Understanding the Role of Connectivity Dynamics of Resting-State Functional MRI in the Diagnosis of Autism Spectrum Disorder: A Comprehensive Study
Abstract
1. Introduction
- The impact of using different atlases, including the automated anatomical labeling (AAL) and Talaraich and Tournoux (TT) atlases.
- The effect of using different preprocessing strategies.
- The effect of using our novel dFC, in comparison to using conventional static FC.
- The dimensionality reduction problem, using two-stage feature selection, with four types of kernels.
- The role of the classification strategy, investigating six different classifiers.
- The ability to highlight the importance of each of the previous choices on the overall performance.
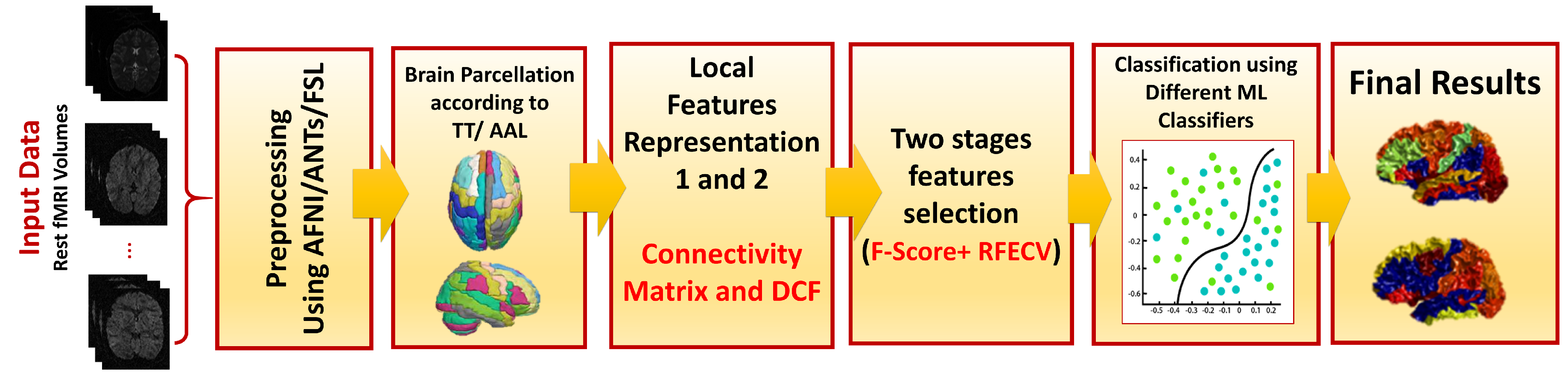
2. Materials and Methods
2.1. Dataset
2.2. Proposed Framework
2.3. Preprocessing
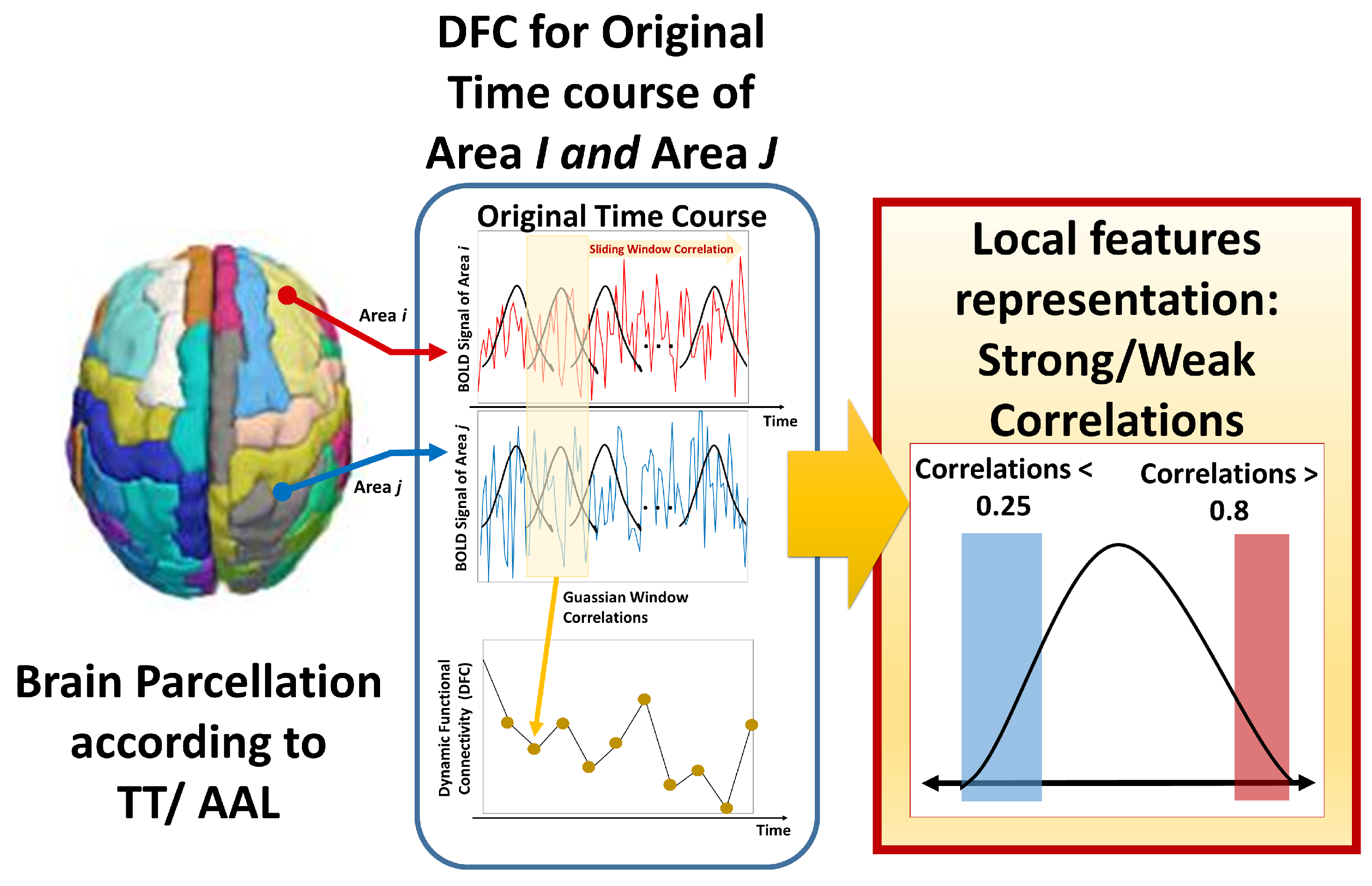
2.4. Feature Representation
2.5. Feature Selection
2.6. Machine Learning
| Algorithm1 Step-by-step rs-fMRI diagnosis algorithm |
|
2.7. Performance Metrics
- Specificity:
- Sensitivity (recall):
- Accuracy:
- Balanced accuracy: average of true positive rate (sensitivity) and true negative rate (specificity).
3. Results
3.1. Significance of Data Representation
3.1.1. Preprocessing Pipeline
3.1.2. Atlas Use
3.1.3. Dynamic Connectivity
3.2. Model Results
Identified Brain Areas
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
| ABIDE | autism brain imaging data exchange |
| MRI | magnetic resonance imaging |
| sMRI | structural MRI |
| fMRI | functional MRI |
| rs-fMRI | resting-state functional MRI |
| ASD | autism spectrum disorder |
| CAD | computer-aided diagnosis |
| ML | machine learning |
References
- Frith, U.; Happé, F. Autism spectrum disorder. Curr. Biol. 2005, 15, R786–R790. [Google Scholar] [CrossRef] [PubMed]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; (DSM-5); American Psychiatric Association: Arlington, VA, USA, 2013. [Google Scholar]
- Casanova, M.F.; El-Baz, A.; Suri, J.S. Autism Imaging and Devices; CRC Press: Boca Raton, FL, USA, 2017. [Google Scholar]
- Ismail, M.M.; Keynton, R.S.; Mostapha, M.M.; ElTanboly, A.H.; Casanova, M.F.; Gimel’farb, G.L.; El-Baz, A. Studying autism spectrum disorder with structural and diffusion magnetic resonance imaging: A survey. Front. Hum. Neurosci. 2016, 10, 211. [Google Scholar] [CrossRef] [PubMed]
- Brieber, S.; Neufang, S.; Bruning, N.; Kamp-Becker, I.; Remschmidt, H.; Herpertz-Dahlmann, B.; Fink, G.R.; Konrad, K. Structural brain abnormalities in adolescents with autism spectrum disorder and patients with attention deficit/hyperactivity disorder. J. Child Psychol. Psychiatry 2007, 48, 1251–1258. [Google Scholar] [CrossRef]
- Dekhil, O.; Ali, M.; Haweel, R.; Elnakib, Y.; Ghazal, M.; Hajjdiab, H.; Fraiwan, L.; Shalaby, A.; Soliman, A.; Mahmoud, A.; et al. A Comprehensive Framework for Differentiating Autism Spectrum Disorder from Neurotypicals by Fusing Structural MRI and Resting State Functional MRI. Semin. Pediatr. Neurol. 2020, 34, 100805. [Google Scholar] [CrossRef] [PubMed]
- Noriuchi, M.; Kikuchi, Y.; Yoshiura, T.; Kira, R.; Shigeto, H.; Hara, T.; Tobimatsu, S.; Kamio, Y. Altered white matter fractional anisotropy and social impairment in children with autism spectrum disorder. Brain Res. 2010, 1362, 141–149. [Google Scholar] [CrossRef]
- ElNakieb, Y.; Ali, M.T.; Elnakib, A.; Shalaby, A.; Soliman, A.; Mahmoud, A.; Ghazal, M.; Barnes, G.N.; El-Baz, A. The Role of Diffusion Tensor MR Imaging (DTI) of the Brain in Diagnosing Autism Spectrum Disorder: Promising Results. Sensors 2021, 21, 8171. [Google Scholar] [CrossRef]
- Cherkassky, V.L.; Kana, R.K.; Keller, T.A.; Just, M.A. Functional connectivity in a baseline resting-state network in autism. Neuroreport 2006, 17, 1687–1690. [Google Scholar] [CrossRef] [PubMed]
- Rausch, A.; Zhang, W.; Haak, K.V.; Mennes, M.; Hermans, E.J.; van Oort, E.; van Wingen, G.; Beckmann, C.F.; Buitelaar, J.K.; Groen, W.B. Altered functional connectivity of the amygdaloid input nuclei in adolescents and young adults with autism spectrum disorder: A resting state fMRI study. Mol. Autism 2016, 7, 13. [Google Scholar] [CrossRef]
- Weng, S.J.; Carrasco, M.; Swartz, J.R.; Wiggins, J.L.; Kurapati, N.; Liberzon, I.; Risi, S.; Lord, C.; Monk, C.S. Neural activation to emotional faces in adolescents with autism spectrum disorders. J. Child Psychol. Psychiatry 2011, 52, 296–305. [Google Scholar] [CrossRef] [PubMed]
- Dekhil, O.; Ismail, M.; Shalaby, A.; Switala, A.; Elmaghraby, A.; Keynton, R.; Gimel’farb, G.; Barnes, G.; El-Baz, A. A novel CAD system for autism diagnosis using structural and functional MRI. In Proceedings of the Biomedical Imaging (ISBI 2017), Melbourne, Australia, 18–21 April 2017. [Google Scholar]
- Kim, J.; Calhoun, V.D.; Shim, E.; Lee, J.H. Deep neural network with weight sparsity control and pre-training extracts hierarchical features and enhances classification performance: Evidence from whole-brain resting-state functional connectivity patterns of schizophrenia. Neuroimage 2016, 124, 127–146. [Google Scholar] [CrossRef]
- Ramzan, F.; Khan, M.U.G.; Rehmat, A.; Iqbal, S.; Saba, T.; Rehman, A.; Mehmood, Z. A deep learning approach for automated diagnosis and multi-class classification of Alzheimer’s disease stages using resting-state fMRI and residual neural networks. J. Med. Syst. 2020, 44, 37. [Google Scholar] [CrossRef] [PubMed]
- Rudie, J.D.; Brown, J.A.; Beck-Pancer, D.; Hernandez, L.M.; Dennis, E.L.; Thompson, P.M.; Bookheimer, S.Y.; Dapretto, M. Altered functional and structural brain network organization in autism. Neuroimage Clin. 2013, 2, 79–94. [Google Scholar] [CrossRef]
- Deshpande, G.; Libero, L.E.; Sreenivasan, K.R.; Deshpande, H.D.; Kana, R.K. Identification of neural connectivity signatures of autism using machine learning. Front. Hum. Neurosci. 2013, 7, 670. [Google Scholar] [CrossRef]
- Itahashi, T.; Yamada, T.; Watanabe, H.; Nakamura, M.; Jimbo, D.; Shioda, S.; Toriizuka, K.; Kato, N.; Hashimoto, R. Altered network topologies and hub organization in adults with autism: A resting-state fMRI study. PLoS ONE 2014, 9, e94115. [Google Scholar] [CrossRef] [PubMed]
- Just, M.A.; Cherkassky, V.L.; Keller, T.A.; Minshew, N.J. Cortical activation and synchronization during sentence comprehension in high-functioning autism: Evidence of underconnectivity. Brain 2004, 127, 1811–1821. [Google Scholar] [CrossRef] [PubMed]
- Alaerts, K.; Swinnen, S.P.; Wenderoth, N. Sex differences in autism: A resting-state fMRI investigation of functional brain connectivity in males and females. Soc. Cogn. Affect. Neurosci. 2016, 11, 1002–1016. [Google Scholar] [CrossRef]
- Tyszka, J.M.; Kennedy, D.P.; Paul, L.K.; Adolphs, R. Largely typical patterns of resting-state functional connectivity in high-functioning adults with autism. Cereb. Cortex 2013, 24, 1894–1905. [Google Scholar] [CrossRef]
- Plitt, M.; Barnes, K.A.; Martin, A. Functional connectivity classification of autism identifies highly predictive brain features but falls short of biomarker standards. Neuroimage Clin. 2015, 7, 359–366. [Google Scholar] [CrossRef]
- Hahamy, A.; Behrmann, M.; Malach, R. The idiosyncratic brain: Distortion of spontaneous connectivity patterns in autism spectrum disorder. Nat. Neurosci. 2015, 18, 302. [Google Scholar] [CrossRef]
- Di Martino, A.; Yan, C.G.; Li, Q.; Denio, E.; Castellanos, F.X.; Alaerts, K.; Anderson, J.S.; Assaf, M.; Bookheimer, S.Y.; Dapretto, M.; et al. The autism brain imaging data exchange: Towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol. Psychiatry 2014, 19, 659. [Google Scholar] [CrossRef]
- Supekar, K.; Uddin, L.Q.; Khouzam, A.; Phillips, J.; Gaillard, W.D.; Kenworthy, L.E.; Yerys, B.E.; Vaidya, C.J.; Menon, V. Brain hyperconnectivity in children with autism and its links to social deficits. Cell Rep. 2013, 5, 738–747. [Google Scholar] [CrossRef] [PubMed]
- Bellman, R. Dynamic programming. Science 1966, 153, 34–37. [Google Scholar] [CrossRef] [PubMed]
- Sen, B.; Borle, N.C.; Greiner, R.; Brown, M.R. A general prediction model for the detection of ADHD and Autism using structural and functional MRI. PLoS ONE 2018, 13, e0194856. [Google Scholar] [CrossRef] [PubMed]
- Haar, S.; Berman, S.; Behrmann, M.; Dinstein, I. Anatomical abnormalities in autism? Cereb. Cortex 2016, 26, 1440–1452. [Google Scholar] [CrossRef]
- Heinsfeld, A.S.; Franco, A.R.; Craddock, R.C.; Buchweitz, A.; Meneguzzi, F. Identification of autism spectrum disorder using deep learning and the ABIDE dataset. Neuroimage Clin. 2018, 17, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Hull, J.V.; Dokovna, L.B.; Jacokes, Z.J.; Torgerson, C.M.; Irimia, A.; Van Horn, J.D. Resting-state functional connectivity in autism spectrum disorders: A review. Front. Psychiatry 2017, 7, 205. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhang, L.; Feng, J.; Lo, C.Y.Z. Tracking the main states of dynamic functional connectivity in resting state. Front. Neurosci. 2019, 13, 685. [Google Scholar] [CrossRef]
- Filippi, M.; Spinelli, E.G.; Cividini, C.; Agosta, F. Resting state dynamic functional connectivity in neurodegenerative conditions: A review of magnetic resonance imaging findings. Front. Neurosci. 2019, 13, 657. [Google Scholar] [CrossRef]
- Hindriks, R.; Adhikari, M.H.; Murayama, Y.; Ganzetti, M.; Mantini, D.; Logothetis, N.K.; Deco, G. Can sliding-window correlations reveal dynamic functional connectivity in resting-state fMRI? Neuroimage 2016, 127, 242–256. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.; Glover, G.H. Time–frequency dynamics of resting-state brain connectivity measured with fMRI. Neuroimage 2010, 50, 81–98. [Google Scholar] [CrossRef]
- Lindquist, M.A.; Xu, Y.; Nebel, M.B.; Caffo, B.S. Evaluating dynamic bivariate correlations in resting-state fMRI: A comparison study and a new approach. NeuroImage 2014, 101, 531–546. [Google Scholar] [CrossRef] [PubMed]
- Craddock, C.; Benhajali, Y.; Chu, C.; Chouinard, F.; Evans, A.; Jakab, A.; Khundrakpam, B.S.; Lewis, J.D.; Li, Q.; Milham, M.; et al. The neuro bureau preprocessing initiative: Open sharing of preprocessed neuroimaging data and derivatives. Front. Neuroinform. 2013, 7, 27. [Google Scholar]
- Craddock, C.; Sikka, S.; Cheung, B.; Khanuja, R.; Ghosh, S.S.; Yan, C.; Li, Q.; Lurie, D.; Vogelstein, J.; Burns, R.; et al. Towards automated analysis of connectomes: The configurable pipeline for the analysis of connectomes (c-pac). Front. Neuroinform. 2013, 42, 10–3389. [Google Scholar]
- Guyon, I.; Weston, J.; Barnhill, S.; Vapnik, V. Gene selection for cancer classification using support vector machines. Mach. Learn. 2002, 46, 389–422. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- ElNakieb, Y.; Ali, M. rs-fMRI-Paper-Code. Available online: https://github.com/ynakieb/rs-fMRI-paper (accessed on 15 December 2022).
- Chen, H.; Duan, X.; Liu, F.; Lu, F.; Ma, X.; Zhang, Y.; Uddin, L.Q.; Chen, H. Multivariate classification of autism spectrum disorder using frequency-specific resting-state functional connectivity—A multi-center study. Prog.-Neuro-Psychopharmacol. Biol. Psychiatry 2016, 64, 1–9. [Google Scholar] [CrossRef]
- Boddaert, N.; Belin, P.; Chabane, N.; Poline, J.B.; Barthélémy, C.; Mouren-Simeoni, M.C.; Brunelle, F.; Samson, Y.; Zilbovicius, M. Perception of complex sounds: Abnormal pattern of cortical activation in autism. Am. J. Psychiatry 2003, 160, 2057–2060. [Google Scholar] [CrossRef] [PubMed]
- Blakemore, S.J. The social brain in adolescence. Nat. Rev. Neurosci. 2008, 9, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Rolls, E.T.; Zhou, Y.; Cheng, W.; Gilson, M.; Deco, G.; Feng, J. Effective connectivity in autism. Autism Res. 2020, 13, 32–44. [Google Scholar] [CrossRef]
- Chen, T.; Chen, Y.; Yuan, M.; Gerstein, M.; Li, T.; Liang, H.; Froehlich, T.; Lu, L. The development of a practical artificial intelligence tool for diagnosing and evaluating autism spectrum disorder: Multicenter study. JMIR Med Informa. 2020, 8, e15767. [Google Scholar] [CrossRef]
- Xu, S.; Li, M.; Yang, C.; Fang, X.; Ye, M.; Wei, L.; Liu, J.; Li, B.; Gan, Y.; Yang, B.; et al. Altered functional connectivity in children with low-function autism spectrum disorders. Front. Neurosci. 2019, 13, 806. [Google Scholar] [CrossRef] [PubMed]
- Philip, R.C.; Dauvermann, M.R.; Whalley, H.C.; Baynham, K.; Lawrie, S.M.; Stanfield, A.C. A systematic review and meta-analysis of the fMRI investigation of autism spectrum disorders. Neurosci. Biobehav. Rev. 2012, 36, 901–942. [Google Scholar] [CrossRef] [PubMed]
- Schulte-Rüther, M.; Greimel, E.; Markowitsch, H.J.; Kamp-Becker, I.; Remschmidt, H.; Fink, G.R.; Piefke, M. Dysfunctions in brain networks supporting empathy: An fMRI study in adults with autism spectrum disorders. Soc. Neurosci. 2011, 6, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Choi, U.S.; Park, S.Y.; Oh, S.H.; Yoon, H.W.; Koh, Y.J.; Im, W.Y.; Park, J.I.; Song, D.H.; Cheon, K.A.; et al. Abnormal activation of the social brain network in children with autism spectrum disorder: An FMRI study. Psychiatry Investig. 2015, 12, 37. [Google Scholar] [CrossRef] [PubMed]
- Rakić, M.; Cabezas, M.; Kushibar, K.; Oliver, A.; Lladó, X. Improving the detection of autism spectrum disorder by combining structural and functional MRI information. Neuroimage Clin. 2020, 25, 102181. [Google Scholar] [CrossRef] [PubMed]
- Abraham, A.; Milham, M.P.; Di Martino, A.; Craddock, R.C.; Samaras, D.; Thirion, B.; Varoquaux, G. Deriving reproducible biomarkers from multi-site resting-state data: An Autism-based example. NeuroImage 2017, 147, 736–745. [Google Scholar] [CrossRef]
- Guo, X.; Dominick, K.C.; Minai, A.A.; Li, H.; Erickson, C.A.; Lu, L.J. Diagnosing autism spectrum disorder from brain resting-state functional connectivity patterns using a deep neural network with a novel feature selection method. Front. Neurosci. 2017, 11, 460. [Google Scholar] [CrossRef]
- Kam, T.E.; Suk, H.I.; Lee, S.W. Multiple functional networks modeling for autism spectrum disorder diagnosis. Hum. Brain Mapp. 2017, 38, 5804–5821. [Google Scholar] [CrossRef]
- Sadeghi, M.; Khosrowabadi, R.; Bakouie, F.; Mahdavi, H.; Eslahchi, C.; Pouretemad, H. Screening of autism based on task-free fmri using graph theoretical approach. Psychiatry Res. Neuroimaging 2017, 263, 48–56. [Google Scholar] [CrossRef]
- Spera, G.; Retico, A.; Bosco, P.; Ferrari, E.; Palumbo, L.; Oliva, P.; Muratori, F.; Calderoni, S. Evaluation of altered functional connections in male children with autism spectrum disorders on multiple-site data optimized with machine learning. Front. Psychiatry 2019, 10, 620. [Google Scholar] [CrossRef]
- Tang, L.; Mostafa, S.; Liao, B.; Wu, F.X. A network clustering based feature selection strategy for classifying autism spectrum disorder. BMC Med. Genom. 2019, 12, 153. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Wu, F.X.; Hayrat, R.; Liu, J. AIMAFE: Autism spectrum disorder identification with multi-atlas deep feature representation and ensemble learning. J. Neurosci. Methods 2020, 343, 108840. [Google Scholar] [CrossRef]
- Subah, F.Z.; Deb, K.; Dhar, P.K.; Koshiba, T. A deep learning approach to predict autism spectrum disorder using multisite resting-state fMRI. Appl. Sci. 2021, 11, 3636. [Google Scholar] [CrossRef]
- Al-Hiyali, M.I.; Yahya, N.; Faye, I.; Khan, Z.; Alsaih, K. Classification of BOLD FMRI signals using wavelet transform and transfer learning for detection of autism spectrum disorder. In Proceedings of the 2020 IEEE-EMBS Conference on Biomedical Engineering and Sciences (IECBES), Langkawi Island, Malaysia, 1–3 March 2021; pp. 94–98. [Google Scholar]
- Yin, W.; Mostafa, S.; Wu, F.X. Diagnosis of autism spectrum disorder based on functional brain networks with deep learning. J. Comput. Biol. 2021, 28, 146–165. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.; Wang, G.; Cao, L.; Qiao, L.; Liu, M. Multi-Scale Graph Representation Learning for Autism Identification With Functional MRI. Front. Neuroinform. 2022, 15, 802305. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhang, N.; Schrader, P. A study of brain networks for autism spectrum disorder classification using resting-state functional connectivity. Mach. Learn. Appl. 2022, 8, 100290. [Google Scholar] [CrossRef]
- Ding, J.; Wang, L.; Yu, L.; Xue, M.; Mei, X.; Wang, X. Low-rank Domain Adaptive Method with Inter-class difference Constraint for Multi-site Autism Spectrum Disorder Identification. In Proceedings of the 2022 7th International Conference on Computational Intelligence and Applications (ICCIA), Nanjing, China, 24–26 June 2022; pp. 237–242. [Google Scholar]

| ASD Group (n = 408) | TD Group (n = 476) | |||
|---|---|---|---|---|
| M = 358, F = 50 | M = 388, F = 88 | |||
| AGE | FIQ | AGE | FIQ | |
| count | 408 | 379 | 476 | 442 |
| mean | 17.69 | 106.19 | 16.79 | 111.28 |
| std | 8.93 | 17.01 | 7.35 | 12.48 |
| min | 7 | 41 | 6.47 | 73 |
| max | 64 | 148 | 56.2 | 146 |
| sum_sq | df | F | PR (>F) | |
|---|---|---|---|---|
| C(Feat, Sum) | 0.941852 | 1.0 | 163.004715 | |
| C(Strat, Sum) | 0.177957 | 3.0 | 10.266230 | |
| C(Atls, Sum) | 0.149164 | 1.0 | 25.815521 | |
| C(Feat, Sum):C(Strat, Sum) | 0.117433 | 3.0 | 6.774646 | |
| C(Feat, Sum):C(Atls, Sum) | 0.013784 | 1.0 | 2.385576 | |
| C(Strat, Sum):C(Atls, Sum) | 0.034962 | 3.0 | 2.016924 | |
| C(Feat, Sum):C(Strat, Sum):C(Atls, Sum) | 0.084563 | 3.0 | 4.878417 |
| sum_sq | df | F | PR (>F) | |
|---|---|---|---|---|
| C(Feat, Sum) | 0.936316 | 1.0 | 160.682470 | |
| C(Filt, Sum) | 0.179172 | 3.0 | 10.249322 | |
| C(Atls, Sum) | 0.148105 | 1.0 | 25.416432 | |
| C(Feat, Sum):C(Filt, Sum) | 0.117208 | 3.0 | 6.704737 |
| Metric | Accuracy | Sensitivity | Specificity | Balanced Accuracy |
|---|---|---|---|---|
| 5-fold value | 0.988 ± 0.004 | 0.987 ± 0.008 | 0.989 ± 0.007 | 0.988 ± 0.004 |
| Index | First Selected Features | Top Frequent Region Names | Frequency |
|---|---|---|---|
| 1 | Precentral_L__Rolandic_Oper_R_wk | Temporal_Sup_L | 24 |
| 2 | Precentral_L__Supp_Motor_Area_L_st | Postcentral_R | 23 |
| 3 | Precentral_L__Fusiform_L_wk | Frontal_Sup_Medial_R | 22 |
| 4 | Precentral_L__Parietal_Inf_L_wk | Precuneus_R | 22 |
| 5 | Precentral_L__SupraMarginal_R_st | SupraMarginal_R | 21 |
| 6 | Precentral_L__Precuneus_R_wk | Frontal_Sup_R | 21 |
| 7 | Precentral_L__Temporal_Sup_L_wk | Cingulum_Ant_R | 21 |
| 8 | Precentral_L__Temporal_Pole_Sup_R_wk | Supp_Motor_Area_L | 21 |
| 9 | Precentral_L__Cerebelum_Crus2_R_wk | Angular_R | 21 |
| 10 | Precentral_R__Frontal_Inf_Tri_L_wk | Hippocampus_R | 21 |
| Article | Used Classifier | Achieved Accuracy |
|---|---|---|
| Abraham et al., 2017 [50] | SVM | 67.0% |
| Guo et al., 2017 [51] | Deep neural networks with feature selection (DNN-FS) | 86.4% |
| Kam et al., 2017 [52] | Discriminative restricted Boltzmann machines (DRBM) | 80.8% |
| Sadeghi et al., 2017 [53] | SVM | 92% |
| Spera et al., 2019 [54] | SVM | 71.0% |
| Tang et al., 2019 [55] | SVM | 62.6% |
| Wang et al., 2020 [56] | MLP and a voting strategy | 74.5% |
| Rakić et al., 2020 [49] | Ensemble of classifiers | 85.0% |
| Subah et al., 2021 [57] | DNN | 87.0% |
| Al-Hiyali et al., 2021 [58] | SVM, K-nearest neighbors (KNN) | 85.9% |
| Yin et al., 2021 [59] | Autoencoders, CNN, DNN | 79.2% |
| Chu et al., 2022 [60] | Multi-scale graph convolutional network (GCN) | 79.5% |
| Yang et al., 2022 [61] | LR, SVM, DNN, supervised learning classifier | 69.4% |
| Ding et al., 2022 [62] | low-rank domain adaptive method with inter-class difference constraint | 75.5% |
| Proposed Work | Best: LSVM | 98.8% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
ElNakieb, Y.; Ali, M.T.; Elnakib, A.; Shalaby, A.; Mahmoud, A.; Soliman, A.; Barnes, G.N.; El-Baz, A. Understanding the Role of Connectivity Dynamics of Resting-State Functional MRI in the Diagnosis of Autism Spectrum Disorder: A Comprehensive Study. Bioengineering 2023, 10, 56. https://doi.org/10.3390/bioengineering10010056
ElNakieb Y, Ali MT, Elnakib A, Shalaby A, Mahmoud A, Soliman A, Barnes GN, El-Baz A. Understanding the Role of Connectivity Dynamics of Resting-State Functional MRI in the Diagnosis of Autism Spectrum Disorder: A Comprehensive Study. Bioengineering. 2023; 10(1):56. https://doi.org/10.3390/bioengineering10010056
Chicago/Turabian StyleElNakieb, Yaser, Mohamed T. Ali, Ahmed Elnakib, Ahmed Shalaby, Ali Mahmoud, Ahmed Soliman, Gregory Neal Barnes, and Ayman El-Baz. 2023. "Understanding the Role of Connectivity Dynamics of Resting-State Functional MRI in the Diagnosis of Autism Spectrum Disorder: A Comprehensive Study" Bioengineering 10, no. 1: 56. https://doi.org/10.3390/bioengineering10010056
APA StyleElNakieb, Y., Ali, M. T., Elnakib, A., Shalaby, A., Mahmoud, A., Soliman, A., Barnes, G. N., & El-Baz, A. (2023). Understanding the Role of Connectivity Dynamics of Resting-State Functional MRI in the Diagnosis of Autism Spectrum Disorder: A Comprehensive Study. Bioengineering, 10(1), 56. https://doi.org/10.3390/bioengineering10010056










