Mice Placental ECM Components May Provide A Three-Dimensional Placental Microenvironment
Abstract
1. Introduction
2. Material and Methods
2.1. Decellularization Process
2.2. Mass Spectrometry Samples
2.3. Data Collection and Bioinformatic Analysis
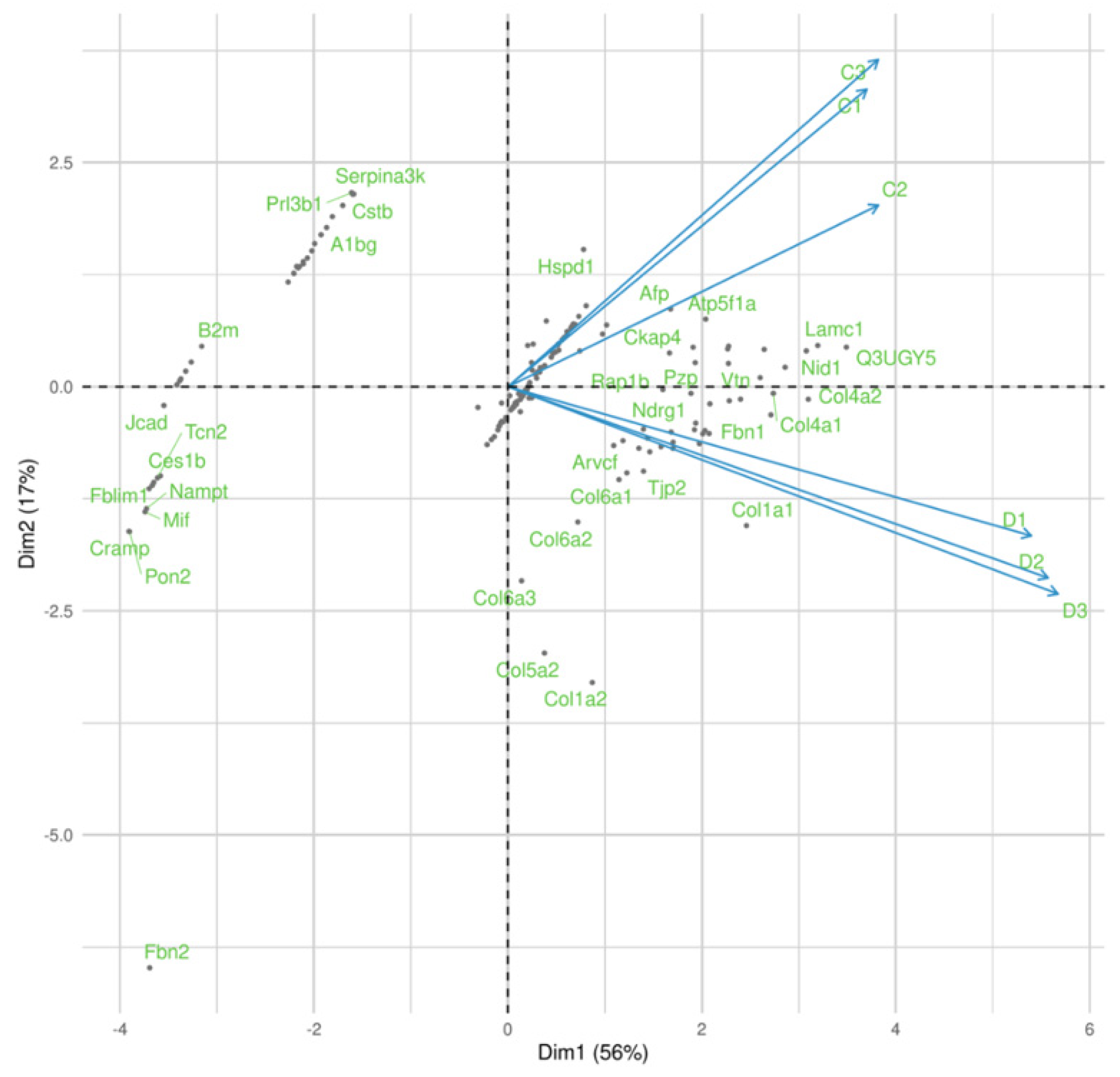
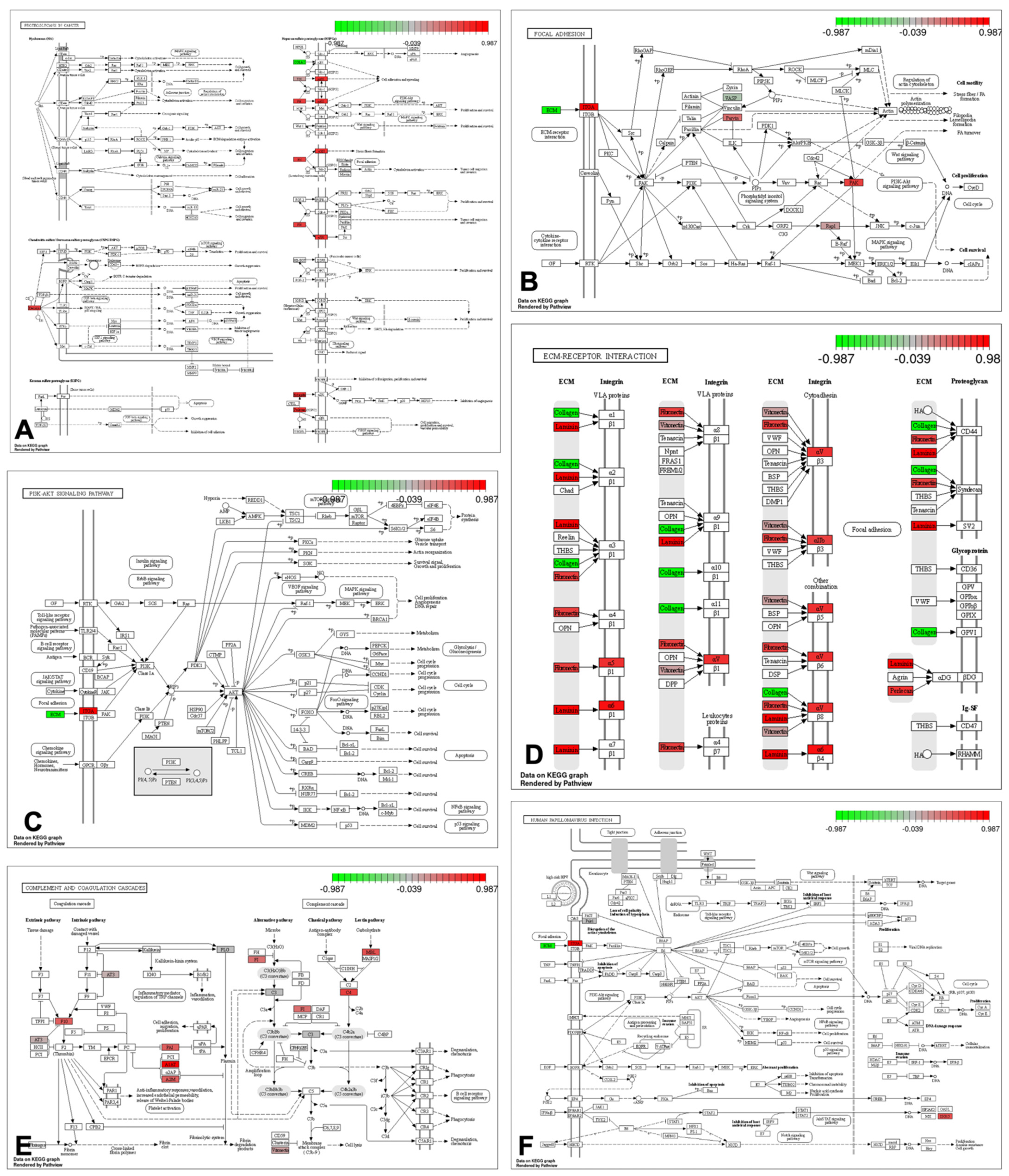
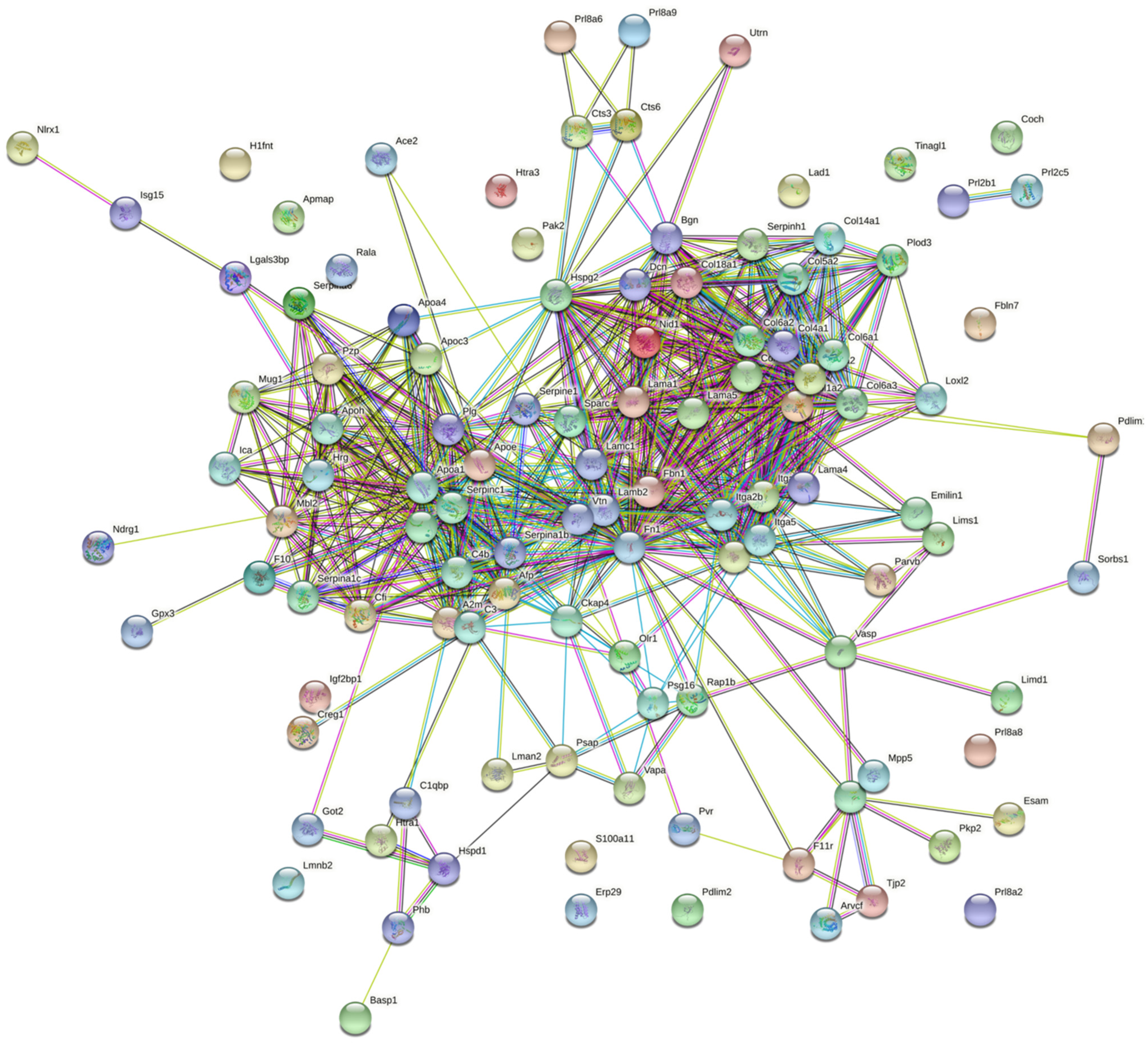
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brosens, I.; Pijnenborg, R.; Vercruysse, L.; Romero, R. The “Great Obstetrical Syndromes” Are Associated with Disorders of Deep Placentation. Am. J. Obstet. Gynecol. 2011, 204, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Schmid, B.P.; Honegger, P.; Kucera, P. Embryonic and Fetal Development: Fundamental Research. Reprod. Toxicol. 1993, 7, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Zohni, K.M.; Gat, I.; Librach, C. Recurrent Implantation Failure: A Comprehensive Review. Minerva Ginecol. 2016, 68, 653–667. [Google Scholar] [PubMed]
- Cherubini, M.; Erickson, S.; Haase, K. Modelling the Human Placental Interface in Vitro—A Review. Micromachines 2021, 12, 884. [Google Scholar] [CrossRef] [PubMed]
- Carter, A.M. Evolution of the Placenta and Fetal Membranes Seen in the Light of Molecular Phylogenetics. Placenta 2001, 22, 800–807. [Google Scholar] [CrossRef]
- Carter, A.M. Animal Models of Human Placentation—A Review. Placenta 2007, 28, S41–S47. [Google Scholar] [CrossRef]
- Hemberger, M.; Hanna, C.W.; Dean, W. Mechanisms of Early Placental Development in Mouse and Humans. Nat. Rev. Genet. 2020, 21, 27–43. [Google Scholar] [CrossRef]
- Moffett, A.; Loke, C. Immunology of Placentation in Eutherian Mammals. Nat. Rev. Immunol. 2006, 6, 584–594. [Google Scholar] [CrossRef]
- Leiser, R.; Kaufmann, P. Placental Structure: In a Comparative Aspect. Exp. Clin. Endocrinol. 1994, 102, 122–134. [Google Scholar] [CrossRef]
- Li, Z.; Kurosawa, O.; Iwata, H. A Novel Human Placental Barrier Model Based on Trophoblast Stem Cells Derived from Human Induced Pluripotent Stem Cells. Tissue Eng.–Part A 2020, 26, 780–791. [Google Scholar] [CrossRef]
- Kuo, C.Y.; Guo, T.; Cabrera-Luque, J.; Arumugasaamy, N.; Bracaglia, L.; Garcia-Vivas, A.; Santoro, M.; Baker, H.; Fisher, J.; Kim, P. Placental Basement Membrane Proteins Are Required for Effective Cytotrophoblast Invasion in a Three-Dimensional Bioprinted Placenta Model. J. Biomed. Mater. Res.–Part A 2018, 106, 1476–1487. [Google Scholar] [CrossRef] [PubMed]
- Carter, A.M. Animal Models of Human Pregnancy and Placentation: Alternatives to the Mouse. Reproduction 2020, 160, R129–R143. [Google Scholar] [CrossRef] [PubMed]
- Arora, N.; Sadovsky, Y.; Dermody, T.S.; Coyne, C.B. Microbial Vertical Transmission during Human Pregnancy. Cell Host Microbe 2017, 21, 561–567. [Google Scholar] [CrossRef]
- Megli, C.J.; Coyne, C.B. Infections at the Maternal–Fetal Interface: An Overview of Pathogenesis and Defence. Nat. Rev. Microbiol. 2022, 20, 67–82. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, M.A.; Zhou, J.; Franz, A.W.E.; Schust, D.J. Modeling the Human Placenta to Investigate Viral Infections during Pregnancy. Front. Virol. 2022, 2, 831754. [Google Scholar] [CrossRef]
- Roberts, D.J.; Edlow, A.G.; Romero, R.J.; Coyne, C.B.; Ting, D.T.; Hornick, J.L.; Zaki, S.R.; Das Adhikari, U.; Serghides, L.; Gaw, S.L.; et al. A Standardized Definition of Placental Infection by SARS-CoV-2, a Consensus Statement from the National Institutes of Health/Eunice Kennedy Shriver National Institute of Child Health and Human Development SARS-CoV-2 Placental Infection Workshop. Am. J. Obstet. Gynecol. 2021, 225, 593.e1–593.e9. [Google Scholar] [CrossRef]
- Jaklin, M.; Zhang, J.D.; Barrow, P.; Ebeling, M.; Clemann, N.; Leist, M.; Kustermann, S. Focus on Germ-Layer Markers: A Human Stem Cell-Based Model for in Vitro Teratogenicity Testing. Reprod. Toxicol. 2020, 98, 286–298. [Google Scholar] [CrossRef]
- Fliedel, L.; Alhareth, K.; Mignet, N.; Fournier, T.; Andrieux, K. Placental Models for Evaluation of Nanocarriers as Drug Delivery Systems for Pregnancy Associated Disorders. Biomedicines 2022, 10, 936. [Google Scholar] [CrossRef]
- Li, M.; Gong, J.; Gao, L.; Zou, T.; Kang, J.; Xu, H. Advanced Human Developmental Toxicity and Teratogenicity Assessment Using Human Organoid Models. Ecotoxicol. Environ. Saf. 2022, 235, 113429. [Google Scholar] [CrossRef]
- Tutar, R.; Çelebi-Saltik, B. Modeling of Artificial 3D Human Placenta. Cells Tissues Organs 2022, 211, 527–536. [Google Scholar] [CrossRef]
- Almeida, G.H.D.; Iglesia, R.P.; Araujo, M.S.; Carreira, A.C.O.; Dos Santos, E.X.; Calomeno, C.V.A.Q.; Miglino, M.A. Uterine Tissue Engineering: Where We Stand and the Challenges Ahead. Tissue Eng. Part B Rev. 2022, 28, 861–890. [Google Scholar] [CrossRef]
- Turco, M.Y.; Gardner, L.; Kay, R.G.; Hamilton, R.S.; Prater, M.; Hollinshead, M.S.; McWhinnie, A.; Esposito, L.; Fernando, R.; Skelton, H.; et al. Trophoblast Organoids as a Model for Maternal–Fetal Interactions during Human Placentation. Nature 2018, 564, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, M.A.; Zhao, X.; Fernando, R.C.; Gardner, L.; Perez-Garcia, V.; Li, Q.; Marsh, S.G.E.; Hamilton, R.; Moffett, A.; Turco, M.Y. Characterization of Primary Models of Human Trophoblast. Development 2021, 148, dev199749. [Google Scholar] [CrossRef]
- Haider, S.; Meinhardt, G.; Saleh, L.; Kunihs, V.; Gamperl, M.; Kaindl, U.; Ellinger, A.; Burkard, T.R.; Fiala, C.; Pollheimer, J.; et al. Self-Renewing Trophoblast Organoids Recapitulate the Developmental Program of the Early Human Placenta. Stem Cell Rep. 2018, 11, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Bischof, P.; Campana, A. Trophoblast Differentiation and Invasion: Its Significance for Human Embryo Implantation. Early Pregnancy 1997, 3, 81–95. [Google Scholar]
- Majewska, M.; Lipka, A.; Paukszto, L.; Jastrzebski, J.P.; Myszczynski, K.; Gowkielewicz, M.; Jozwik, M.; Majewski, M.K. Transcriptome Profile of the Human Placenta. Funct. Integr. Genom. 2017, 17, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Zhao, K.; Jiang, P.; Lu, Z.-X.; Wang, J.; Murray, J.C.; Xing, Y. Transcriptome Landscape of the Human Placenta. BMC Genom. 2012, 13, 115. [Google Scholar] [CrossRef] [PubMed]
- Abdulghani, M.; Song, G.; Kaur, H.; Walley, J.W.; Tuteja, G. Comparative Analysis of the Transcriptome and Proteome during Mouse Placental Development. J. Proteome Res. 2019, 18, 2088–2099. [Google Scholar] [CrossRef]
- Abbas, Y.; Carnicer-Lombarte, A.; Gardner, L.; Thomas, J.; Brosens, J.J.; Moffett, A.; Sharkey, A.M.; Franze, K.; Burton, G.J.; Oyen, M.L. Tissue Stiffness at the Human Maternal-Fetal Interface. Hum. Reprod. 2019, 34, 1999–2008. [Google Scholar] [CrossRef] [PubMed]
- Benedictus, L.; Thomas, A.J.; Jorritsma, R.; Davies, C.J.; Koets, A.P. Two-Way Calf to Dam Major Histocompatibility Class I Compatibility Increases Risk for Retained Placenta in Cattle. Am. J. Reprod. Immunol. 2012, 67, 224–230. [Google Scholar] [CrossRef]
- Barreto, R.S.N.; Romagnolli, P.; Fratini, P.; Mess, A.M.; Miglino, M.A. Mouse Placental Scaffolds: A Three-Dimensional Environment Model for Recellularization. J. Tissue Eng. 2019, 10, 2041731419867962. [Google Scholar] [CrossRef] [PubMed]
- Pfarrer, C.D. Characterization of the Bovine Placenta by Cytoskeleton, Integrin Receptors, and Extracellular Matrix. Methods Mol. Med. 2006, 121, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Hedrick, V.E.; LaLand, M.N.; Nakayasu, E.S.; Paul, L.N. Digestion, Purification, and Enrichment of Protein Samples for Mass Spectrometry. Curr. Protoc. Chem. Biol. 2015, 7, 201–222. [Google Scholar] [CrossRef] [PubMed]
- Matias, G.S.S.; Barreto, R.d.S.N.; Carreira, A.C.O.; Nishiyama-Junior, M.Y.; Ferreira, C.R.; Fratini, P.; Miglino, M.A. Proteomic Profile of Extracellular Matrix from Native and Decellularized Chorionic Canine Placenta. J. Proteom. 2022, 256, 104497. [Google Scholar] [CrossRef]
- Barreto, R.d.S.N.; Matias, G.d.S.S.; Junior, M.Y.N.; Carreira, A.C.O.; Miglino, M.A. ECM Proteins Involved in Cell Migration and Vessel Formation Compromise Bovine Cloned Placentation. Theriogenology 2022, 188, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.J.; Washburn, M.P.; Florens, L. Probing the Sensitivity of the Orbitrap Lumos Mass Spectrometer Using a Standard Reference Protein in a Complex Background. J. Proteome Res. 2018, 17, 3586–3592. [Google Scholar] [CrossRef]
- Piehowski, P.D.; Petyuk, V.A.; Orton, D.J.; Xie, F.; Moore, R.J.; Ramirez-Restrepo, M.; Engel, A.; Lieberman, A.P.; Albin, R.L.; Camp, D.G.; et al. Sources of Technical Variability in Quantitative LC-MS Proteomics: Human Brain Tissue Sample Analysis. J. Proteome Res. 2013, 12, 3586–3592. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant Enables High Peptide Identification Rates, Individualized p.p.b.-Range Mass Accuracies and Proteome-Wide Protein Quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. ClusterProfiler: An R Package for Comparing Biological Themes among Gene Clusters. Omi. A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor Package for Pathway-Based Data Integration and Visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Soufan, O.; Ewald, J.; Hancock, R.E.W.; Basu, N.; Xia, J. NetworkAnalyst 3.0: A Visual Analytics Platform for Comprehensive Gene Expression Profiling and Meta-Analysis. Nucleic Acids Res. 2019, 47, W234–W241. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.W.; Lai, Z.Z.; Yang, H.L.; Yang, S.L.; Wang, C.J.; Ao, D.; Ruan, L.Y.; Shen, H.H.; Zhou, W.J.; Mei, J.; et al. Collagen at the Maternal-Fetal Interface in Human Pregnancy. Int. J. Biol. Sci. 2020, 16, 2220–2234. [Google Scholar] [CrossRef] [PubMed]
- Favaron, P.O.; Borghesi, J.; Mess, A.M.; Castelucci, P.; Schiavo Matias, G.d.S.; Barreto, R.D.S.N.; Miglino, M.A. Establishment of 3-Dimensional Scaffolds from Hemochorial Placentas. Placenta 2019, 81, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Katz, S.G. Extracellular Breakdown of Collagen by Mice Decidual Cells. A Cytochemical and Ultrastructural Study. Biocell 2005, 29, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Amenta, P.S.; Gay, S.; Vaheri, A.; Martinez-Hernandez, A. The Extracellular Matrix Is an Integrated Unit: Ultrastructural Localization of Collagen Types I, III, IV, V, VI, Fibronectin, and Laminin in Human Term Placenta. Coll. Relat. Res. 1986, 6, 125–152. [Google Scholar] [CrossRef]
- Barreto, R.d.S.N.; Romagnolli, P.; Mess, A.M.; Miglino, M.A. Decellularized Bovine Cotyledons May Serve as Biological Scaffolds with Preserved Vascular Arrangement. J. Tissue Eng. Regen. Med. 2018, 12, e1880–e1888. [Google Scholar] [CrossRef]
- Matias, G.d.S.S.; Carreira, A.C.O.; Batista, V.F.; de Carvalho, H.J.C.; Miglino, M.A. Paula Fratini In Vivo Biocompatibility Analysis of the Recellularized Canine Tracheal Scaffolds with Canine Epithelial and Endothelial Progenitor Cells. Bioengineered 2021, 13, 3551–3565. [Google Scholar] [CrossRef]
- Matias, G.d.S.S.; Rigoglio, N.N.; Carreira, A.C.O.; Romagnolli, P.; Barreto, R.d.S.N.; Mess, A.M.; Miglino, M.A.; Fratini, P. Optimization of Canine Placenta Decellularization: An Alternative Source of Biological Scaffolds for Regenerative Medicine. Cells Tissues Organs 2018, 205, 217–225. [Google Scholar] [CrossRef]
- Kiyozumi, D.; Nakano, I.; Sato-Nishiuchi, R.; Tanaka, S.; Sekiguchi, K. Laminin Is the ECM Niche for Trophoblast Stem Cells. Life Sci. Alliance 2020, 3, e201900515. [Google Scholar] [CrossRef]
- Seguin, L.; Desgrosellier, J.S.; Weis, S.M.; Cheresh, D.A. Integrins and Cancer: Regulators of Cancer Stemness, Metastasis, and Drug Resistance. Trends Cell Biol. 2015, 25, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Weitzner, O.; Seraya-Bareket, C.; Biron-Shental, T.; Fishamn, A.; Yagur, Y.; Tzadikevitch-Geffen, K.; Farladansky-Gershnabel, S.; Kidron, D.; Ellis, M.; Ashur-Fabian, O. Enhanced Expression of AVβ3 Integrin in Villus and Extravillous Trophoblasts of Placenta Accreta. Arch. Gynecol. Obstet. 2021, 303, 1175–1183. [Google Scholar] [CrossRef]
- Nguyen, S.L.; Ahn, S.H.; Greenberg, J.W.; Collaer, B.W.; Agnew, D.W.; Arora, R.; Petroff, M.G. Integrins Mediate Placental Extracellular Vesicle Trafficking to Lung and Liver in Vivo. Sci. Rep. 2021, 11, 4217. [Google Scholar] [CrossRef] [PubMed]
- Ruoslahti, E.; Reed, J.C. Anchorage Dependence, Integrins, and Apoptosis. Cell 1994, 77, 477–478. [Google Scholar] [CrossRef] [PubMed]
- Humphries, J.D.; Chastney, M.R.; Askari, J.A.; Humphries, M.J. Signal Transduction via Integrin Adhesion Complexes. Curr. Opin. Cell Biol. 2019, 56, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Herdl, S.; Huebner, H.; Volkert, G.; Marek, I.; Menendez-Castro, C.; Noegel, S.C.; Ruebner, M.; Rascher, W.; Hartner, A.; Fahlbusch, F.B. Integrin A8 Is Abundant in Human, Rat, and Mouse Trophoblasts. Reprod. Sci. 2017, 24, 1426–1437. [Google Scholar] [CrossRef]
- Hill, A.B.T.; Alves, A.A.S.; Barreto, R.d.S.N.; Bressan, F.F.; Miglino, M.A.; Garcia, J.M. Placental Scaffolds Have the Ability to Support Adipose-Derived Cells Differentiation into Osteogenic and Chondrogenic Lineages. J. Tissue Eng. Regen. Med. 2020, 14, 1161–1172. [Google Scholar] [CrossRef]
- Yañez, M.J.; Leiva, A. Human Placental Intracellular Cholesterol Transport: A Focus on Lysosomal and Mitochondrial Dysfunction and Oxidative Stress. Antioxidants 2022, 11, 500. [Google Scholar] [CrossRef]
- Friedl, P.; Bröcker, E.B. The Biology of Cell Locomotion within Three-Dimensional Extracellular Matrix. Cell. Mol. Life Sci. 2000, 57, 41–64. [Google Scholar] [CrossRef]
- Chapman, H.A. Plasminogen Activators, Integrins, and the Coordinated Regulation of Cell Adhesion and Migration. Curr. Opin. Cell Biol. 1997, 9, 714–724. [Google Scholar] [CrossRef]
- Smith, L.R.; Pichika, R.; Meza, R.C.; Gillies, A.R.; Baliki, M.N.; Chambers, H.G.; Lieber, R.L. Contribution of Extraellular Matrix Components to the Stiffness of Skeletal Muscle Contractures in Patients with Cerebral Palsy. Connect. Tissue Res. 2021, 62, 287–298. [Google Scholar] [CrossRef] [PubMed]
- López, B.; Querejeta, R.; González, A.; Larman, M.; Díez, J. Collagen Cross-Linking but Not Collagen Amount Associates with Elevated Filling Pressures in Hypertensive Patients with Stage C Heart Failure: Potential Role of Lysyl Oxidase. Hypertension 2012, 60, 677–683. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.N.; Brinkworth, A.J. Manipulation of Focal Adhesion Signaling by Pathogenic Microbes. Int. J. Mol. Sci. 2021, 22, 1358. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.J.; Li, D.Z.; Lin, B.Y.; Geng, L.; Yang, Z.; Zheng, S.S. SNHG16 Promotes Hepatocellular Carcinoma Development via Activating ECM Receptor Interaction Pathway. Hepatobiliary Pancreat. Dis. Int. 2022, 21, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Iozzo, R.V.; Sanderson, R.D. Proteoglycans in Cancer Biology, Tumour Microenvironment and Angiogenesis. J. Cell. Mol. Med. 2011, 15, 1013–1031. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Chen, L.; Zhuang, Y.; Han, Y.; Tang, W.; Xia, F. Fibronectin 1 Inhibits the Apoptosis of Human Trophoblasts by Activating the PI3K/Akt Signaling Pathway. Int. J. Mol. Med. 2020, 46, 1908–1922. [Google Scholar] [CrossRef]
- Xu, Y.; Sui, L.; Qiu, B.; Yin, X.; Liu, J.; Zhang, X. ANXA4 Promotes Trophoblast Invasion via the PI3K/Akt/ENOS Pathway in Preeclampsia. Am. J. Physiol.—Cell Physiol. 2019, 316, C481–C491. [Google Scholar] [CrossRef]
- Ducat, A.; Vargas, A.; Doridot, L.; Bagattin, A.; Lerner, J.; Vilotte, J.L.; Buffat, C.; Pontoglio, M.; Miralles, F.; Vaiman, D. Low-Dose Aspirin Protective Effects Are Correlated with Deregulation of HNF Factor Expression in the Preeclamptic Placentas from Mice and Humans. Cell Death Discov. 2019, 5, 94. [Google Scholar] [CrossRef]
- Pinheiro, M.B.; Gomes, K.B.; Dusse, L.M.S. Fibrinolytic System in Preeclampsia. Clin. Chim. Acta 2013, 416, 67–71. [Google Scholar] [CrossRef]
- Chen, J.; Khalil, R.A. Matrix Metalloproteinases in Normal Pregnancy and Preeclampsia. Prog. Mol. Biol. Transl. Sci. 2017, 148, 87–165. [Google Scholar] [CrossRef]
- Qu, H.; Khalil, R.A. Vascular Mechanisms and Molecular Targets in Hypertensive Pregnancy and Preeclampsia. Am. J. Physiol.—Heart Circ. Physiol. 2020, 319, H661–H681. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.; Gumina, D.; McPeak, K.; Moldovan, R.; Post, M.D.; Su, E.J. Human Placental Villous Stromal Extracellular Matrix Regulates Fetoplacental Angiogenesis in Severe Fetal Growth Restriction. Clin. Sci. 2021, 135, 1127–1143. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.H.; Murphy, H.A.; Chapman, H.; George, E.M. Syncytialization Alters the Extracellular Matrix and Barrier Function of Placental Trophoblasts. Am. J. Physiol.—Cell Physiol. 2021, 321, C694–C703. [Google Scholar] [CrossRef] [PubMed]
- Cartwright, J.E.; Keogh, R.J.; Patot, M.C.T. Hypoxia and Placental Remodelling. Adv. Exp. Med. Biol. 2007, 618, 113–126. [Google Scholar] [CrossRef]
- Barreto, R.D.S.N.; Miglino, M.A.; Meirelles, F.V.; Visintin, J.A.; da Silva, S.M.; Burioli, K.C.; da Fonseca, R.; Bertan, C.; de Assis Neto, A.C.; Pereira, F.T.V. Caracterização Da Fusão Caruncular Em Gestações Naturais e de Conceptos Bovinos Clonados. Pesqui. Veterinária Bras. 2009, 29, 779–787. [Google Scholar] [CrossRef]
- Mess, A.; Carter, A.M. Evolution of the Placenta during the Early Radiation of Placental Mammals. Comp. Biochem. Physiol.—A Mol. Integr. Physiol. 2007, 148, 769–779. [Google Scholar] [CrossRef]
- Aplin, J.D. Developmental Cell Biology of Human Villous Trophoblast: Current Research Problems. Int. J. Dev. Biol. 2010, 54, 323–329. [Google Scholar] [CrossRef]
- Carter, A.M. Factors Affecting Gas Transfer across the Placenta and the Oxygen Supply to the Fetus. J. Dev. Physiol. 1989, 12, 305–322. [Google Scholar]
- Barreto, R.S.N.; Romagnolli, P.; Cereta, A.D.; Coimbra-Campos, L.M.C.; Birbrair, A.; Miglino, M.A. Pericytes in the Placenta: Role in Placental Development and Homeostasis. In Advances in Experimental Medicine and Biology; Springer: Berlin/Heidelberg, Germany, 2019; Volume 1122, pp. 125–151. [Google Scholar]
- Fallon, B.P.; Mychaliska, G.B. Development of an Artificial Placenta for Support of Premature Infants: Narrative Review of the History, Recent Milestones, and Future Innovation. Transl. Pediatr. 2021, 10, 1470–1485. [Google Scholar] [CrossRef]
- Myllyharju, J.; Kivirikko, K.I. Collagens and Collagen-Related Diseases. Ann. Med. 2001, 33, 7–21. [Google Scholar] [CrossRef]
- Ben Hamouda, S.; Vargas, A.; Boivin, R.; Miglino, M.A.; da Palma, R.K.; Lavoie, J.-P. Recellularization of Bronchial Extracellular Matrix With Primary Bronchial Smooth Muscle Cells. J. Equine Vet. Sci. 2021, 96, 103313. [Google Scholar] [CrossRef] [PubMed]
- Evangelista-Leite, D.; Oliveira Carreira, A.C.; Gilpin, S.E.; Miglino, M.A. Protective Effects of Extracellular Matrix Derived Hydrogels in Idiopathic Pulmonary Fibrosis. Tissue Eng. Part B Rev. 2021, 28, 517–530. [Google Scholar] [CrossRef] [PubMed]
- Walker, C.K.; Krakowiak, P.; Baker, A.; Hansen, R.L.; Ozonoff, S.; Hertz-Picciotto, I. Preeclampsia, Placental Insufficiency, and Autism Spectrum Disorder or Developmental Delay. JAMA Pediatr. 2015, 169, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Walker, N.; Filis, P.; Soffientini, U.; Bellingham, M.; O’Shaughnessy, P.J.; Fowler, P.A. Placental Transporter Localization and Expression in the Human: The Importance of Species, Sex, and Gestational Age Difference. Biol. Reprod. 2017, 96, 733–742. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barreto, R.d.S.N.; Carreira, A.C.O.; Silva, M.D.d.; Fernandes, L.A.; Ribeiro, R.R.; Almeida, G.H.D.R.; Pantoja, B.T.d.S.; Nishiyama Junior, M.Y.; Miglino, M.A. Mice Placental ECM Components May Provide A Three-Dimensional Placental Microenvironment. Bioengineering 2023, 10, 16. https://doi.org/10.3390/bioengineering10010016
Barreto RdSN, Carreira ACO, Silva MDd, Fernandes LA, Ribeiro RR, Almeida GHDR, Pantoja BTdS, Nishiyama Junior MY, Miglino MA. Mice Placental ECM Components May Provide A Three-Dimensional Placental Microenvironment. Bioengineering. 2023; 10(1):16. https://doi.org/10.3390/bioengineering10010016
Chicago/Turabian StyleBarreto, Rodrigo da Silva Nunes, Ana Claudia Oliveira Carreira, Mônica Duarte da Silva, Leticia Alves Fernandes, Rafaela Rodrigues Ribeiro, Gustavo Henrique Doná Rodrigues Almeida, Bruna Tassia dos Santos Pantoja, Milton Yutaka Nishiyama Junior, and Maria Angelica Miglino. 2023. "Mice Placental ECM Components May Provide A Three-Dimensional Placental Microenvironment" Bioengineering 10, no. 1: 16. https://doi.org/10.3390/bioengineering10010016
APA StyleBarreto, R. d. S. N., Carreira, A. C. O., Silva, M. D. d., Fernandes, L. A., Ribeiro, R. R., Almeida, G. H. D. R., Pantoja, B. T. d. S., Nishiyama Junior, M. Y., & Miglino, M. A. (2023). Mice Placental ECM Components May Provide A Three-Dimensional Placental Microenvironment. Bioengineering, 10(1), 16. https://doi.org/10.3390/bioengineering10010016









