1. Introduction
Stroke is one of the leading causes of both death and disability worldwide, resulting in 6.55 million deaths and 143 million disability-adjusted life-years (DALYs) in 2019 [
1]. With the aging of the population and the epidemiological transition from infectious to non-communicable diseases in low-income and middle-income countries, the global burden of stroke and subsequent post-stroke care will significantly increase in the future [
2]. In addition to the serious social, emotional, and financial repercussions on the individuals who suffer stroke and their families, stroke also has a substantial impact on health and social care services. The direct and indirect cost of stroke in the USA was estimated to be
$103.5 billion per year [
3]. Most strokes are ischemic [
4], and the main culprit is the blockage of the cerebral artery by an embolus. Carotid atherosclerosis is a chronic inflammatory disease involving the gradual accumulation of plaque within the inner wall of the carotid arteries. The disease is characterized by endothelial dysfunction, mononuclear accumulation, smooth muscle cell and fibrous matrix proliferation, leading to the formation of atherosclerotic plaques [
5], which is the major source of emboli in the form of platelet aggregates and plaque debris. Interventions with dietary and medical treatments are effective in preventing most vascular events associated with carotid atherosclerosis [
6,
7,
8]. Therefore, sensitive and cost-effective assessment tools or biomarkers for carotid atherosclerosis are essential for measuring carotid atherosclerosis burden, managing patients exposed to high risk of vascular events, and validating treatment strategies in clinical trials.
Ultrasound imaging technique has been used for quantitative assessment of carotid atherosclerosis burden. Carotid intima-media thickness (IMT) measured from the longitudinal view of two-dimensional ultrasound (2DUS) images has been used for more than 30 years. However, IMT is a weak predictor of cardiovascular events [
9], and the annual change of IMT (∼0.015 mm) is too small to be detected within a clinically affordable time frame [
10]. In addition, IMT measurements assess the intima-media thickening measured in the common carotid artery (CCA), which is more often caused by hypertension-induced medial hypertrophy and therefore does not directly indicate atherosclerosis [
11]. Total plaque area (TPA) directly quantifies plaque burden from 2DUS images and is more sensitive than IMT in predicting stroke [
12]. However, both IMT and TPA are obtained from 2DUS examinations that require an operator to locate a 2D imaging plane to be scanned. The operator variability in selecting the imaging plane makes 2DUS suboptimal for serial monitoring of atherosclerosis.
Mechanical three-dimensional ultrasound (3DUS) imaging technique has been developed that uses a motorized mechanical device to translate the transducer along a patient’s neck, capturing a series of successive 2D images, which are then reconstructed into 3D images [
13]. The availability of 3DUS obviates the need for the clinician to mentally transform multiple 2D ultrasound images to form a 3D impression of the complex structure of the carotid vessel and plaque and allows for objective and quantitative 3D volume measurements to be made. These measurements include total plaque volume (TPV) and vessel wall volume (VWV), which were shown to be more sensitive to changes in carotid atherosclerosis than IMT and TPA [
9,
14], as the thickness, length and circumferential extent of plaque burden can be measured by 3DUS. TPV and VWV were shown to be sensitive to medical treatment effect and reduce the sample size and duration required to establish the treatment effectiveness [
15,
16,
17], thereby making clinical trials more cost-effective and shortening the time required to withhold effective treatments from patients who need them. The progression of TPV was shown to predict cardiovascular events in a study involving over 300 patients, whereas TPA progression did not prevent events [
9], providing evidence to support the increased sensitivity of TPV in risk stratification. VWV shows higher inter-scan reproducibility than TPV as media-adventitia boundaries (MAB) and lumen-intima boundaries (LIB) are easier to be segmented from 3DUS than plaque [
18], and can be used to assess population without measurable plaques [
19]. The vessel-wall-plus-plaque thickness (VWT) is the point-by-point measurement of the distance between points on the MAB and the corresponding points on the LIB; it can be mapped onto a 2D L-shaped carotid map to show the spatial distribution of the vessel wall and plaque changes over the carotid artery and for quantitative analysis [
20,
21]. VWT-based metrics were demonstrated to be more sensitive than VWV and TPV to the effect of dietary intervention [
22,
23].
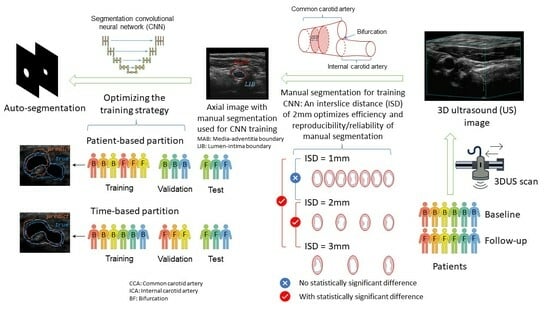
The pre-requisite for VWT and VWV measurements is the availability of LIB and MAB. Although they can be segmented from 3DUS images manually and manual segmentation has been performed for previous clinical studies [
14,
16,
18], manual segmentation is prohibitively time-consuming even for small-scale clinical studies involving tens of patients. Observers trained in our group take five hours to segment the MAB and LIB in a 3DUS volume with a 1 mm interslice distance (ISD) for a coverage of 25 mm (15 mm of CCA and 10 mm of ICA). The small-scale study involving 56 patients in a placebo-controlled study of the effect of Vitamin B supplement [
22] requires the segmentation of 224 volumes (4 volumes per patient: left and right carotids at baseline and follow-up image sessions) and would take more than a thousand hours of manual segmentation time. In addition, the reliability of manual segmentation depends heavily on the expertise of individual observers. For this reason, observers are required to be trained for the ability to provide reproducible measurements for VWV [
24] and VWT [
23] for the same set of images on different segmentation sessions, which are separated for a period to minimize the effect of memory. These trainings typically take weeks to complete. Without sufficient training on manual segmentation, the observer would not be able to produce the reliable segmentation required to train a segmentation CNN. The need for more efficient ways to delineate LIB and MAB reproducibly motivates the development of semi-automatic or automatic segmentation algorithms designed to minimize human intervention and improve observer reproducibility while substantially reducing the segmentation time required.
Early approaches are based on deformable models that require extensive interaction (2–14 min) to initialize the models [
25,
26,
27]. These methods have been phased out by recently developed deep learning algorithms that require fewer human interactions to generate segmentation results. Menchón-Lara et al. [
28] used multi-layer perceptrons trained under the scaled conjugate gradient algorithm to delineate MAB and LIB of the common carotid arteries (CCA) from 2D longitudinal carotid US images. Zhou et al. [
29] proposed a semiautomatic CCA segmentation method using the dynamic convolutional neural network and U-Net to segment MAB and LIB, respectively, from 3D carotid US images. The same group [
30] proposed another segmentation network combining a 3D deep convolutional neural network and a continuous max-flow module to segment CCA from a manually identified region of interest (ROI). Lin et al. [
31] used the cross-shaped window (CSWin) transformer to optimize a standard U-shaped neural network and proposed a U-CSWT model to segment the LIB and MAB of CCA in 3DUS images. However, the above deep learning approaches made no effort in segmenting the internal carotid arteries (ICA), thereby precluding vessel wall quantification at the ICA. VWV and VWT measured only at CCA may be less sensitive to treatment effect than those measured in previous studies that involved both CCA and ICA [
16,
22], with carotid plaques typically more prevalent at ICA [
32]. Jiang et al. [
33] previously developed a two-channel U-Net, driven by a novel loss function called the adaptive triple Dice loss (ATDL) function that can segment LIB and MAB simultaneously. The segmentation performed on CCA slices was fully automatic, whereas the ICA was enclosed by a manually identified bounding box that ensured the ICA was segmented, instead of the external carotid artery (ECA), which are less relevant to cardiovascular events [
34] and typically not measured for atherosclerosis [
9,
22,
35]. As the application of this algorithm allows automated quantification of VWV and VWT at the CCA and ICA, the segmentation strategies proposed in the current study were built upon this algorithm.
The goal of the current study is to evaluate and optimize the efficiency, reproducibility, and accuracy of the 3DUS carotid segmentation and quantification workflow. The major bottleneck in the efficiency of the deep-learning framework is the requirement to train the network by manual segmentation of MAB and LIB. One strategy to reduce the manual segmentation time is to increase the inter-slice distance (ISD) between adjacent segmented boundaries (so that fewer axial images are needed to be segmented), but doing so may reduce the reproducibility of carotid measurements. Although the effect of ISD on mean plaque volume has been investigated [
36], the current study is the first to investigate the effect of different ISD settings on the reproducibility of MAB and LIB segmentations and the associated VWV and VWT measurements.
Secondly, images used to train deep networks are typically randomly partitioned in the training, validation, and testing sets. An alternative way to partition data for studies involving serial monitoring of patients that may lead to higher segmentation performance would be to train the network on baseline images of patients with manual segmentation and test it on follow-up images. The network trained by baseline images had been exposed to the geometry of the carotid arteries in the entire cohort. While segmenting the follow-up images of the same cohort, we hypothesize that the segmentation accuracy of the network would be higher than a network trained by a randomly selected training set as the network had seen the images of the same set of patients, although at a different time point. This hypothesis, if validated, would suggest a clinically feasible workflow in which the network is trained by manually segmented baseline images for automatic segmentation of the follow-up images. This new workflow would be even more advantageous in serial monitoring of atherosclerosis development in which multiple images of the same patient acquired over time are assessed, as the model trained at baseline can be applied in multiple follow-up imaging sessions. In this study, we compared the segmentation performance of the time-based partition strategy, in which manually segmented boundaries are provided for baseline volumes of all patients investigated for network training, with the patient-based partition strategy, in which manually segmented boundaries are provided for the baseline and follow-up volumes of half of the patient population for training. Specifically, we compared the segmentation accuracy of the time-based and patient-based partition strategies and the VWV and VWT measured from the boundaries segmented by these two partitioning approaches. Quantified assessment of VWT and VWT measurements generated by the two partitioning strategies were evaluated by comparison with the corresponding measurements obtained from manual segmentation. VWV and VWT measured from manual segmentation were shown to be sensitive to medical [
16,
21] and dietary interventions [
22,
23]; if comparable VWV and VWT measurements can be made with the proposed automated workflow, it would give rise to more efficient analysis of the progression/regression of carotid atherosclerosis in clinical studies involving treatment effect monitoring and evaluation.
The remaining sections of the paper are organized as follows.
Section 2 is the method section that includes a description of the data set used in this study, the methodology with which we evaluated manual segmentation variability, a description of the segmentation algorithm, the conventional and the proposed data partitioning strategies, and the statistical analyses involved. The results of the study are presented in
Section 3. A discussion and the conclusion of the study are provided in
Section 4 and
Section 5, respectively.
4. Discussion
VWV and VWT are quantitative 3DUS measurements useful for monitoring changes of carotid atherosclerosis and they were shown to be sensitive in detecting the effects of medical therapies and dietary interventions [
16,
22,
43]. A major bottleneck in previous clinical studies involving 3D carotid US assessment is the requirement to segment the MAB and LIB manually. While CNN segmentation networks have been developed to provide automatic segmentation, they still require supervision by manually segmented boundaries. Therefore, there is a critical requirement to design a 3DUS carotid image analysis workflow to improve the efficiency of generating manual segmentation. While segmentation time can be reduced by increasing the interval at which axial slices are segmented (i.e., increasing ISD), segmentation reliability may be compromised as adjacent axial slices with a larger ISD may appear less continuous and more difficult to segment. The first question addressed in this study is to what extent we could increase the ISD (therefore reducing segmentation time) while maintaining an acceptable level of the reliability of the boundaries used to train the network. The current study assessed the effect of ISD on the intra-observer reproducibility of MAB and LIB segmentations and the VWV and VWT measured from the segmented MAB and LIB. We established that the intra-observer reproducibility of MAB and LIB segmentations, quantified by DSC, at ISDs of 1 mm and 2 mm was not statistically significantly different, whereas the reproducibilities at ISD = 1 mm and 2 mm were both statistically significantly higher than that attained at ISD = 3 mm. The ICCs of VWV and VWT measurements obtained at an ISD of 3 mm were also substantially lower than those attained at the ISD of 1 mm and 2 mm. Our result suggests that manual segmentation reliability at ISD = 2 mm is similar to that at ISD = 1 mm, and the reliability substantially reduces with the ISD increasing to 3 mm. As setting the ISD at 2 mm saves half of the segmentation time compared to 1 mm and we attempt to optimize for efficiency of the automated 3DUS analysis workflow, we applied an ISD of 2 mm for generating training boundaries when evaluating the CNN segmentation network.
Random partitioning of the available data into training and testing sets allows a machine learning model to generalize to new patient data. Furthermore, for a fair evaluation of the machine learning model, different data items associated with a patient are partitioned into the same set. However, the considerations involved in designing a segmentation model suitable for
serial monitoring of patients are slightly different from a typical machine learning model. Instead of being able to generalize to new patient data, a more important consideration is higher segmentation accuracy in a batch of data acquired at a follow-up time point for the same cohort of patients involved in training. For this reason, we proposed training our segmentation network with the baseline images of the entire cohort of patients and testing the network with the follow-up images of the same cohort of patients. In such a partitioning scheme we call
time-based partitioning, the network is exposed to the geometry of the carotid vessel in the entire cohort of patients, and therefore, is expected to perform better in segmenting the follow-up images for the same cohort of patients. We compared time-based partitioning with the more conventional patient-based partitioning in evaluating our segmentation model. Our result shows that the segmentation model under the time-based partitioning scheme was better at segmenting more complex structures, such as the carotid bifurcation, than under the patient-based partitioning scheme. As the artery is about to divide from the CCA into the ICA and ECA at the bifurcation, the carotid geometry is highly elongated at the carotid bifurcation; additionally, as plaques are prone to develop at the bifurcation, the shape of the LIB here can be highly irregular (e.g.,
Figure 7a). As the bifurcation is a key location to be characterized in carotid atherosclerosis monitoring and assessment, higher segmentation performance in this location by the time-based partitioning scheme suggests that a better workflow for carotid analysis is to train the network using baseline images of the entire cohort and use the trained network for automatic segmentation for follow-up images of the same cohort. This workflow would be particularly cost-effective if serial monitoring is carried out at multiple follow-up time points.
We reported that the DSCs of manual MAB and LIB segmentations at ISD = 1 and 2 mm were statistically different from the DSCs at ISD = 3 mm. Additionally, the standard deviations of DSC and HD of LIB segmentation at 3 mm ISD setting were substantially larger than at 1 and 2 mm, as shown in
Table 1. There are two possible explanations for these observations. The first is related to the presence of calcified and hypo-echoic plaques, as demonstrated in
Figure 10. The presence of calcified plaques leads to acoustic shadowing. The use of compounding imaging mitigated the effect of acoustic shadowing [
44] and in this study, acoustic shadowing mostly appears beyond the arteries and does not substantially obscure the wall and lumen. In acquiring 3DUS images, the sonographer identifies plaques based on Doppler images and attempts to adjust the contrast and level of the ultrasound images before an observer or an algorithm segments boundaries. This adjustment mitigates the difficulty in segmenting hypo-echoic plaques. Despite the use of these strategies, the presence of these two types of plaques is expected to increase observer variability. The availability of 3DUS images allows observers to review adjacent slices, thereby providing visual clues that better equip them to trace out obscured boundaries more reproducibly, as demonstrated in the segmentation of the image shown in
Figure 10d that has a calcified plaque in the near wall and a hypo-echoic plaque in the far wall. However, the observer variability in segmenting the hypo-echoic plaque was substantially larger at ISD = 3 mm than at ISD = 1 and 2 mm. The distal images associated with the ISD of 3, 2 and 1 mm are shown in
Figure 10a–c, respectively, whereas the proximal images associated with the ISD of 1, 2 and 3 mm are shown in
Figure 10e–g, respectively. At ISD = 3 mm, the distal (
Figure 10a) and the proximal (
Figure 10g) provided limited visual clues for LIB segmentation, thereby leading to a decrease in the DSC of the repeated LIB segmentations by approximately 5% compared to when the ISD is set to 1 or 2 mm. The second explanation of the increased variability at ISD = 3 mm is related to the existence of plaques with small longitudinal lengths as shown in
Figure 6. With ISD = 3 mm, there were either one or two resliced planes intersecting these plaques. The variability in the number of planes intersecting them in different segmentation sessions contributed to a large variability in their longitudinal coverage, which in turn reduced the reproducibility in LIB segmentation.
Related to the reflection of the ultrasound beam by calcified plaques discussed above, the literature has described that highly reflective structures, such as calcification, may expand beyond their true boundaries and appear larger in an image [
45]. This effect, commonly referred to as the blooming effect, is less of an issue in our study. The image intensity adjustment based on Doppler imaging by the sonographer ensures the size of a calcified plaque is appropriately displayed in the ultrasound image. The image intensity adjustment also benefited from compounding imaging, which combines images scanned at different angles. Compounding imaging has been shown to improve the definition of plaque surfaces [
44]. As demonstrated in
Figure 10, the observer segmenting MAB and LIB was also helped by the ability to observe adjacent axial images in a 3DUS image.
In this study, we applied the segmentation model we previously developed [
33] to evaluate the proposed time-based partitioning approach. The DSC attained in the current study are
and
for the MAB and LIB in CCA segmentation and
and
for the MAB and LIB in ICA segmentation. The results are similar to those attained for a cohort studied in Jiang et al. [
33] with asymptomatic carotid stenosis involved in a placebo-controlled trial evaluating the effect of atorvastatin (CCA:
for MAB and
for LIB; ICA:
for MAB and
for LIB). The quantification of the intra-observer variability in MAB and LIB segmentations in the current study (
Table 1) affords us an opportunity to assess the performance of the segmentation network in relation to observer variability. The DSC attainable by the segmentation framework is close to the DSC attributable to intra-observer variability for ICA segmentation, whereas the DSC of the segmentation framework for CCA segmentation is 2–3% lower than the DSC associated with intra-observer variability. The ICA was enclosed by a manually identified ROI before the application of the segmentation network, and therefore, localization of the ICA was less of an issue for the segmentation network. On the other hand, the segmentation network is not provided an ROI for CCA segmentation and there is a chance that the CCA is mislocalized as demonstrated in
Figure 7.
Figure 7 also shows that the artery at the bifurcation typically has a more complex shape and the network sometimes fails to segment the entire artery. These two factors contributed to the reduced DSC attained by the segmentation network as compared to the DSC associated with intra-observer variability.
When segmenting a given axial image, the observer has knowledge of the arterial shape on adjacent axial slices. The segmentation network evaluated in this study, however, is a 2D segmentation network processing each slice independently. We applied the 2D network in this study because the major focus of this paper is to evaluate how training and inference should be made to improve segmentation accuracy in series assessment of 3D carotid US images. Further improvement to the segmentation performance may be provided by segmentation networks that consider neighboring slices as well as the current axial slice being segmentation (2.5D network) or 3D segmentation network [
46] and improvement of the time-based partitioning approach in these networks are required to be evaluated in a future study. We did not include an inter-observer reproducibility analysis when assessing the effect of different ISDs on segmentation reproducibility. Chan et al. [
47] performed an extensive study to evaluate the inter-observer reproducibility in measuring VWV from 3DUS and found that the coefficients of variance of VWV measured by five observers were similar. In the current study, the focus was more on assessing the impact of ISD on manual segmentation reliability; with the results from Chan’s study indicating the similarity of VWV reproducibility measured by multiple observers under the same protocol, we decided not to perform an intra-observer analysis, as manual segmentation by multiple observers would require long training and implementation times. Additionally, a central reading center involving a standard grading protocol is typically used for multi-center clinical trials involving imaging endpoints [
48].