum stress caused by loss of the co-chaperone DNAJC3. Front. Cell Dev. Biol. 2021, 9, 710247. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.; Zhu, Y. Metagenomic analysis of the soil microbial composition and salt tolerance mechanism in Yuncheng Salt Lake, Shanxi Province. Front. Microbiol. 2022, 13, 1004556. [Google Scholar] [CrossRef] [PubMed]
- Deshnium, P.; Los, D.A. Transformation of Synechococcus with a gene for choline oxidase enhances tolerance to salt stress. Plant Mol. Biol. 1995, 29, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Backofen, B.; Leeb, T. Genomic organization of the murine aminomethyltransferase gene (Amt). DNA Seq. 2002, 13, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Wang, J. IrrE, a global regulator of extreme radiation resistance in Deinococcus radiodurans, enhances salt tolerance in Escherichia coli and Brassica napus. PLoS ONE 2009, 4, e4422. [Google Scholar] [CrossRef]
- Swapnil, P.; Rai, A.K. Physiological responses to salt stress of salt-adapted and directly salt (NaCl and NaCl + Na2SO4 mixture)-stressed cyanobacterium Anabaena fertilissima. Protoplasma 2018, 255, 963–976. [Google Scholar] [CrossRef]
- Diao, Y.; Xu, H. Cloning a glutathione peroxidase gene from Nelumbo nucifera and enhanced salt tolerance by overexpressing in rice. Mol. Biol. Rep. 2014, 41, 4919–4927. [Google Scholar] [CrossRef]
- Upadhyay, V.A.; Brunner, A.M. Isocitrate dehydrogenase (IDH) inhibition as treatment of myeloid malignancies: Progress and future directions. Pharmacol. Ther. 2017, 177, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Webb, D.C.; Rosenberg, H. Mutational analysis of the Escherichia coli phosphate-specific transport system, a member of the traffic ATPase (or ABC) family of membrane transporters. A role for proline residues in transmembrane helices. J. Biol. Chem. 1992, 267, 24661–24668. [Google Scholar] [CrossRef] [PubMed]
- Long, Q.; Shen, L. Synthesis and application of organic phosphonate salts as draw solutes in forward osmosis for oil-water separation. Environ. Sci. Technol. 2016, 50, 12022–12029. [Google Scholar] [CrossRef]
- Rao, N.N.; Torriani, A. Molecular aspects of phosphate transport in Escherichia coli. Mol. Microbiol. 1990, 4, 1083–1090. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, L.; Sanchez-Hevia, D. A new family of nitrate/nitrite transporters involved in denitrification. Int. Microbiol. 2019, 22, 19–28. [Google Scholar] [CrossRef]
- Ramírez, S.; Moreno, R. Two nitrate/nitrite transporters are encoded within the mobilizable plasmid for nitrate respiration of Thermus thermophilus HB8. J. Bacteriol. 2000, 182, 2179–2183. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vitt, S.; Ma, K. The F420-reducing [NiFe]-hydrogenase complex from Methanothermobacter marburgensis, the first X-ray structure of a group 3 family member. J. Mol. Biol. 2014, 426, 2813–2826. [Google Scholar] [CrossRef]
- Graham, D.E.; Xu, H. Identification of the 7, 8-didemethyl-8-hydroxy-5-deazariboflavin synthase required for coenzyme F 420 biosynthesis. Arch. Microbiol. 2003, 180, 455–464. [Google Scholar] [CrossRef]
- El Kadmiri, N.; Slassi, I. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and Alzheimer’s disease. Pathol. Biol. 2014, 62, 333–336. [Google Scholar] [CrossRef] [PubMed]
- Neznansky, A.; Opatowsky, Y. Expression, purification and crystallization of the phosphate-binding PstS protein from Pseudomonas aeruginosa. Acta Crystallogr. F Struct. Biol. Commun. 2014, 70, 906–910. [Google Scholar] [CrossRef]
- Aguilar-Barajas, E.; Díaz-Pérez, C. Bacterial transport of sulfate, molybdate, and related oxyanions. Biometals 2011, 24, 687–707. [Google Scholar] [CrossRef]
- Hosie, A.H.F.; Allaway, D. Rhizobium leguminosarum has a second general amino acid permease with unusually broad substrate specificity and high similarity to branched-chain amino acid transporters (Bra/LIV) of the ABC Fam. J. Bacteriol. 2002, 184, 4071–4080. [Google Scholar] [CrossRef] [PubMed]
- Omata, T.; Gohta, S. Involvement of a CbbR homolog in low CO2-induced activation of the bicarbonate transporter operon in cyanobacteria. J. Bacteriol. 2001, 183, 1891–1898. [Google Scholar] [CrossRef]
- Kaku, N.; Hibino, T. Effects of overexpression of Escherichia coli katE and bet genes on the tolerance for salt stress in a freshwater cyanobacterium Synechococcus sp. PCC 7942. Plant Sci. 2000, 159, 281–288. [Google Scholar] [CrossRef]
- Schäfer, G.; Kardinahl, S. Iron superoxide dismutases: Structure and function of an archaic enzyme. Biochem. Soc. Trans. 2003, 31, 1330–1334. [Google Scholar] [CrossRef]
- Kees, E.D.; Murugapiran, S.K. Distribution and genomic variation of thermophilic cyanobacteria in diverse microbial mats at the upper temperature limits of photosynthesis. mSystems 2022, 7, e0031722. [Google Scholar] [CrossRef]
- Pedersen, D.; Miller, S.R. Photosynthetic temperature adaptation during niche diversification of the thermophilic cyanobacterium Synechococcus A/B clade. Isme J. 2017, 11, 1053–1057. [Google Scholar] [CrossRef]
- Adalsteinsson, B.T.; Kristjansdottir, T. Efficient genome editing of an extreme thermophile, Thermus thermophilus, using a thermostable Cas9 variant. Sci. Rep. 2021, 11, 9586. [Google Scholar] [CrossRef]
- Kim, Y.; Jeon, J. Photosynthetic functions of Synechococcus in the ocean microbiomes of diverse salinity and seasons. PLoS ONE 2018, 13, e0190266. [Google Scholar] [CrossRef]
- Hebert, P.D.; Ratnasingham, S. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc. Biol. Sci. 2003, 270 (Suppl. S1), 96–99. [Google Scholar] [CrossRef]
- Ruffing, A.M.; Jensen, T.J. Genetic tools for advancement of Synechococcus sp. PCC 7002 as a cyanobacterial chassis. Microb. Cell Fact. 2016, 15, 190. [Google Scholar] [CrossRef]
- Włodarczyk, A.; Selão, T.T. Newly discovered Synechococcus sp. PCC 11901 is a robust cyanobacterial strain for high biomass production. Commun. Biol. 2020, 3, 215. [Google Scholar] [CrossRef] [PubMed]
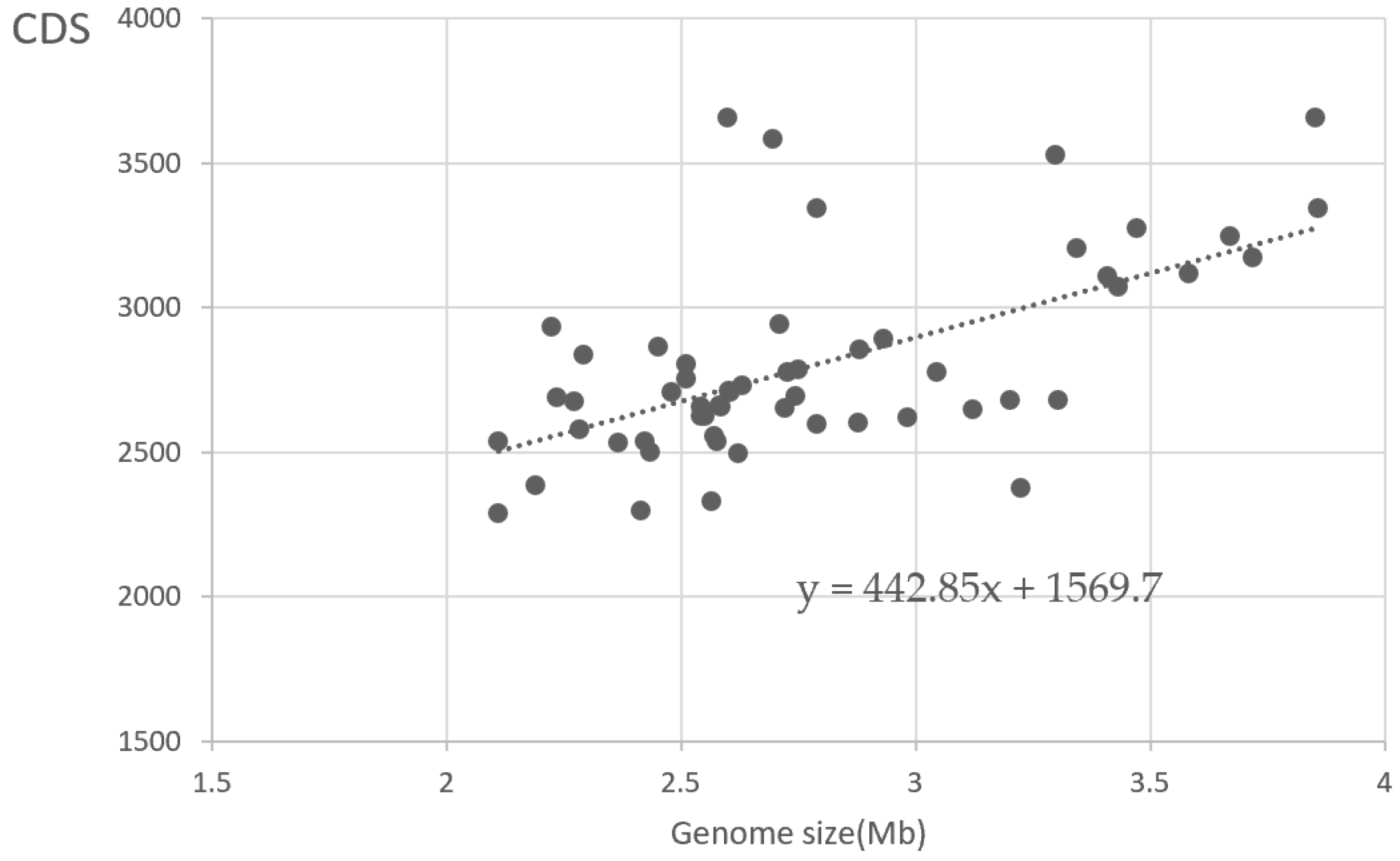


| Organism Name | Accession | Ecosystem | Size (Mb) | GC% | CDS |
|---|---|---|---|---|---|
| Synechococcus sp. A15-127 | NZ_CP047948.1 | Marine | 2.54 | 60.6 | 2653 |
| Synechococcus sp. A15-44 | NZ_CP047938.1 | Marine | 2.62 | 60.8 | 2496 |
| Synechococcus sp. A15-60 | NZ_CP047933.1 | Marine | 2.54 | 58.2 | 2623 |
| Synechococcus sp. A15-62 | NZ_CP047950.1 | Marine | 2.29 | 59.9 | 2837 |
| Synechococcus sp. A18-25c | NZ_CP047957.1 | Marine | 2.51 | 58.2 | 2803 |
| Synechococcus sp. BIOS-U3-1 | NZ_CP047936.1 | Marine | 2.71 | 55 | 2943 |
| Synechococcus sp. BMK-MC-1 | NZ_CP047939.1 | Marine | 2.60 | 60.4 | 3654 |
| Synechococcus sp. CB0101 | NZ_CP039373.1 | Marine | 2.79 | 64.1 | 3343 |
| Synechococcus sp. CBW1002 | NZ_CP060398.1 | Marine | 3.85 | 64.6 | 3653 |
| Synechococcus sp. CBW1004 | NZ_CP060397.1 | Marine | 3.67 | 67.1 | 3247 |
| Synechococcus sp. CBW1006 | NZ_CP060396.1 | Marine | 3.86 | 64.6 | 3340 |
| Synechococcus sp. CBW1107 | NZ_CP064908.1 | Marine | 3.2 | 66.3 | 2678 |
| Synechococcus sp. CBW1108 | NZ_CP060395.1 | Marine | 3.22 | 63.7 | 2373 |
| Synechococcus sp. CC9311 | NC_008319.1 | Marine | 2.60 | 52.4 | 2707 |
| Synechococcus sp. CC9605 | NC_007516.1 | Marine | 2.51 | 59.2 | 2753 |
| Synechococcus sp. CC9902 | NC_007513.1 | Marine | 2.23 | 54.2 | 2689 |
| Synechococcus sp. JA-2-3B′a(2-13) | NC_007776.1 | Thermal springs | 3.05 | 58.5 | 2774 |
| Synechococcus sp. JA-3-3Ab | NC_007775.1 | Thermal springs | 2.93 | 60.2 | 2890 |
| Synechococcus sp. KORDI-100 | NZ_CP006269.1 | Marine | 2.80 | 57.5 | 2594 |
| Synechococcus sp. KORDI-49 | NZ_CP006270.1 | Marine | 2.59 | 61.4 | 2654 |
| Synechococcus sp. KORDI-52 | NZ_CP006271.1 | Marine | 2.57 | 59.1 | 2555 |
| Synechococcus sp. LTW-R | NZ_CP059060.1 | Marine | 2.41 | 62.6 | 2297 |
| Synechococcus sp. M16.1 | NZ_CP047954.1 | Marine | 2.11 | 61.4 | 2537 |
| Synechococcus sp. MEDNS5 | NZ_CP047952.1 | Marine | 2.44 | 59.1 | 2500 |
| Synechococcus sp. Minos11 | NZ_CP047953.1 | Marine | 2.28 | 60.5 | 2577 |
| Synechococcus sp. MIT S9220 | NZ_CP047958.1 | Marine | 2.42 | 56.4 | 2537 |
| Synechococcus sp. MVIR-18-1 | NZ_CP047942.1 | Marine | 2.45 | 53.4 | 2864 |
| Synechococcus sp. NIES-970 | NZ_AP017959.1 | Marine | 3.12 | 49.4 | 2648 |
| Synechococcus sp. NOUM97013 | NZ_CP047941.1 | Marine | 2.55 | 58.8 | 2625 |
| Synechococcus sp. PCC 11901 | NZ_CP040360.1 | Marine | 3.47 | 49.1 | 3272 |
| Synechococcus elongatus PCC 6301 | NC_006576.1 | Freshwater | 2.70 | 55.5 | 3581 |
| Synechococcus sp. PCC 6312 | NC_019680.1 | Freshwater | 3.72 | 48.5 | 3171 |
| Synechococcus sp. PCC 7002 | NC_010475.1 | Marine | 3.41 | 49.2 | 3109 |
| Synechococcus sp. PCC 7003 | NZ_CP016474.1 | Marine | 3.35 | 49.3 | 3204 |
| Synechococcus sp. PCC 7117 | NZ_CP016477.1 | Marine | 3.4 | 49.1 | 3071 |
| Synechococcus sp. PCC 73109 | NZ_CP013998.1 | Marine | 3.30 | 49.3 | 3524 |
| Synechococcus sp. PCC 7502 | NC_019702.1 | Freshwater | 3.58 | 40.6 | 3118 |
| Synechococcus elongatus PCC 7942 | NC_007604.1 | Freshwater | 2.72 | 46.3 | 2649 |
| Synechococcus sp. PCC 8807 | NZ_CP016483.1 | Marine | 3.30 | 49.3 | 2679 |
| Synechococcus sp. PROS-7-1 | NZ_CP047945.1 | Marine | 2.57 | 59.3 | 2328 |
| Synechococcus sp. PROS-9-1 | NZ_CP047961.1 | Marine | 2.27 | 53.9 | 2675 |
| Synechococcus sp. PROS-U-1 | NZ_CP047951.1 | Marine | 2.57 | 58.4 | 2535 |
| Synechococcus sp. RCC307 | CT978603.1 | Marine | 2.22 | 60.8 | 2931 |
| Synechococcus sp. ROS8604 | NZ_CP047946.1 | Marine | 2.88 | 54 | 2600 |
| Synechococcus sp. RS9902 | NZ_CP047949.1 | Marine | 2.48 | 59.8 | 2706 |
| Synechococcus sp. RS9907 | NZ_CP047944.1 | Marine | 2.58 | 60.4 | 2659 |
| Synechococcus sp. RS9909 | NZ_CP047943.1 | Marine | 2.60 | 64.4 | 2711 |
| Synechococcus sp. RSCCF101 | NZ_CP035632.1 | Marine | 2.98 | 68 | 2618 |
| Synechococcus sp. SYN20 | NZ_CP047959.1 | Marine | 2.72 | 53.4 | 2773 |
| Synechococcus sp. SynAce01 | NZ_CP018091.1 | Marine | 2.75 | 63.9 | 2784 |
| Synechococcus sp. TAK9802 | NZ_CP047937.1 | Marine | 2.19 | 60.8 | 2382 |
| Synechococcus sp. UTEX 2973 | NZ_CP006471.1 | Freshwater | 2.74 | 55.5 | 2691 |
| Synechococcus elongatus UTEX 3055 | NZ_CP033061.1 | Freshwater | 2.88 | 55.1 | 2853 |
| Synechococcus sp. WH 7803 | CT971583.1 | Marine | 2.36 | 60.2 | 2533 |
| Synechococcus sp. WH 8101 | NZ_CP035914.1 | Marine | 2.63 | 63.3 | 2730 |
| Synechococcus sp. WH 8109 | NZ_CP006882.1 | Marine | 2.11 | 60.1 | 2288 |
| Strain Name | Geographic Origin | Isolation Latitude | Isolation Longitude | Depth (m) | Isolation Date |
|---|---|---|---|---|---|
| A15-127 | North Atlantic Ocean | −31.12 | −3.92 | 45 | 24 October 2004 |
| A15-44 | North Atlantic Ocean | 21.68 | −17.83 | 20 | 1 October 2004 |
| A15-60 | North Atlantic Ocean | 17.62 | −20.95 | 10 | 4 October 2004 |
| A15-62 | North Atlantic Ocean | 17.62 | −20.95 | 15 | 4 October 2004 |
| A18-25c | Pacific Ocean | 27.63 | −37.03 | 74 | 14 September 2008 |
| BIOS-U3-1 | Pacific Ocean | −33.86 | −73.34 | 5 | 28 November 2004 |
| BMK-MC-1 | Mediterranean Sea | 40.8 | 14.15 | 23 | 13 October 2009 |
| CB0101 | Chesapeake Bay | 39.28 | −76.6 | 0 | 17 July 2004 |
| CBW1002 | Chesapeake Bay | 39.28 | −76.6 | 0 | 21 December 2010 |
| CBW1004 | Chesapeake Bay | 39.28 | −76.6 | 0 | 28 December 2010 |
| CBW1006 | Chesapeake Bay | 39.28 | −76.6 | 0 | 28 December 2010 |
| CBW1107 | Chesapeake Bay | 39.28 | −76.6 | 0 | 1 February 2011 |
| CBW1108 | Chesapeake Bay | 39.28 | −76.6 | 0 | 1 February 2011 |
| CC9311 | North Pacific, California | 31.91 | −124.17 | 95 | 8 April 1993 |
| CC9605 | North Pacific, California | 30.42 | −124 | 51 | 1 January 1993 |
| CC9902 | North Pacific, California | 32.87 | −117.26 | 5 | 1 January 1996 |
| JA-2-3B′a(2-13) | Octopus Spring | 22.45 | −158 | 400 | 10 July 2002 |
| JA-3-3Ab | Octopus Spring | 44.55 | −110.82 | 400 | 25 July 2002 |
| KORDI-100 | Pacific Ocean | 9.15 | 158.4 | 0 | September 2007 |
| KORDI-49 | Pacific Ocean | 32.3 | 126 | 20 | 1 March 2004 |
| KORDI-52 | Pacific Ocean | 32 | 126.45 | 30 | 1 May 2005 |
| LTW-R | Pearl river, China | 22.22 | 114.129 | - | July 2014 |
| M16.1 | North Atlantic Ocean | 27.7 | −91.3 | 275 | 9 February 2004 |
| MEDNS5 | Mediterranean Sea | 41 | 6 | 80 | 1 July 1993 |
| MINOS11 | Mediterranean Sea | 34 | 18 | 20 | 2 November 2010 |
| MIT S9220 | Pacific Ocean | 0 | −140 | 0 | 1 October 1992 |
| MVIR-18-1 | North Atlantic Ocean | 61 | 1.59 | 25 | 23 July 2007 |
| NIES-970 | Rikuhama Beach, Japan | - | - | - | 28 August 1998 |
| NOUM97013 | Pacific Ocean | −22.33 | 166.33 | 0 | 19 November 1996 |
| PCC 11901 | Singapore | 1.25 | 103.57 | 20 | 6 May 2017 |
| PCC 6301 | Austin, Texas, USA | 30.41 | −97.76 | - | 1 January 1952 |
| PCC 6312 | California, USA | - | - | - | 1 January 1963 |
| PCC 7002 | Magueyes, Puerto Rico | 17.96 | −67.04 | - | 1 January 1961 |
| PCC 7003 | Greenwich, USA | - | - | - | 1 January 1960 |
| PCC 7117 | Port Hedland, Australia | - | - | - | 1 January 1971 |
| PCC 73109 | City Island, New York | - | - | - | 1 January 1961 |
| PCC 7502 | Lake, Switzerland | - | - | - | 1 January 1972 |
| PCC 7942 | California, USA | - | - | - | 1 January 1973 |
| PCC 8807 | Lagoon, Gabon | - | - | - | 1 January 1979 |
| PROS-7-1 | Mediterranean Sea | 37.4 | 15.62 | 5 | 26 September 1999 |
| PROS-9-1 | Mediterranean Sea | 41.9 | 10.43 | 30 | 28 September 1999 |
| PROS-U-1 | North Atlantic Ocean | 71.22 | −136.72 | 5 | 12 September 1999 |
| RCC307 | Mediterranean Sea | 39.17 | 6.18 | 15 | 25 May 1996 |
| ROS8604 | North Atlantic | 48.73 | −3.98 | 1 | 24 November 1986 |
| RS9902 | Indian ocean, Red Sea | 29.47 | 34.92 | 1 | 23 March 1999 |
| RS9907 | Indian ocean, Red Sea | 29.47 | 34.92 | 10 | 29 August 1999 |
| RS9909 | Indian ocean, Red Sea | 29.47 | 34.92 | 10 | 7 September 1999 |
| RSCCF101 | Indian ocean, Red Sea | 29.47 | 34.92 | 20 | July 2014 |
| SYN20 | North Atlantic Ocean | 60.27 | 5.21 | 2 | 1 June 2004 |
| SynAce01 | Ace Lake, Antarctica | −65.31 | 78.5 | 15 | 1992 |
| TAK9802 | South Pacific Ocean | 34.92 | 34.92 | 7 | 6 February 1998 |
| UTEX 2973 | Austin, Texas, USA | - | - | 0 | 7 December 2011 |
| UTEX 3055 | Austin, Texas, USA | 30.17 | −97.44 | - | 13 December 2013 |
| WH7803 | North Atlantic Ocean | 33.75 | −67.5 | 25 | 3 July 1978 |
| WH8101 | North Atlantic Ocean | 41.52 | −70.67 | 0 | 1 January 1981 |
| WH8109 | North Atlantic Ocean | 39.28 | −70.46 | - | 1 June 1981 |
| Strain Name | Culture Collection Number | Culture Medium | Culture Temperature (°C) | Growth Temperature (°C) | Optimum Temperature (°C) | Ecotype |
|---|---|---|---|---|---|---|
| A15-127 | RCC 2378 | PCR-S11 | 22 | - | - | - |
| A15-44 | RCC 2527 | PCR-S11 | 22 | 25–40 | 30 | Warm |
| A15-60 | RCC 2554 | PCR-S11 | 22 | - | - | - |
| A15-62 | RCC 2374 | PCR-S11 | 22 | 16–32 | 30 | Warm |
| A18-25c | - | PCR-S11 | 22 | - | - | - |
| BIOS-U3-1 | RCC 2533 | PCR-S11 | 22 | 12–29 | 25 | Cold |
| BMK-MC-1 | - | PCR-S11 | 22 | - | - | - |
| CB0101 | - | SN | 25 | 4–30 | 25 | Cold |
| CBW1002 | SN15 | 23 | 4–10 | 6.5 | Cold | |
| CBW1004 | - | SN15 | 23 | 4–10 | 6.2 | Cold |
| CBW1006 | - | SN15 | 23 | 4–10 | 6.2 | Cold |
| CBW1107 | - | SN15 | 23 | 4–10 | 2.5 | Cold |
| CBW1108 | - | SN15 | 23 | 4–10 | 2.5 | Cold |
| CC9311 | RCC 1086 | SN35 | 23 | - | - | Warm |
| CC9605 | RCC 753 | PCR-S11 | 22 | - | - | - |
| CC9902 | RCC 2673 | SN | 22 | 10–30 | 24 | Cold |
| JA-2-3B′a(2-13) | - | BG11 | 25 | 45–65 | 51–61 | Thermophile |
| JA-3-3Ab | - | BG11 | 25 | 45–70 | 58–65 | Thermophile |
| KORDI-100 | - | BG11 | 28 | - | 29.5 | - |
| KORDI-49 | - | ASN III | 20 | - | 13.5 | - |
| KORDI-52 | - | BG11 | 25 | - | 14.5 | - |
| LTW-R | - | BG11 | 25 | - | - | - |
| M16.1 | RCC 791 | PCR-S11 | 22 | 18–35 | 32 | Warm |
| MEDNS5 | RCC 2368 | PCR-S11 | 22 | - | - | - |
| MINOS11 | RCC 2319 | PCR-S11 | 22 | - | - | - |
| MIT S9220 | RCC 2571 | PCR-S11 | 22 | 16–31 | 30 | Warm |
| MVIR-18-1 | RCC 2385 | PCR-S11 | 15 | 4–25 | 22 | Cold |
| NIES-970 | NIES 970 | ESM | 20 | - | - | - |
| NOUM97013 | RCC 2433 | PCR-S11 | 22 | - | - | - |
| PCC 11901 | PCC 11901 | 1536 | 30 | 20–43 | 38–41 | Warm |
| PCC 6301 | PCC 6301 | 1540 | 22 | 20–40 | 38 | Warm |
| PCC 6312 | PCC 6312 | 1539 | 22 | 20–40 | 38 | Warm |
| PCC 7002 | PCC 7002 | 1550 | 22 | 20–40 | 38 | Warm |
| PCC 7003 | PCC 7003 | 1536 | 22 | - | - | - |
| PCC 7117 | PCC 7117 | 1536 | 22 | - | - | - |
| PCC 73109 | PCC73109 | 1535 | 22 | - | - | - |
| PCC 7502 | PCC 7502 | 1539 | 22 | - | - | - |
| PCC 7942 | PCC 7942 | 1539 | 22 | 20–45 | 38 | Warm |
| PCC 8807 | PCC 8807 | 1546 | 22 | - | - | - |
| PROS-7-1 | RCC 2381 | PCR-S11 | 22 | - | - | - |
| PROS-9-1 | RCC 328 | PCR-S11 | 22 | - | - | - |
| PROS-U-1 | RCC 2369 | PCR-S11 | 22 | - | - | - |
| RCC307 | RCC 307 | PCR-S11 | 20 | 10–40 | 30 | Cold |
| ROS8604 | RCC 2380 | PCR-S11 | 20 | 16–30 | 26 | Warm |
| RS9902 | RCC 2376 | PCR-S11 | 22 | - | - | - |
| RS9907 | RCC 2382 | PCR-S11 | 22 | 18–35 | 30–32 | Warm |
| RS9909 | RCC 2383 | PCR-S11 | 22 | - | - | - |
| RSCCF101 | - | PCR-S11 | 30–38 | 25–38 | 30 | Warm |
| SYN20 | RCC 2035 | PCR-S11 | 22 | 10–40 | 30 | Cold |
| SynAce01 | - | PCR-S11 | 10 | −17–29.5 | 20 | Cold |
| TAK9802 | RCC 2528 | PCR-S11 | 22 | - | - | - |
| UTEX 2973 | 2973 | BG11 | 20 | 15–41 | 38–41 | Warm |
| UTEX 3055 | 3055 | BG11 | 20 | 10–42 | 30 | Warm |
| WH7803 | RCC 752 | PCR-S11 | 20 | 16–34 | 33 | Warm |
| WH8101 | RCC 2555 | PCR-S11 | 22 | 16–35 | 25 | Warm |
| WH8109 | RCC 2033 | PCR-S11 | 22 | 16–35 | 28 | Warm |
| Strain Name | Size (Mb) | GC (%) | Salinity Tolerance (ppt) | Optimal Salinity (ppt) | Category |
|---|---|---|---|---|---|
| CB0101 | 2.78965 | 64.1 | 0–35 | 15 | euryhaline |
| CBW1002 | 3.85412 | 64.6 | 5–22 | 17 | euryhaline |
| CBW1004 | 3.67232 | 67.1 | 5–22 | 19 | euryhaline |
| CBW1006 | 3.86013 | 64.6 | 5–22 | 19 | euryhaline |
| CBW1107 | 3.20209 | 66.3 | 5–22 | 8 | euryhaline |
| CBW1108 | 3.22622 | 63.7 | 5–22 | 8 | euryhaline |
| CC9311 | 2.60675 | 52.4 | 24–44 | 33.3 | strictly marine |
| CC9605 | 2.51066 | 59.2 | 24–44 | 34 | strictly marine |
| CC9902 | 2.23483 | 54.2 | 30–44 | 35 | strictly marine |
| KORDI-100 | 2.789 | 57.5 | - | 34.2 | strictly marine |
| KORDI-49 | 2.58581 | 61.4 | - | 34.2 | strictly marine |
| KORDI-52 | 2.57207 | 59.1 | - | 33.8 | strictly marine |
| LTW-R | 2.41553 | 62.6 | 14–44 | 24–34 | euryhaline |
| PCC 11901 | 3.47181 | 49.1 | 0–100 | 25 | euryhaline |
| PCC 6301 | 2.69625 | 55.5 | - | - | freshwater |
| PCC 6312 | 3.7205 | 48.5 | - | - | freshwater |
| PCC 7002 | 3.40993 | 49.2 | 0–90 | 0–25 | euryhaline |
| PCC 7003 | 3.34509 | 49.3 | - | - | strictly marine |
| PCC 7117 | 3.4321 | 49.1 | - | - | euryhaline |
| PCC 73109 | 3.29868 | 49.3 | - | - | euryhaline |
| PCC 7502 | 3.58373 | 40.6 | - | - | freshwater |
| PCC 7942 | 2.72155 | 46.3 | 0–29 | 23 | freshwater |
| RCC307 | 2.22491 | 60.8 | - | 37 | strictly marine |
| SynAce01 | 2.75063 | 63.9 | 10–50 | 20–30 | euryhaline |
| UTEX 2973 | 2.74463 | 55.5 | 6–12 | 9 | freshwater |
| UTEX 3055 | 2.88122 | 55.1 | - | - | freshwater |
| WH7803 | 2.36698 | 60.2 | 24–44 | 34 | strictly marine |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qian, M.; Han, X.; Liu, J.; Xu, P.; Tao, F. Genomic Insights on the Carbon-Negative Workhorse: Systematical Comparative Genomic Analysis on 56 Synechococcus Strains. Bioengineering 2023, 10, 1329. https://doi.org/10.3390/bioengineering10111329
Qian M, Han X, Liu J, Xu P, Tao F. Genomic Insights on the Carbon-Negative Workhorse: Systematical Comparative Genomic Analysis on 56 Synechococcus Strains. Bioengineering. 2023; 10(11):1329. https://doi.org/10.3390/bioengineering10111329
Chicago/Turabian StyleQian, Meiwen, Xiao Han, Jiongqin Liu, Ping Xu, and Fei Tao. 2023. "Genomic Insights on the Carbon-Negative Workhorse: Systematical Comparative Genomic Analysis on 56 Synechococcus Strains" Bioengineering 10, no. 11: 1329. https://doi.org/10.3390/bioengineering10111329
APA StyleQian, M., Han, X., Liu, J., Xu, P., & Tao, F. (2023). Genomic Insights on the Carbon-Negative Workhorse: Systematical Comparative Genomic Analysis on 56 Synechococcus Strains. Bioengineering, 10(11), 1329. https://doi.org/10.3390/bioengineering10111329






