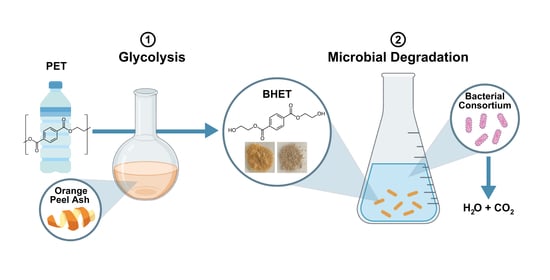
Two-Step Chemo-Microbial Degradation of Post-Consumer Polyethylene Terephthalate (PET) Plastic Enabled by a Biomass-Waste Catalyst
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of OPA Catalyst
2.3. PET Glycolysis
2.4. Characterization of Glycolysis Products and OPA Catalyst
2.4.1. Nuclear Magnetic Resonance (NMR)
2.4.2. Fourier-Transform Infrared Spectroscopy–Attenuated Total Reflectance (FTIR-ATR)
2.4.3. High-Performance Liquid Chromatography (HPLC)
2.4.4. Powder X-ray Diffraction (PXRD)
2.4.5. Single-Crystal X-ray Diffraction (SCXRD)
2.4.6. Inductively Coupled Plasma Spectrometry (ICP)
2.5. Microbial BHET Degradation
2.6. HPLC Analysis of Microbial BHET Degradation
2.7. Degradation Rate Calculation
3. Results and Discussion
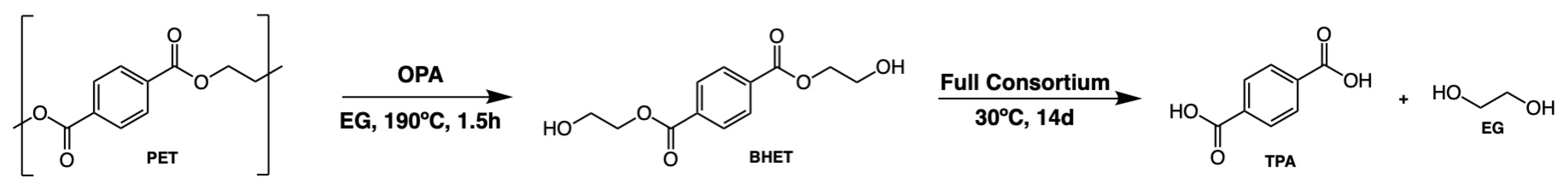
3.1. PET Glycolysis
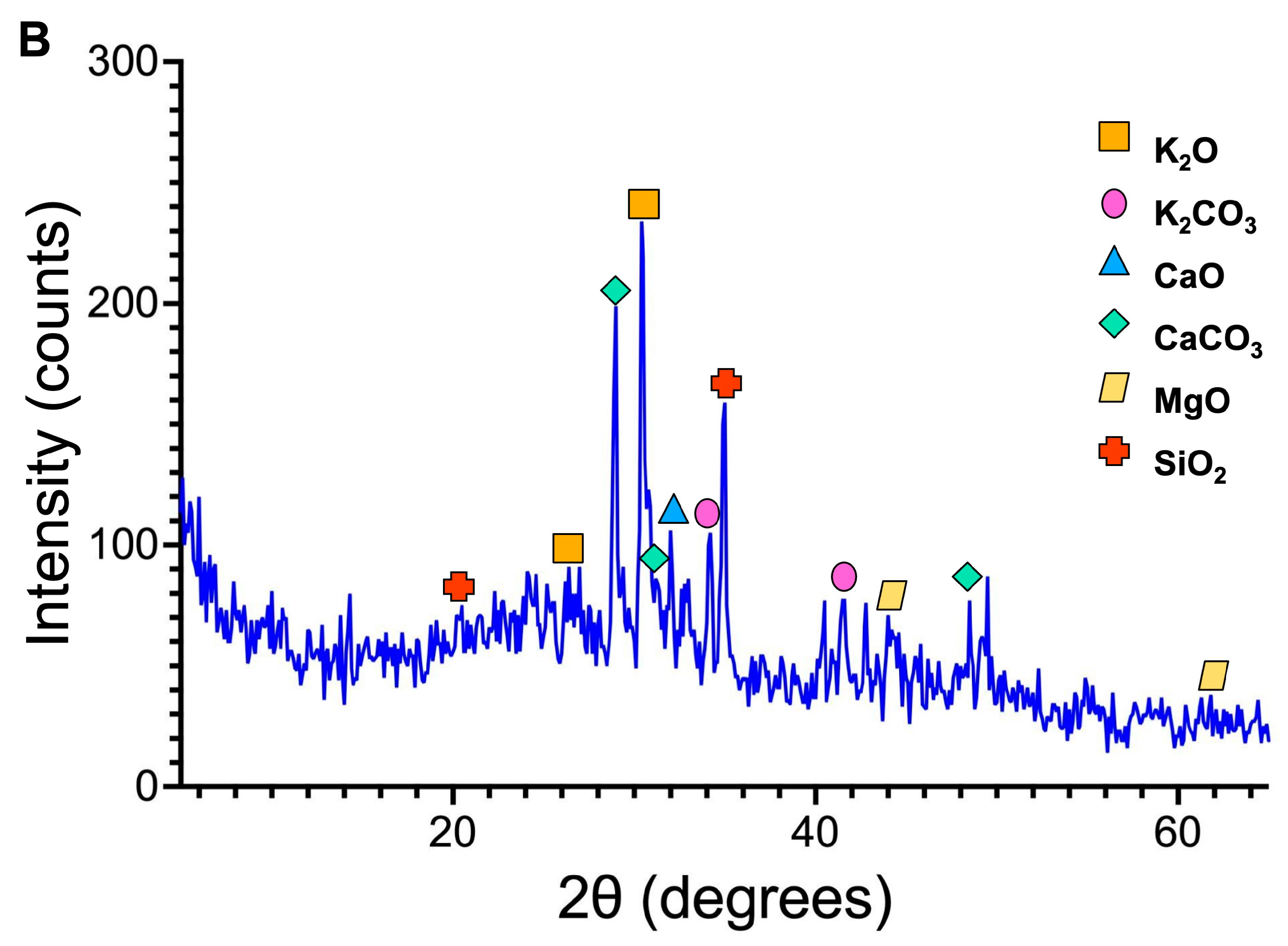
3.1.1. OPA Characterization
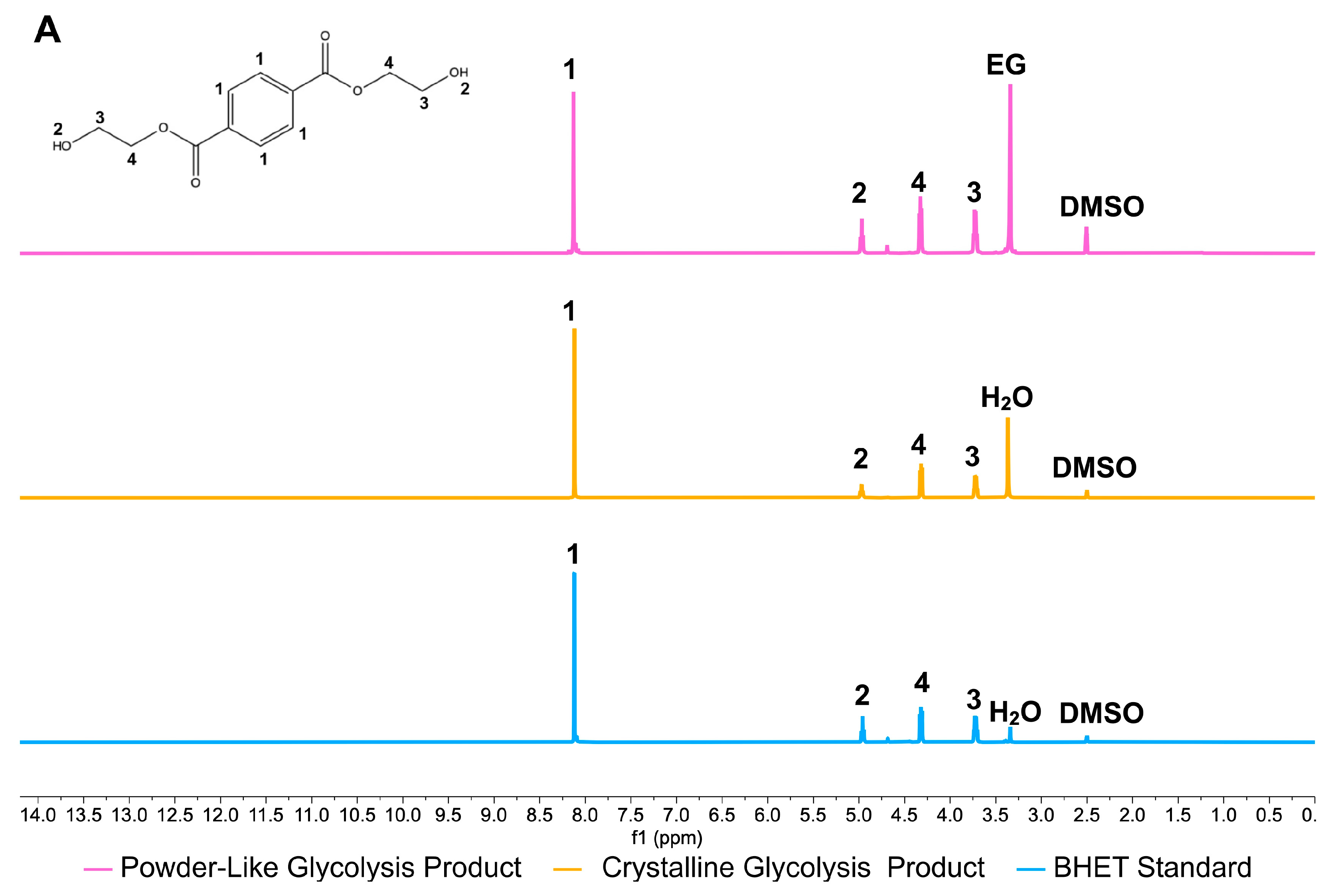
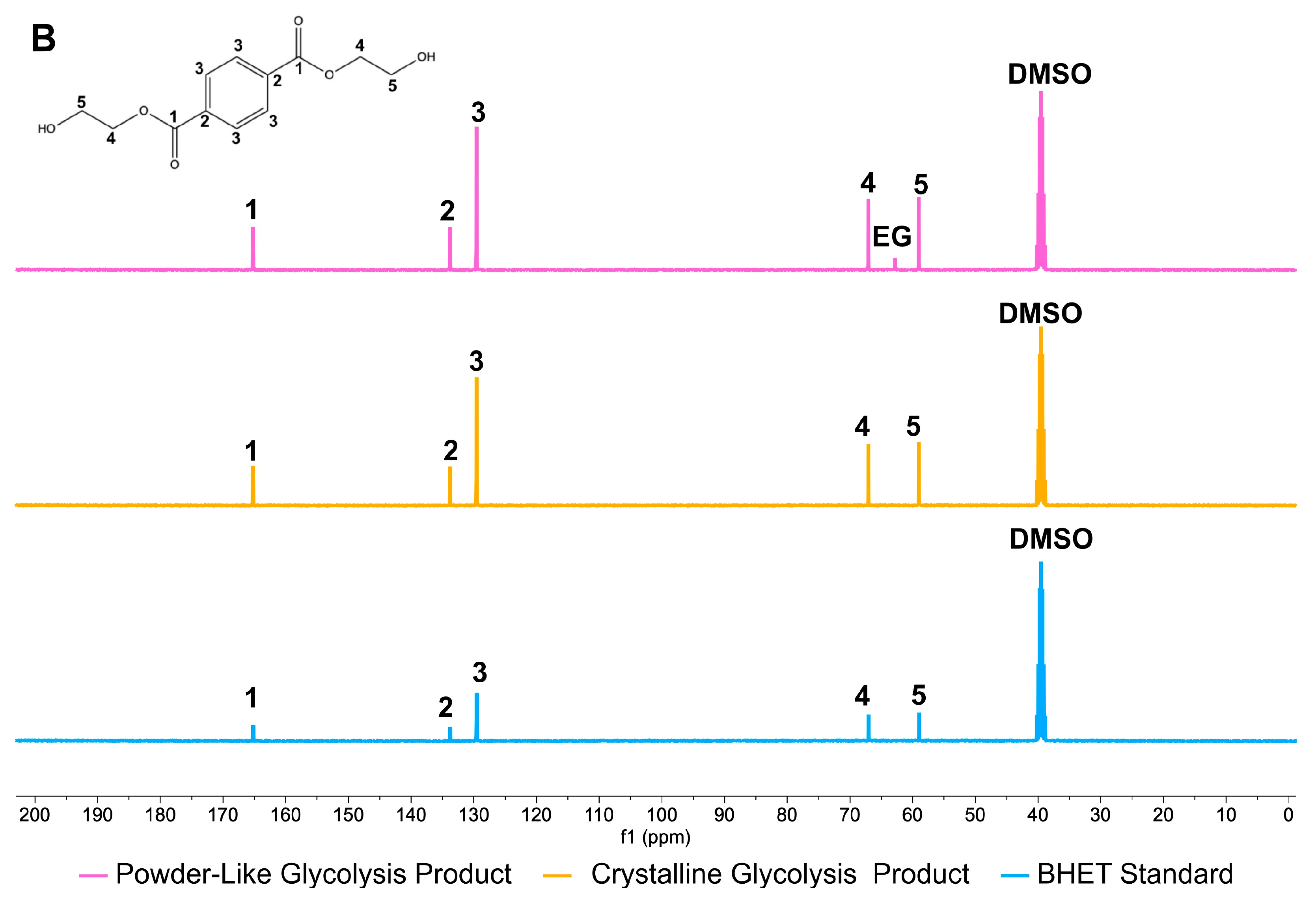
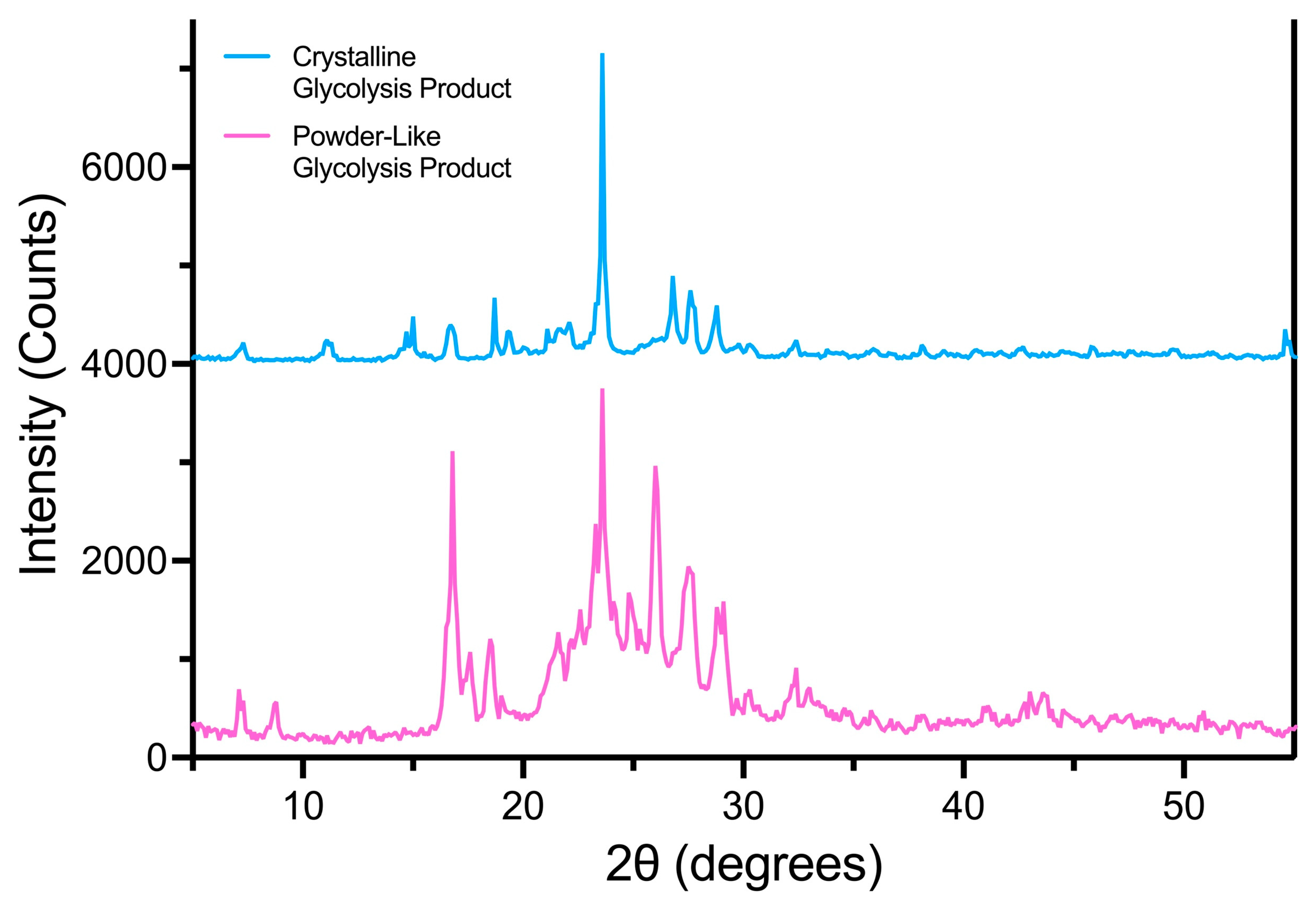
3.1.2. Yield and Characterization of the Glycolysis Products
3.2. Biodegradation of Synthesized BHET
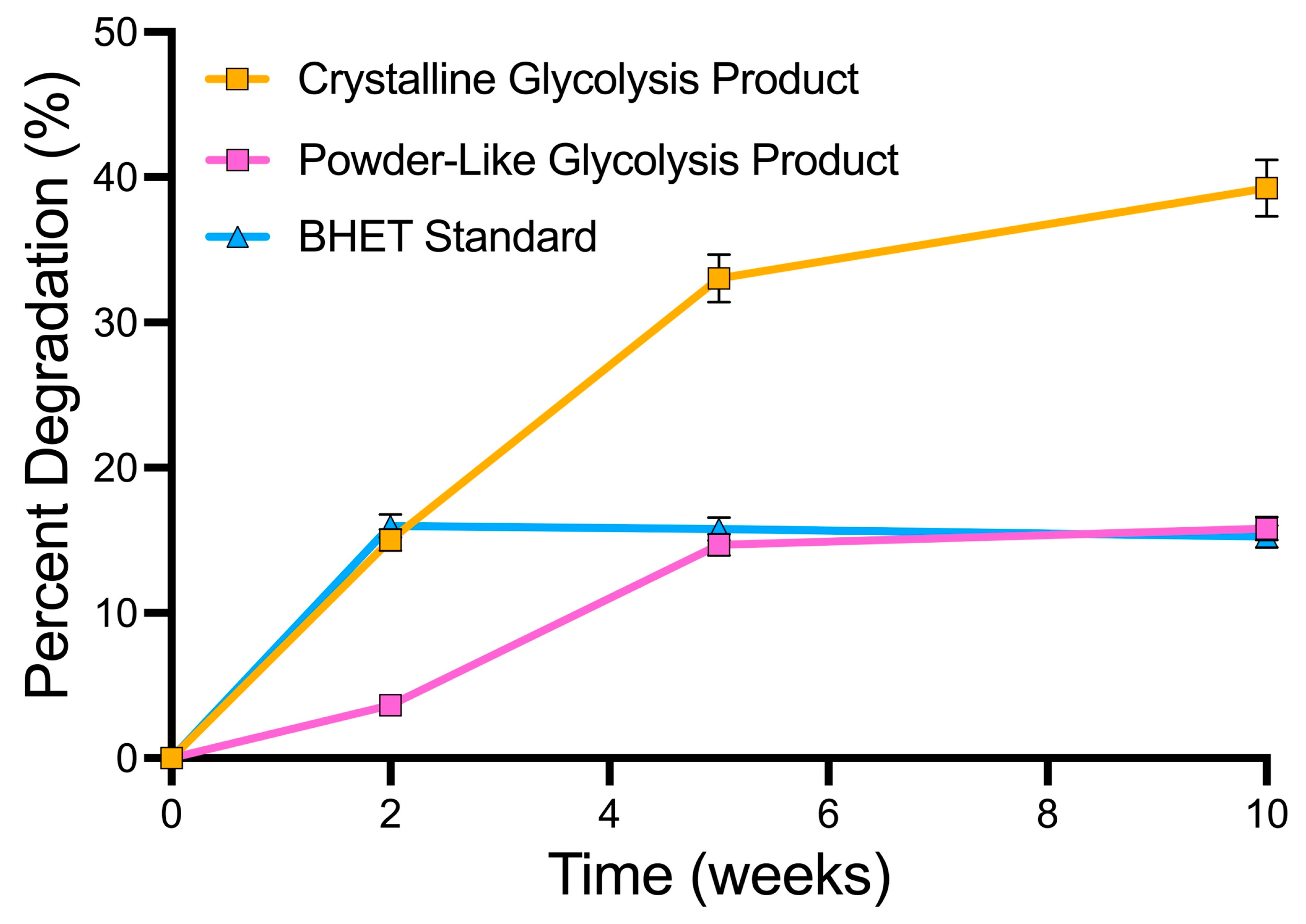
3.3. Effect of BHET Solid Structure on Biodegradation
3.4. Efficacy of the Two-Step Degradation Process
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Geyer, R.; Jambeck, J.R.; Law, K.L. Production, Use, and Fate of All Plastics Ever Made. Sci. Adv. 2017, 3, e1700782. [Google Scholar] [CrossRef] [PubMed]
- Shams, M.; Alam, I.; Mahbub, M.S. Plastic Pollution during COVID-19: Plastic Waste Directives and Its Long-Term Impact on the Environment. Environ. Adv. 2021, 5, 100119. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Wu, P.; Schartup, A.T.; Zhang, Y. Plastic Waste Release Caused by COVID-19 and Its Fate in the Global Ocean. Proc. Natl. Acad. Sci. USA 2021, 118, e2111530118. [Google Scholar] [CrossRef]
- Danso, D.; Chow, J.; Streit, W.R. Plastics: Microbial Degradation, Environmental and Biotechnological Perspectives. Appl. Environ. Microbiol. 2019, 85, e01095-19. [Google Scholar] [CrossRef] [PubMed]
- Thompson, R.C.; Moore, C.J.; Vom Saal, F.S.; Swan, S.H. Plastics, the Environment and Human Health: Current Consensus and Future Trends. Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 2153–2166. [Google Scholar] [CrossRef] [PubMed]
- Kosloski-Oh, S.C.; Wood, Z.A.; Manjarrez, Y.; De Los Rios, J.P.; Fieser, M.E. Catalytic Methods for Chemical Recycling or Upcycling of Commercial Polymers. Mater. Horiz. 2021, 8, 1084–1129. [Google Scholar] [CrossRef]
- Laldinpuii, T.Z.; Lalmuanpuia, C.; Lalhmangaihzuala, S.; Khiangte, V.; Pachuau, Z.; Vanlaldinpuia, K. Biomass Waste-Derived Recyclable Heterogeneous Catalyst for Aqueous Aldol Reaction and Depolymerization of PET Waste. New J. Chem. 2021, 45, 19542–19552. [Google Scholar] [CrossRef]
- Lalhmangaihzuala, S.; Laldinpuii, Z.; Khiangte, V.; Lallawmzuali, G.; Thanhmingliana; Vanlaldinpuia, K. Orange Peel Ash Coated Fe3O4 Nanoparticles as a Magnetically Retrievable Catalyst for Glycolysis and Methanolysis of PET Waste. Adv. Powder Technol. 2023, 34, 104076. [Google Scholar] [CrossRef]
- Shirazimoghaddam, S.; Amin, I.; Faria Albanese, J.A.; Shiju, N.R. Chemical Recycling of Used PET by Glycolysis Using Niobia-Based Catalysts. ACS Eng. Au 2023, 3, 37–44. [Google Scholar] [CrossRef]
- Kuete, M.A.; Van Velthem, P.; Ballout, W.; Nysten, B.; Devaux, J.; Ndikontar, M.K.; Pardoen, T.; Bailly, C. Integrated Approach to Eco-Friendly Thermoplastic Composites Based on Chemically Recycled PET Co-Polymers Reinforced with Treated Banana Fibres. Polymers 2022, 14, 4791. [Google Scholar] [CrossRef]
- Kim, Y.; Kim, M.; Hwang, J.; Im, E.; Moon, G.D. Optimizing PET Glycolysis with an Oyster Shell-Derived Catalyst Using Response Surface Methodology. Polymers 2022, 14, 656. [Google Scholar] [CrossRef] [PubMed]
- Lalhmangaihzuala, S.; Laldinpuii, Z.; Lalmuanpuia, C.; Vanlaldinpuia, K. Glycolysis of Poly(Ethylene Terephthalate) Using Biomass-Waste Derived Recyclable Heterogeneous Catalyst. Polymers 2021, 13, 37. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Chandra, R. Ligninolytic Enzymes and Its Mechanisms for Degradation of Lignocellulosic Waste in Environment. Heliyon 2020, 6, e03170. [Google Scholar] [CrossRef] [PubMed]
- Mohanan, N.; Montazer, Z.; Sharma, P.K.; Levin, D.B. Microbial and Enzymatic Degradation of Synthetic Plastics. Front. Microbiol. 2020, 11, 580709. [Google Scholar] [CrossRef]
- Carr, C.M.; Clarke, D.J.; Dobson, A.D.W. Microbial Polyethylene Terephthalate Hydrolases: Current and Future Perspectives. Front. Microbiol. 2020, 11, 571265. [Google Scholar] [CrossRef]
- Benavides Fernández, C.D.; Guzmán Castillo, M.P.; Quijano Pérez, S.A.; Carvajal Rodríguez, L.V. Microbial Degradation of Polyethylene Terephthalate: A Systematic Review. SN Appl. Sci. 2022, 4, 263. [Google Scholar] [CrossRef]
- Roberts, C.; Edwards, S.; Vague, M.; León-Zayas, R.; Scheffer, H.; Chan, G.; Swartz, N.A.; Mellies, J.L. Environmental Consortium Containing Pseudomonas and Bacillus Species Synergistically Degrades Polyethylene Terephthalate Plastic. mSphere 2020, 5, e01151-20. [Google Scholar] [CrossRef]
- Edwards, S.; León-Zayas, R.; Ditter, R.; Laster, H.; Sheehan, G.; Anderson, O.; Beattie, T.; Mellies, J.L. Microbial Consortia and Mixed Plastic Waste: Pangenomic Analysis Reveals Potential for Degradation of Multiple Plastic Types via Previously Identified PET Degrading Bacteria. Int. J. Mol. Sci. 2022, 23, 5612. [Google Scholar] [CrossRef]
- Sulaiman, S.; Yamato, S.; Kanaya, E.; Kim, J.-J.; Koga, Y.; Takano, K.; Kanaya, S. Isolation of a Novel Cutinase Homolog with Polyethylene Terephthalate-Degrading Activity from Leaf-Branch Compost by Using a Metagenomic Approach. Appl. Environ. Microbiol. 2012, 78, 1556–1562. [Google Scholar] [CrossRef]
- Tanasupawat, S.; Takehana, T.; Yoshida, S.; Hiraga, K.; Oda, K. Ideonella Sakaiensis Sp. Nov., Isolated from a Microbial Consortium That Degrades Poly(Ethylene Terephthalate). Int. J. Syst. Evol. Microbiol. 2016, 66, 2813–2818. [Google Scholar] [CrossRef]
- Yoshida, S.; Hiraga, K.; Takehana, T.; Taniguchi, I.; Yamaji, H.; Maeda, Y.; Toyohara, K.; Miyamoto, K.; Kimura, Y.; Oda, K. A Bacterium That Degrades and Assimilates Poly(Ethylene Terephthalate). Science 2016, 351, 1196–1199. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Han, D.O.; In Shim, K.; Kim, J.K.; Pelton, J.G.; Ryu, M.H.; Joo, J.C.; Han, J.W.; Kim, H.T.; Kim, K.H. One-Pot Chemo-Bioprocess of PET Depolymerization and Recycling Enabled by a Biocompatible Catalyst, Betaine. ACS Catal. 2021, 11, 3996–4008. [Google Scholar] [CrossRef]
- Quartinello, F.; Vajnhandl, S.; Volmajer Valh, J.; Farmer, T.J.; Vončina, B.; Lobnik, A.; Herrero Acero, E.; Pellis, A.; Guebitz, G.M. Synergistic Chemo-enzymatic Hydrolysis of Poly(Ethylene Terephthalate) from Textile Waste. Microb. Biotechnol. 2017, 10, 1376–1383. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.T.; Hee Ryu, M.; Jung, Y.J.; Lim, S.; Song, H.M.; Park, J.; Hwang, S.Y.; Lee, H.; Yeon, Y.J.; Sung, B.H.; et al. Chemo-Biological Upcycling of Poly(Ethylene Terephthalate) to Multifunctional Coating Materials. ChemSusChem 2021, 14, 4251–4259. [Google Scholar] [CrossRef]
- Neves Ricarte, G.; Lopes Dias, M.; Sirelli, L.; Antunes Pereira Langone, M.; Machado De Castro, A.; Zarur Coelho, M.A.; Dias Ribeiro, B. Chemo-enzymatic Depolymerization of Industrial and Assorted Post-consumer Poly(Ethylene Terephthalate) (PET) Wastes Using a Eutectic-based Catalyst. J. Chem. Technol. Biotechnol. 2021, 96, 3237–3244. [Google Scholar] [CrossRef]
- Tournier, V.; Topham, C.M.; Gilles, A.; David, B.; Folgoas, C.; Moya-Leclair, E.; Kamionka, E.; Desrousseaux, M.-L.; Texier, H.; Gavalda, S.; et al. An Engineered PET Depolymerase to Break down and Recycle Plastic Bottles. Nature 2020, 580, 216–219. [Google Scholar] [CrossRef]
- Ghaemy, M.; Mossaddegh, K. Depolymerisation of Poly(Ethylene Terephthalate) Fibre Wastes Using Ethylene Glycol. Polym. Degrad. Stab. 2005, 90, 570–576. [Google Scholar] [CrossRef]
- Furukawa, M.; Kawakami, N.; Oda, K.; Miyamoto, K. Acceleration of Enzymatic Degradation of Poly(Ethylene Terephthalate) by Surface Coating with Anionic Surfactants. ChemSusChem 2018, 11, 4018–4025. [Google Scholar] [CrossRef]
- Merkys, A.; Vaitkus, A.; Butkus, J.; Okulič-Kazarinas, M.; Kairys, V.; Gražulis, S. COD::CIF::Parser: An Error-Correcting CIF Parser for the Perl Language. J. Appl. Crystallogr. 2016, 49, 292–301. [Google Scholar] [CrossRef]
- Gražulis, S.; Merkys, A.; Vaitkus, A.; Okulič-Kazarinas, M. Computing Stoichiometric Molecular Composition from Crystal Structures. J. Appl. Crystallogr. 2015, 48, 85–91. [Google Scholar] [CrossRef]
- Gražulis, S.; Daškevič, A.; Merkys, A.; Chateigner, D.; Lutterotti, L.; Quirós, M.; Serebryanaya, N.R.; Moeck, P.; Downs, R.T.; Le Bail, A. Crystallography Open Database (COD): An Open-Access Collection of Crystal Structures and Platform for World-Wide Collaboration. Nucleic Acids Res. 2012, 40, D420–D427. [Google Scholar] [CrossRef] [PubMed]
- Merkys, A.; Vaitkus, A.; Grybauskas, A.; Konovalovas, A.; Quirós, M.; Gražulis, S. Graph Isomorphism-Based Algorithm for Cross-Checking Chemical and Crystallographic Descriptions. J. Cheminform. 2023, 15, 25. [Google Scholar] [CrossRef] [PubMed]
- Quirós, M.; Gražulis, S.; Girdzijauskaitė, S.; Merkys, A.; Vaitkus, A. Using SMILES Strings for the Description of Chemical Connectivity in the Crystallography Open Database. J. Cheminform. 2018, 10, 23. [Google Scholar] [CrossRef] [PubMed]
- Vaitkus, A.; Merkys, A.; Gražulis, S. Validation of the Crystallography Open Database Using the Crystallographic Information Framework. J. Appl. Crystallogr. 2021, 54, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Putz, H.; Brandenburg, K. Match!—Phase Analysis Using Powder Diffraction, Version 3.15, Crystal Impact. Available online: https://www.crystalimpact.de/match (accessed on 28 August 2023).
- Macrae, C.F.; Sovago, I.; Cottrell, S.J.; Galek, P.T.A.; McCabe, P.; Pidcock, E.; Platings, M.; Shields, G.P.; Stevens, J.S.; Towler, M.; et al. Mercury 4.0: From Visualization to Analysis, Design and Prediction. J. Appl. Crystallogr. 2020, 53, 226–235. [Google Scholar] [CrossRef]
- León-Zayas, R.; Roberts, C.; Vague, M.; Mellies, J.L. Draft Genome Sequences of Five Environmental Bacterial Isolates That Degrade Polyethylene Terephthalate Plastic. Microbiol. Resour. Announc. 2019, 8, e00237-19. [Google Scholar] [CrossRef]
- Changmai, B.; Sudarsanam, P.; Rokhum, S.L. Biodiesel Production Using a Renewable Mesoporous Solid Catalyst. Ind. Crops Prod. 2020, 145, 111911. [Google Scholar] [CrossRef]
- Taghavi, F.; Gholizadeh, M.; Saljooghi, A.S.; Ramezani, M. Metal Free Synthesis of Tetrahydrobenzo[a]Xanthenes Using Orange Peel as a Natural and Low Cost Efficient Heterogeneous Catalyst. RSC Adv. 2016, 6, 87082–87087. [Google Scholar] [CrossRef]
- Yunita, I.; Putisompon, S.; Chumkaeo, P.; Poonsawat, T.; Somsook, E. Effective Catalysts Derived from Waste Ostrich Eggshells for Glycolysis of Post-Consumer PET Bottles. Chem. Pap. 2019, 73, 1547–1560. [Google Scholar] [CrossRef]
- Scé, F.; Cano, I.; Martin, C.; Beobide, G.; Castillo, Ó.; De Pedro, I. Comparing Conventional and Microwave-Assisted Heating in PET Degradation Mediated by Imidazolium-Based Halometallate Complexes. New J. Chem. 2019, 43, 3476–3485. [Google Scholar] [CrossRef]
- Steinbach, A.; Schulz, S.; Giebler, J.; Schulz, S.; Pronk, G.J.; Kögel-Knabner, I.; Harms, H.; Wick, L.Y.; Schloter, M. Clay Minerals and Metal Oxides Strongly Influence the Structure of Alkane-Degrading Microbial Communities during Soil Maturation. ISME J. 2015, 9, 1687–1691. [Google Scholar] [CrossRef] [PubMed]
- Pereira, J.D.C.; Giese, E.C.; Moretti, M.M.D.S.; Gomes, A.C.D.S.; Perrone, O.M.; Boscolo, M.; Da Silva, R.; Gomes, E.; Martins, D.A.B. Effect of Metal Ions, Chemical Agents and Organic Compounds on Lignocellulolytic Enzymes Activities. In Enzyme Inhibitors and Activators; Senturk, M., Ed.; InTech: London, UK, 2017; ISBN 978-953-51-3057-4. [Google Scholar]
- Beamson, G.; Clark, D.T.; Hayes, N.W.; Law, D.S.-L. Effect of Crystallinity on the XPS Spectrum of Poly(Ethylene Terephthalate). Surf. Sci. Spectra 1994, 3, 357–365. [Google Scholar] [CrossRef]
- Wei, R.; Breite, D.; Song, C.; Gräsing, D.; Ploss, T.; Hille, P.; Schwerdtfeger, R.; Matysik, J.; Schulze, A.; Zimmermann, W. Biocatalytic Degradation Efficiency of Postconsumer Polyethylene Terephthalate Packaging Determined by Their Polymer Microstructures. Adv. Sci. 2019, 6, 1900491. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, M.; Kawakami, N.; Tomizawa, A.; Miyamoto, K. Efficient Degradation of Poly(Ethylene Terephthalate) with Thermobifida Fusca Cutinase Exhibiting Improved Catalytic Activity Generated Using Mutagenesis and Additive-Based Approaches. Sci. Rep. 2019, 9, 16038. [Google Scholar] [CrossRef]
- Thomsen, T.B.; Hunt, C.J.; Meyer, A.S. Influence of Substrate Crystallinity and Glass Transition Temperature on Enzymatic Degradation of Polyethylene Terephthalate (PET). New Biotechnol. 2022, 69, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Urbanek, A.K.; Rymowicz, W.; Mirończuk, A.M. Degradation of Plastics and Plastic-Degrading Bacteria in Cold Marine Habitats. Appl. Microbiol. Biotechnol. 2018, 102, 7669–7678. [Google Scholar] [CrossRef] [PubMed]
- Urbanek, A.K.; Kosiorowska, K.E.; Mirończuk, A.M. Current Knowledge on Polyethylene Terephthalate Degradation by Genetically Modified Microorganisms. Front. Bioeng. Biotechnol. 2021, 9, 771133. [Google Scholar] [CrossRef]
- Wallace, N.E.; Adams, M.C.; Chafin, A.C.; Jones, D.D.; Tsui, C.L.; Gruber, T.D. The Highly Crystalline PET Found in Plastic Water Bottles Does Not Support the Growth of the PETase -producing Bacterium Ideonella sakaiensis. Environ. Microbiol. Rep. 2020, 12, 578–582. [Google Scholar] [CrossRef]




| Workup Method Used | Yield of Crystalline Product (%) 1 | Yield of Crystalline and Water-Insoluble Product (%) 1 |
|---|---|---|
| Concentration 2 | 16.8 | 86.8 |
| Agitation 3 | 78.4 | 90.2 |
| Concentration and Agitation 4 | 26.6 | 85.3 |
| Carbon Source | 2-Week Yeast-Extract-Supplemented Degradation Rate Including Hydrolysis (%) 1 | 2-Week Yeast-Supplemented Degradation Rate without Hydrolysis (%) 2 |
|---|---|---|
| Crystalline glycolysis product | 62.80 | 19.81 |
| Powder-like glycolysis product | 61.55 | 12.29 |
| BHET standard | 61.32 | 8.69 |
| Degradation Assay | Biodegradation Rate of Glycolysis Products (%) 1 | Two-Step Degradation Rate (%) 1 |
|---|---|---|
| 10-week degradation assay 2 | 35.30 | 32.63 |
| 2-week degradation assay not including hydrolysis | 18.82 | 16.98 |
| 2-week degradation assay including hydrolysis 3 | 62.63 | 56.50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shingwekar, D.; Laster, H.; Kemp, H.; Mellies, J.L. Two-Step Chemo-Microbial Degradation of Post-Consumer Polyethylene Terephthalate (PET) Plastic Enabled by a Biomass-Waste Catalyst. Bioengineering 2023, 10, 1253. https://doi.org/10.3390/bioengineering10111253
Shingwekar D, Laster H, Kemp H, Mellies JL. Two-Step Chemo-Microbial Degradation of Post-Consumer Polyethylene Terephthalate (PET) Plastic Enabled by a Biomass-Waste Catalyst. Bioengineering. 2023; 10(11):1253. https://doi.org/10.3390/bioengineering10111253
Chicago/Turabian StyleShingwekar, Deepika, Helen Laster, Hannah Kemp, and Jay L. Mellies. 2023. "Two-Step Chemo-Microbial Degradation of Post-Consumer Polyethylene Terephthalate (PET) Plastic Enabled by a Biomass-Waste Catalyst" Bioengineering 10, no. 11: 1253. https://doi.org/10.3390/bioengineering10111253
APA StyleShingwekar, D., Laster, H., Kemp, H., & Mellies, J. L. (2023). Two-Step Chemo-Microbial Degradation of Post-Consumer Polyethylene Terephthalate (PET) Plastic Enabled by a Biomass-Waste Catalyst. Bioengineering, 10(11), 1253. https://doi.org/10.3390/bioengineering10111253