Applied Methods to Assess the Antimicrobial Activity of Metallic-Based Nanoparticles
Abstract
:1. Background
2. Methods and Materials
2.1. General Materials and nanoparticle Preparations
2.2. Microbial Cultures
2.3. Agar Well Diffusion
2.4. Resazurin Broth Assay
2.5. Resazurin Microtitre MIC Assay
2.6. Spectrometer Growth Rate
2.7. Fluorescent Cell Viability
3. Results
3.1. Evaluation of Zone Inhibitory Effect on Solid Agar
3.2. Resazurin Broth Assay
3.3. Resazurin Microtitre MIC Plate Assay
3.4. Spectrometer Growth Rate
3.5. Determination of Viable Cells
4. Discussion
4.1. Antimicrobial Nanoparticles Screening Approaches
4.2. Quantitative and Qualitative Evaluations of Antimicrobial Nanoparticles
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mahira, S.; Jain, A.; Khan, W.; Domb, A.J. Chapter 1: Antimicrobial Materials—An Overview. In Antimicrobial Materials for Biomedical Applications; Royal Society of Chemistry: London, UK, 2019; pp. 1–37. [Google Scholar]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Cameron, R.; Smith, K. Virus clearance methods applied in bioprocessing operations: An overview of selected inactivation and removal methods. Pharm. Bioprocess. 2014, 2, 75–83. [Google Scholar] [CrossRef]
- Kourmouli, A.; Valenti, M.; van Rijn, E.; Beaumont, H.J.E.; Kalantzi, O.I.; Schmidt-Ott, A.; Biskos, G. Can disc diffusion susceptibility tests assess the antimicrobial activity of engineered nanoparticles? J. Nanoparticle Res. 2018, 20, 62. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Hu, C.; Shao, L. The antimicrobial activity of nanoparticles: Present situation and prospects for the future. Int. J. Nanomed. 2017, 12, 1227–1249. [Google Scholar] [CrossRef] [PubMed]
- Baptista, P.V.; McCusker, M.P.; Carvalho, A.; Ferreira, D.A.; Mohan, N.M.; Martins, M.; Fernandes, A.R. Nano-Strategies to Fight Multidrug Resistant Bacteria—“A Battle of the Titans”. Front. Microbiol. 2018, 8, 1441. [Google Scholar] [CrossRef] [PubMed]
- Kurantowicz, N.; Sawosz, E.; Halik, G.; Strojny, B.; Hotowy, A.; Grodzik, M.; Piast, R.; Pasanphan, W.; Chwalibog, A. Toxicity studies of six types of carbon nanoparticles in a chicken-embryo model. Int. J. Nanomed. 2017, 12, 2887–2898. [Google Scholar] [CrossRef] [PubMed]
- Talapko, J.; Matijevic, T.; Juzbasic, M.; Antolovic-Pozgain, A.; Skrlec, I. Antibacterial Activity of Silver and Its Application. Microorganisms 2020, 8, 1400. [Google Scholar] [CrossRef] [PubMed]
- Loo, Y.Y.; Rukayadi, Y.; Nor-Khaizura, M.A.; Kuan, C.H.; Chieng, B.W.; Nishibuchi, M.; Radu, S. In Vitro Antimicrobial Activity of Green Synthesized Silver Nanoparticles Against Selected Gram-negative Foodborne Pathogens. Front. Microbiol. 2018, 9, 1555. [Google Scholar] [CrossRef]
- Ruparelia, J.P.; Chatterjee, A.K.; Duttagupta, S.P.; Mukherji, S. Strain specificity in antimicrobial activity of silver and copper nanoparticles. Acta Biomater. 2008, 4, 707–716. [Google Scholar] [CrossRef]
- Lozhkomoev, A.S.; Lerner, M.I.; Pervikov, A.V.; Kazantsev, S.O.; Fomenko, A.N. Development of Fe/Cu and Fe/Ag Bimetallic Nanoparticles for Promising Biodegradable Materials with Antimicrobial Effect. Nanotechnologies Russ. 2018, 13, 18–25. [Google Scholar] [CrossRef]
- Ramakritinan, C.M.; .Kaarunya, E.; Shankar, S.; Kumaraguru, A.K. Antibacterial Effects of Ag, Au and Bimetallic (Ag-Au) Nanoparticles Synthesized from Red Algae. Solid State Phenom. 2013, 201, 211–230. [Google Scholar]
- Ren, G.; Hu, D.; Cheng, E.W.; Vargas-Reus, M.A.; Reip, P.; Allaker, R.P. Characterisation of Copper Oxide Nanoparticles for Antimicrobial Applications. Int. J. Antimicrob. Agents 2009, 33, 587–590. [Google Scholar] [CrossRef]
- Lakshmi, S.D.; Avti, P.K.; Hegde, G. Activated carbon nanoparticles from biowaste as new generation antimicrobial agents: A review. Nano-Struct. Nano-Objects 2018, 16, 306–321. [Google Scholar] [CrossRef]
- Matharu, R.K.; Cheong, Y.-K.; Ren, G.; Edirisinghe, M.; Ciric, L. Exploiting the antiviral potential of intermetallic nanoparticles. Emergent Mater. 2022, 5, 1251–1260. [Google Scholar] [CrossRef]
- Cheong, Y.-K.; Yang, X.; Wilson, R.M.; Ren, G. Structure Activity Relationship of Antimicrobial Nanoparticle Formulations to Selective Microbes. In Proceedings of the NanoBio&Med, Barcelona, Spain, 22–24 November 2017; pp. 105–106. [Google Scholar]
- Guerin, T.F.; Mondido, M.; McClenn, B.; Peasley, B. Application of resazurin for estimating abundance of contaminant-degrading micro-organisms. Lett. Appl. Microbiol. 2001, 32, 340–345. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Ingudam, S.; Reed, S.; Gehring, A.; Strobaugh, T.P., Jr.; Irwin, P. Study on the mechanism of antibacterial action of magnesium oxide nanoparticles against foodborne pathogens. J. Nanobiotechnology 2016, 14, 54. [Google Scholar] [CrossRef]
- World Heath Organisation (WHO). No Time to Wait: Securing the Future from Drug-Resistant Infections; Interagency Coordination Group (IACG) on Antimicrobial Resistance: New York, NY, USA, 2019. [Google Scholar]
- Bankier, C.; Cheong, Y.-K.; Mahalingam, S.; Edirisinghe, M.; Ren, G.; Cloutman-Green, E.; Ciric, L. A comparison of methods to assess the antimicrobial activity of nanoparticle combinations on bacterial cells. PLoS ONE 2018, 13, e0192093. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Sae-Tia, S.; Fries, B.C. Candidiasis and Mechanisms of Antifungal Resistance. Antibiotics 2020, 9, 312. [Google Scholar] [CrossRef]
- Hu, C.-H.; Ren, L.-Q.; Zhou, Y.; Ye, B.-C. Characterization of antimicrobial activity of three Lactobacillus plantarum strains isolated from Chinese traditional dairy food. Food Sci. Nutr. 2019, 7, 1997–2005. [Google Scholar] [CrossRef]
- Jain, D.; Shivani; Bhojiya, A.A.; Singh, H.; Daima, H.K.; Singh, M.; Mohanty, S.R.; Stephen, B.J.; Singh, A. Microbial Fabrication of Zinc Oxide Nanoparticles and Evaluation of Their Antimicrobial and Photocatalytic Properties. Front. Chem. 2020, 8, 778. [Google Scholar] [CrossRef]
- Yin, I.X.; Zhang, J.; Zhao, I.S.; Mei, M.L.; Li, Q.; Chu, C.H. The Antibacterial Mechanism of Silver Nanoparticles and Its Application in Dentistry. Int. J. Nanomed. 2020, 15, 2555–2562. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Zhu, Y.; Yang, L.; Yang, D.-Q.; Sacher, E. Ag NP catalysis of Cu ions in the preparation of AgCu NPs and the mechanism of their enhanced antibacterial efficacy. Colloids Surf. A Physicochem. Eng. Asp. 2022, 632, 127831. [Google Scholar] [CrossRef]
- Magaldi, S.; Mata-Essayag, S.; Hartung de Capriles, C.; Perez, C.; Colella, M.T.; Olaizola, C.; Ontiveros, Y. Well Diffusion for Antifungal Susceptibility Testing. Int. J. Infect. Dis. 2004, 8, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Bonev, B.; Hooper, J.; Parisot, J. Principles of assessing bacterial susceptibility to antibiotics using the agar diffusion method. J. Antimicrob. Chemother. 2008, 61, 1295–1301. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Tak, Y.K.; Song, J.M. Does the Antibacterial Activity of Silver Nanoparticles Depend on the Shape of the Nanoparticle? A Study of the Gram-Negative Bacterium Escherichia Coli. Appl. Environ. Microbiol. 2007, 73, 1712–1720. [Google Scholar] [CrossRef] [PubMed]
- Cheon, J.Y.; Kim, S.J.; Rhee, Y.H.; Kwon, O.H.; Park, W.H. Shape-dependent antimicrobial activities of silver nanoparticles. Int. J. Nanomed. 2019, 14, 2273–2780. [Google Scholar] [CrossRef] [PubMed]
- Cha, S.H.; Hong, J.; McGuffie, M.; Yeom, B.; VanEpps, J.S.; Kotov, N.A. Shape-Dependent Biomimetic Inhibition of Enzyme by Nanoparticles and Their Antibacterial Activity. ACS Nano 2015, 9, 9097–91105. [Google Scholar] [CrossRef]
- El Badawy, A.M.; Silva, R.G.; Morris, B.; Sheckel, K.G.; Suidan, M.T.; Tolaymat, T.M. Surface Charge-Dependent Toxcitity of Silver Nanoparticles. Environ. Sci. Technol. 2011, 45, 283–287. [Google Scholar] [CrossRef]
- Wilson, W.W.; Wade, M.M.; Holman, S.C.; Champlin, F.R. Status of methods for assessing bacterial cell surface charge properties based on zeta potential measurements. J. Microbiol. Methods 2001, 43, 153–164. [Google Scholar] [CrossRef]
- Liao, C.; Li, Y.; Tjong, S. Bactericidal and Cytotoxic Properties of Silver Nanoparticles. Int. J. Mol. Sci. 2019, 20, 449. [Google Scholar] [CrossRef]
- Kowalska-Krochmal, B.; Dudek-Wicher, R. The Minimum Inhibitory Concentration of Antibiotics: Methods, Interpretation, Clinical Relevance. Pathogens 2021, 10, 165. [Google Scholar] [CrossRef] [PubMed]
- Costa, P.; Gomes, A.T.P.C.; Braz, M.; Pereira, C.; Almeida, A. Application of the Resazurin Cell Viability Assay to Monitor Escherichia coli and Salmonella Typhimurium Inactivation Mediated by Phages. Antibiotics 2021, 10, 974. [Google Scholar] [CrossRef] [PubMed]
- Hawser, S.P.; Norris, H.; Jessup, C.J.; Ghannoum, M.A. Comparison of a 2,3-Bis(2-Methoxy-4-Nitro-5-Sulfophenyl)-5-[(Phenylamino)Carbonyl]-2H-Tetrazolium Hydroxide (XTT) Colorimetric Method with the Standardized National Committee for Clinical Laboratory Standards Method of Testing Clinical Yeast Isolates for Susceptibility to Antifungal Agents. J. Clin. Microbiol. 1998, 36, 1450–1452. [Google Scholar] [CrossRef] [PubMed]
- Alonso, B.; Cruces, R.; Pérez, A.; Sánchez-Carrillo, C.; Guembe, M. Comparison of the XTT and resazurin assays for quantification of the metabolic activity of Staphylococcus aureus biofilm. J. Microbiol. Methods 2017, 139, 135–137. [Google Scholar] [CrossRef] [PubMed]
- Ramyadevi, J.; Jeyasubramanian, K.; Marikani, A.; Rajakumar, G.; Rahuman, A.A. Synthesis and antimicrobial activity of copper nanoparticles. Mater. Lett. 2012, 71, 114–116. [Google Scholar] [CrossRef]
- Chatterjee, A.; Chakraborty, R.; Basu, T. Mechanism of antibacterial activity of copper nanoparticles. Nanotechnology 2014, 25, 135101. [Google Scholar] [CrossRef] [PubMed]
- Raffi, M.; Mehrwan, S.; Bhatti, T.M.; Akhter, J.I.; Hameed, A.; Yawar, W.; Hasan, M.M.U. Investigations into the Antibacterial Behavior of Copper Nanoparticles against Escherichia Coli. Ann. Microbiol. 2010, 60, 75–80. [Google Scholar] [CrossRef]
- Mayer, F.L.; Wilson, D.; Hube, B. Candida Albicans Pathogenicity Mechanisms. Virulence 2013, 4, 119–128. [Google Scholar] [CrossRef]
- Cooksey, D.A. Copper uptake and resistance in bacteria. Mol. Microbiol. 1994, 7, 1–5. [Google Scholar] [CrossRef]
- Jorgensen, H.L.; Schulz, E. Turbidimetric measurement as a rapid method for the determination of the bacteriological quality of minced meat. Int. J. Food Microbiol. 1985, 2, 177–183. [Google Scholar] [CrossRef]
- Lack, W.K.; Becker, B.; Kraemer, J.; Holzapfel, W.H. Turbidimetry as a rapid method for enumeration of microorganisms in raw vegetables. Arch. Leb. 1999, 50, 136–140. [Google Scholar]
- Taner, M.; Sayar, N.; Yulug, I.G.; Suzer, S. Synthesis, characterization and antibacterial investigation of silver–copper nanoalloys. J. Mater. Chem. 2011, 21, 13150–13154. [Google Scholar] [CrossRef]
- Pan, H.; Zhang, Y.; He, G.-X.; Katagori, N.; Chen, H. A comparison of conventional methods for the quantification of bacterial cells after exposure to metal oxide nanoparticles. BMC Microbiol. 2014, 14, 222. [Google Scholar] [CrossRef] [PubMed]
- Hecht, A.; Endy, D.; Salit, M.; Munson, M.S. When Wavelengths Collide: Bias in Cell Abundance Measurements Due to Expressed Fluorescent Proteins. ACS Synth. Biol. 2016, 5, 1024–1027. [Google Scholar] [CrossRef] [PubMed]
- Beal, J.; Farny, N.G.; Haddock-Angelli, T.; Selvarajah, V.; Baldwin, G.S.; Buckley-Taylor, R.; Gershater, M.; Kiga, D.; Marken, J.; Sanchania, V.; et al. Robust estimation of bacterial cell count from optical density. Commun. Biol. 2020, 3, 512. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Wang, L.; Chen, Y.; Long, Y. Optimization of staining with SYTO 9/propidium iodide: Interplay, kinetics and impact on Brevibacillus brevis. BioTechniques 2020, 69, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Boulos, L.; Prevost, M.; Barbeau, B.; Coallier, J.; Desjardins, R. LIVE/DEAD®® BacLight™: Application of a new rapid staining method for direct enumeration of viable and total bacteria in drinking water. J. Microbiol. Methods 1999, 37, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Berney, M.; Hammes, F.; Bosshard, F.; Weilenmann, H.-U.; Egli, T. Assessment and Interpretation of Bacterial Viability by Using the LIVE/DEAD BacLight Kit in Combination with Flow Cytometry. Appl. Environ. Microbiol. 2007, 73, 3283–3290. [Google Scholar] [CrossRef]
- Paszkiewicz, M.; Gołąbiewska, A.; Rajski, Ł.; Kowal, E.; Sajdak, A.; Zaleska-Medynska, A. Synthesis and Characterization of Monometallic (Ag, Cu) and Bimetallic Ag-Cu Particles for Antibacterial and Antifungal Applications. J. Nanomater. 2016, 2016, 2187940. [Google Scholar] [CrossRef]
- Pinho, E.; Magalhães, L.; Henriques, M.; Oliveira, R. Antimicrobial activity assessment of textiles: Standard methods comparison. Ann. Microbiol. 2011, 61, 493–498. [Google Scholar] [CrossRef]
- Slavin, Y.N.; Asnis, J.; Häfeli, U.O.; Bach, H. Metal nanoparticles: Understanding the mechanisms behind antibacterial activity. J. Nanobiotech. 2017, 15, 65. [Google Scholar] [CrossRef]
- Reyes-Blas, M.; Maldonado-Luna, N.M.; Rivera-Quiñones, C.M.; Vega-Avila, A.L.; Roman-Velazquez, F.R.; Perales-Perez, O.J. Single Step Microwave Assisted Synthesis and Antimicrobial Activity of Silver, Copper and Silver-Copper Nanoparticles. J. Mater. Sci. Chem. Eng. 2020, 08, 13–29. [Google Scholar] [CrossRef]
- Gow, N.A.R.; Hube, B. Importance of the Candida albicans cell wall during commensalism and infection. Curr. Opin. Microbiol. 2012, 15, 406–412. [Google Scholar] [CrossRef]
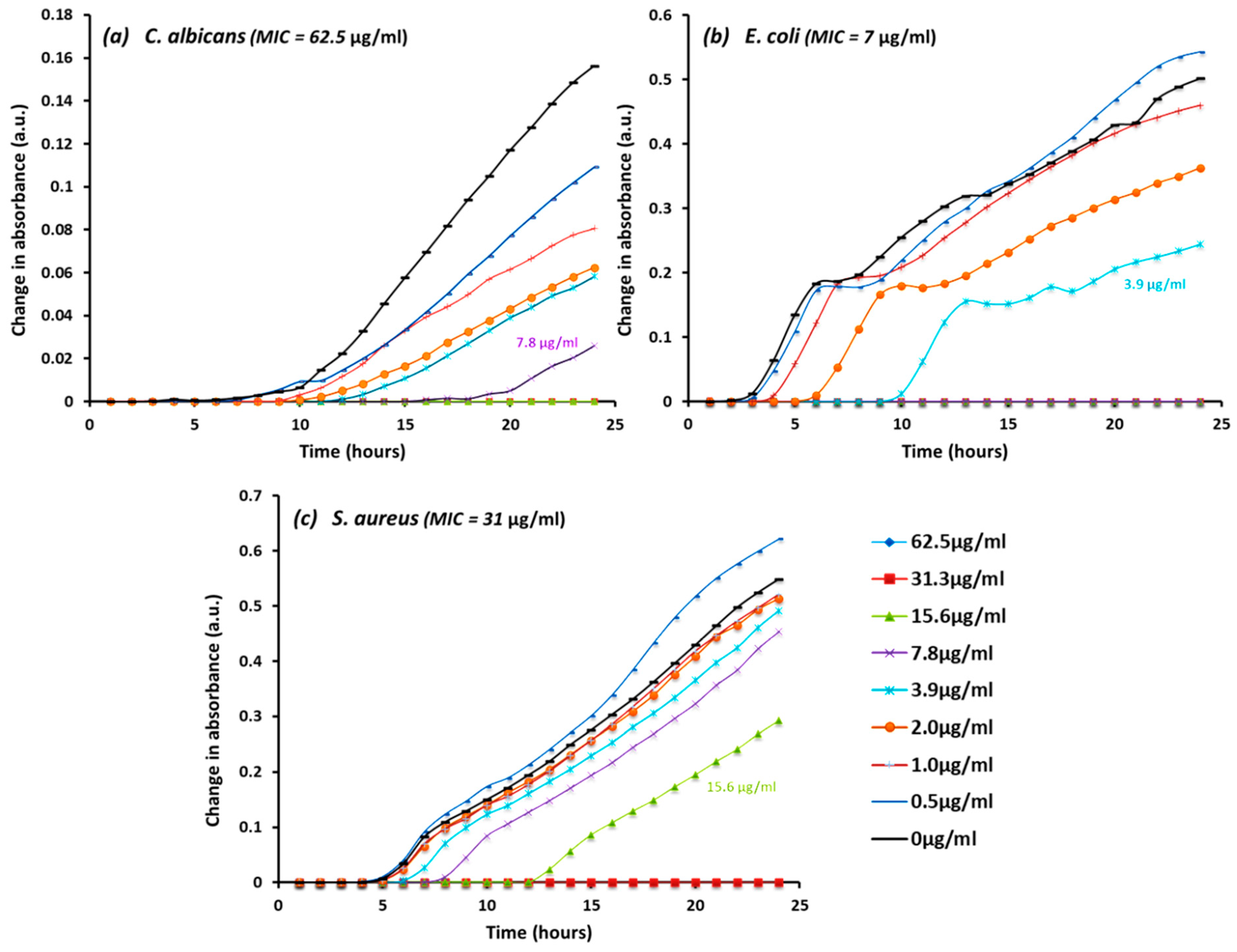
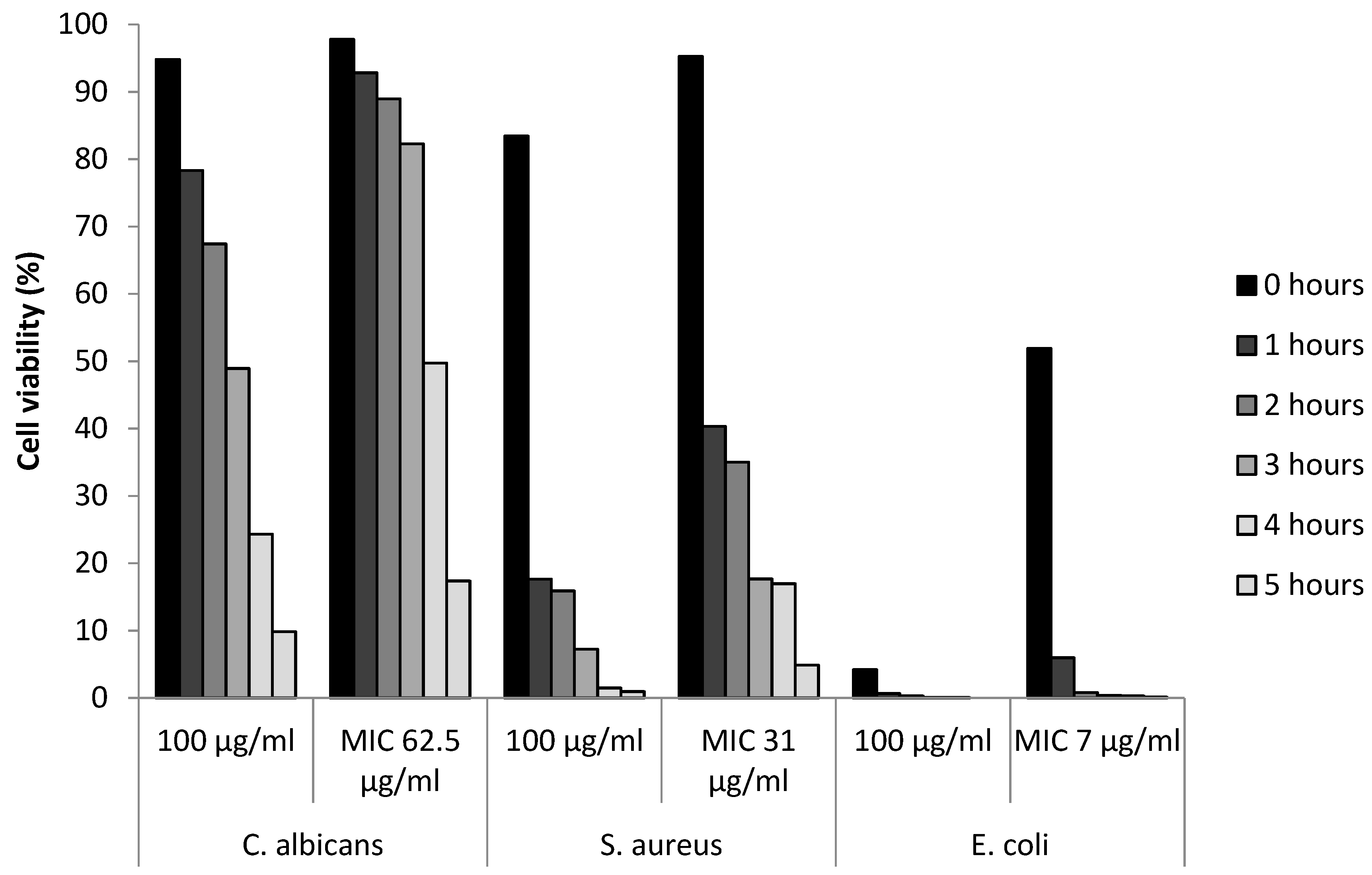
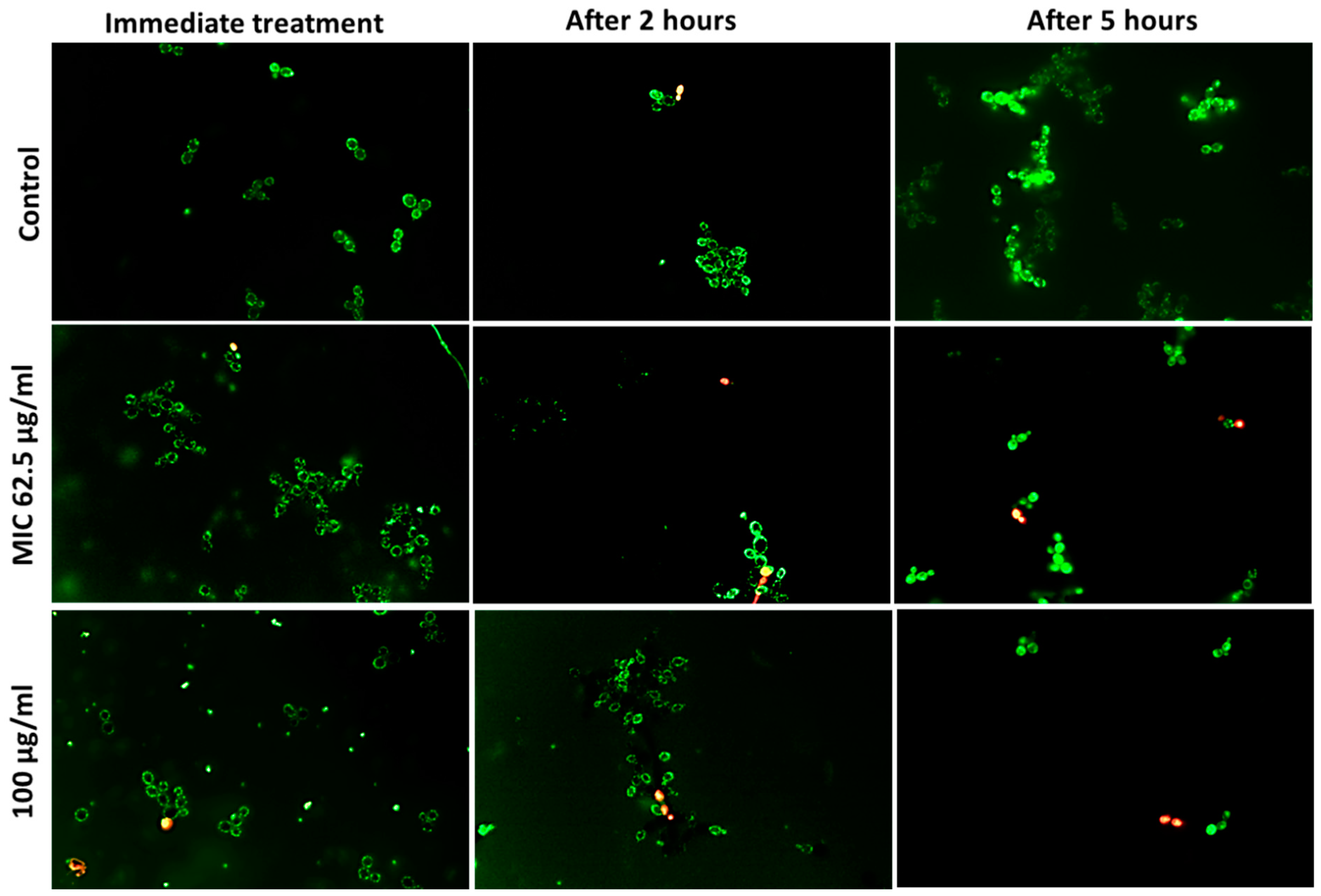
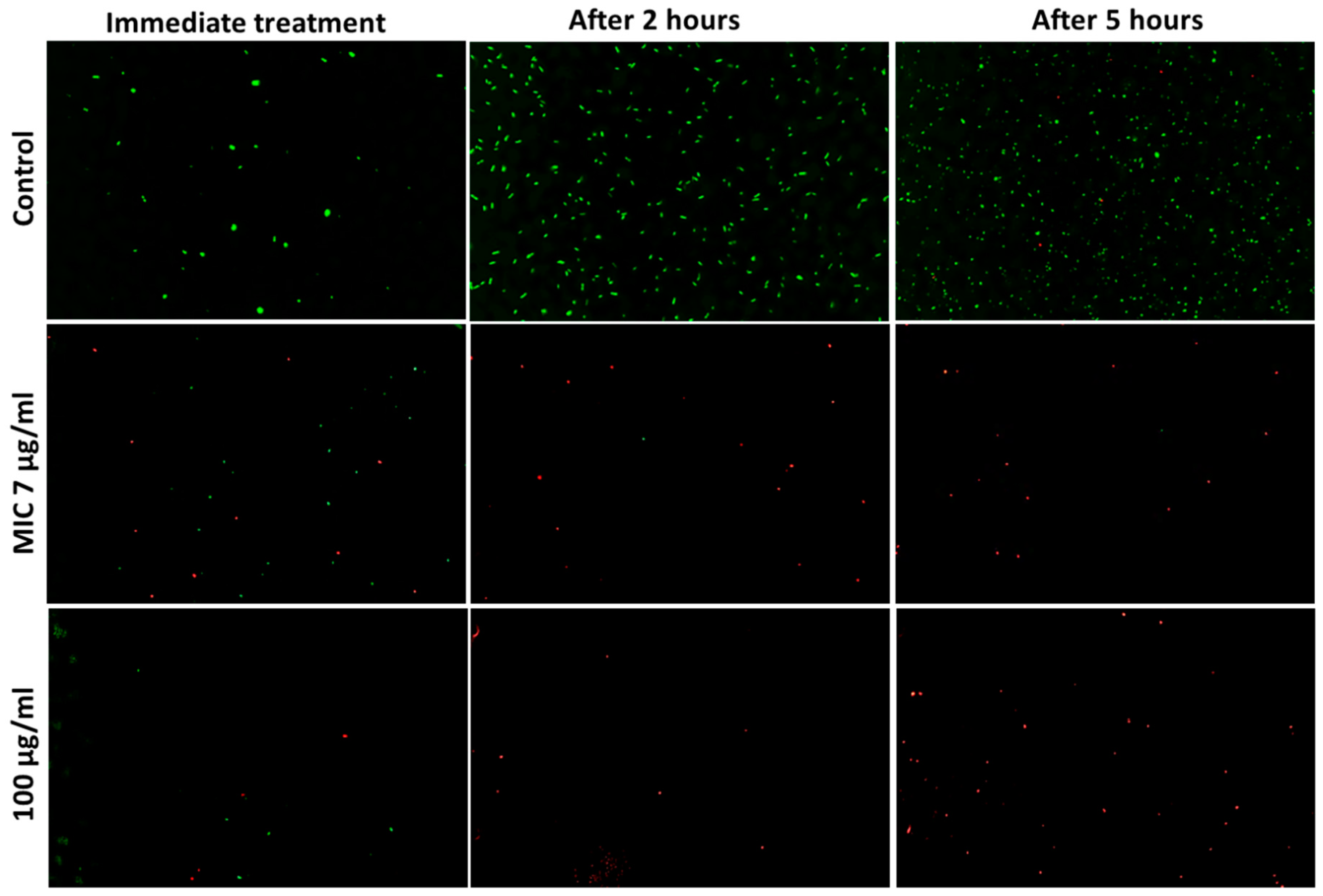

| Microbe | Ag20 | Ag100 | AgCu | Cu10 | Cu60 | Cu90 | CuO10 | CuO40 | CuZn | ZnO | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fungi | C. albicans | 0 | 0.85 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| C. tropicalis | 0 | 0.65 | 0.7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Gram negative | A. baumanii | 0.9 | 0 | 1.85 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| P. aeruginosa | 0 | 0 | 1.2 | 2.35 | 2.35 | 2.7 | 2.15 | 1.95 | 0 | 0 | |
| K. pneumonia | 0.6 | 0 | 1.4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| E. coli | 0.55 | 0 | 0.8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| S. typhimurium | 0.6 | 0 | 1.4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Gram positive | E. faecium | 0 | 0 | 1.4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| S. aureus | 0 | 0 | 1.7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| S. pyogenes | 0.8 | 0 | 1.9 | 0 | 0 | 0 | 0 | 0 | 1.4 | 1.4 |
| Inhibition % Per Strain | Microbe | Ag20 | Ag100 | AgCu | Cu10 | Cu60 | Cu90 | CuO10 | CuO40 | CuZn | ZnO | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fungi | 60 | C. albicans | − | + | + | + | + | + | + | − | − | − |
| 40 | C. tropicalis | − | + | + | + | − | − | + | − | − | − | |
| Gram negative | 80 | A. baumannii | + | + | + | + | + | + | − | + | + | − |
| 80 | P. aeruginosa | + | + | + | + | + | + | − | + | + | − | |
| 90 | K. pneumonia | + | + | + | + | + | + | − | + | + | + | |
| 90 | E. coli | + | + | + | + | + | + | − | + | + | + | |
| 80 | S. typhimurium | + | + | + | + | + | + | − | + | + | − | |
| Gram positive | 60 | E. faecium | + | − | + | + | + | + | + | − | − | − |
| 80 | S. aureus | + | + | + | + | + | + | − | + | + | − | |
| 90 | S. pyogenes | + | + | + | + | + | + | + | − | + | + | |
| % of microbes susceptible | 80 | 90 | 100 | 100 | 90 | 90 | 40 | 60 | 70 | 30 | ||
| Ag20 | AgCu | Cu10 | Cu60 | CuO40 | |
|---|---|---|---|---|---|
| C. albicans | 31 | 62.5 | 250 | 250 | 250 |
| C. tropicalis | 31 | 31 | 125 | 125 | 125 |
| A. baumannii | 15 | 31 | 31 | 31 | 31 |
| P. aeruginosa | 7 | 7 | 250 | 250 | 250 |
| K. pneumonia | 15 | 15 | 250 | 250 | 250 |
| E. coli | 250 | 7 | 250 | 250 | >250 |
| S. aureus | 15 | 31 | 125 | 125 | 125 |
| Method | Resources | Time (hours) | Antimicrobial Validation | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Agar Plate | 96 Well Plate | Broth | Resazurin | PI & SYTO9 | Spectrophotometer | Fluorescent Microscope | |||
| Agar well diffusion | X | X | 24 | Visible inhibitory zone (qualitative) | |||||
| Resazurin broth assay | X | X | X | 48 | Positive/negative antimicrobial activity (qualitative) | ||||
| Resazurin microtitre MIC assay | X | X | X | 48 | Minimum effective concentration of tested reagent (quantitative) | ||||
| Spectrometer growth rate | X | X | X | 24 | Monitoring of kinetic growth (semi-quantitative) | ||||
| Live/Dead assay | X | X | X | X | 6 | Cell viability (qualitative and semi-quantitative) | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chung, E.; Ren, G.; Johnston, I.; Matharu, R.K.; Ciric, L.; Walecka, A.; Cheong, Y.-K. Applied Methods to Assess the Antimicrobial Activity of Metallic-Based Nanoparticles. Bioengineering 2023, 10, 1259. https://doi.org/10.3390/bioengineering10111259
Chung E, Ren G, Johnston I, Matharu RK, Ciric L, Walecka A, Cheong Y-K. Applied Methods to Assess the Antimicrobial Activity of Metallic-Based Nanoparticles. Bioengineering. 2023; 10(11):1259. https://doi.org/10.3390/bioengineering10111259
Chicago/Turabian StyleChung, Etelka, Guogang Ren, Ian Johnston, Rupy Kaur Matharu, Lena Ciric, Agnieszka Walecka, and Yuen-Ki Cheong. 2023. "Applied Methods to Assess the Antimicrobial Activity of Metallic-Based Nanoparticles" Bioengineering 10, no. 11: 1259. https://doi.org/10.3390/bioengineering10111259
APA StyleChung, E., Ren, G., Johnston, I., Matharu, R. K., Ciric, L., Walecka, A., & Cheong, Y.-K. (2023). Applied Methods to Assess the Antimicrobial Activity of Metallic-Based Nanoparticles. Bioengineering, 10(11), 1259. https://doi.org/10.3390/bioengineering10111259









