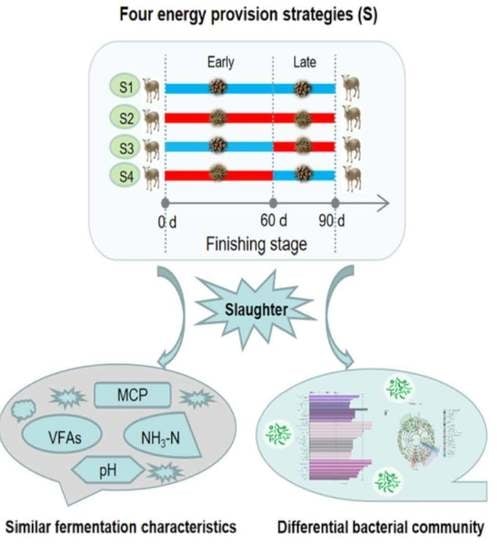
Effect of Energy Provision Strategy on Rumen Fermentation Characteristics, Bacterial Diversity and Community Composition
Abstract
1. Introduction
2. Materials and Methods
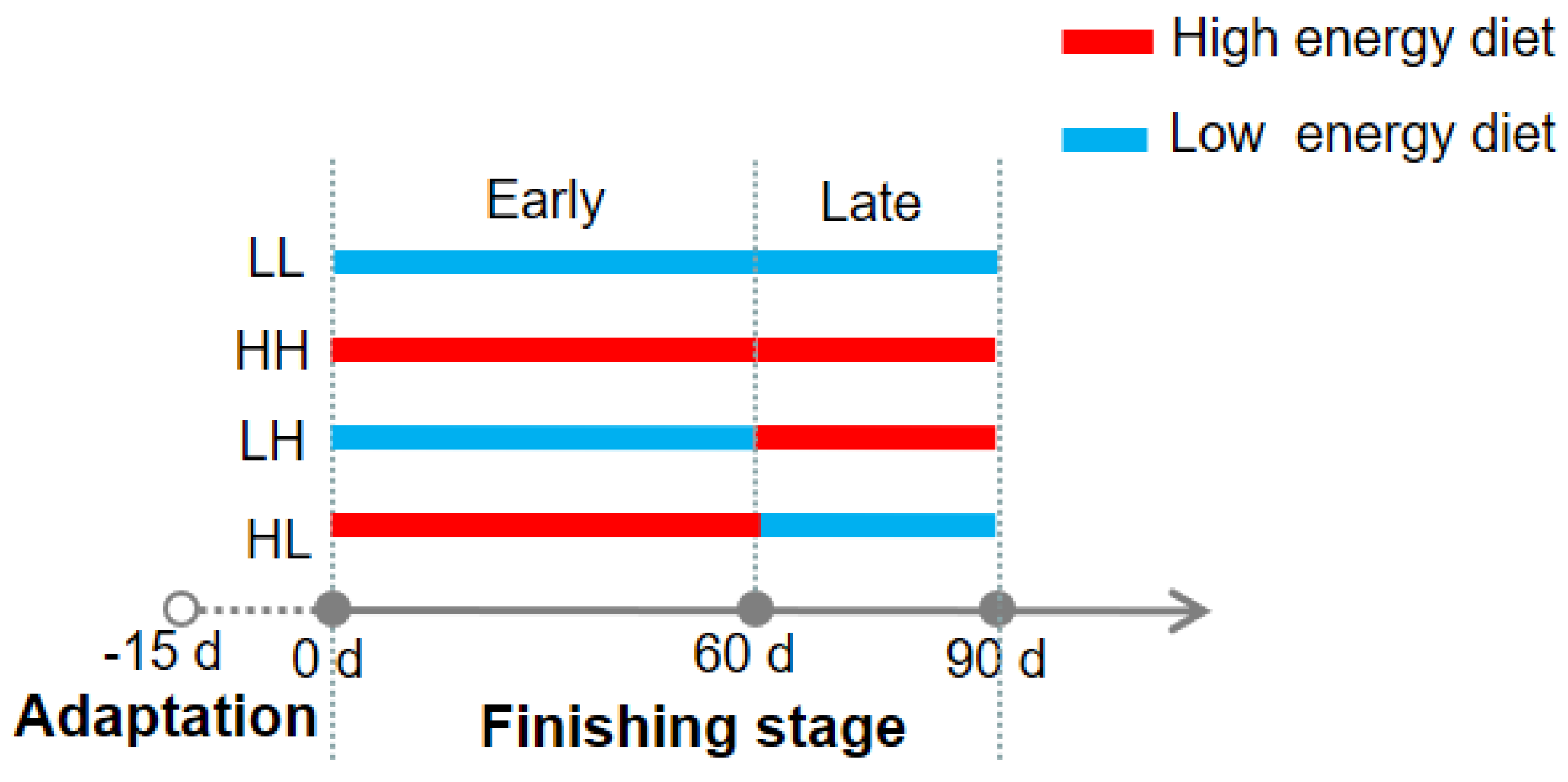
2.1. Animals and Experimental Design
2.2. Sample Collection
2.3. Rumen Fermentation Characteristics Determination
2.4. DNA Extraction, Sequencing, and Data Analysis
2.5. Statistical Analysis
3. Results
3.1. Rumen Fermentation Characteristics
3.2. Alpha-Diversity Metrics of Rumen Bacteria
3.3. Rumen Bacterial Community Composition
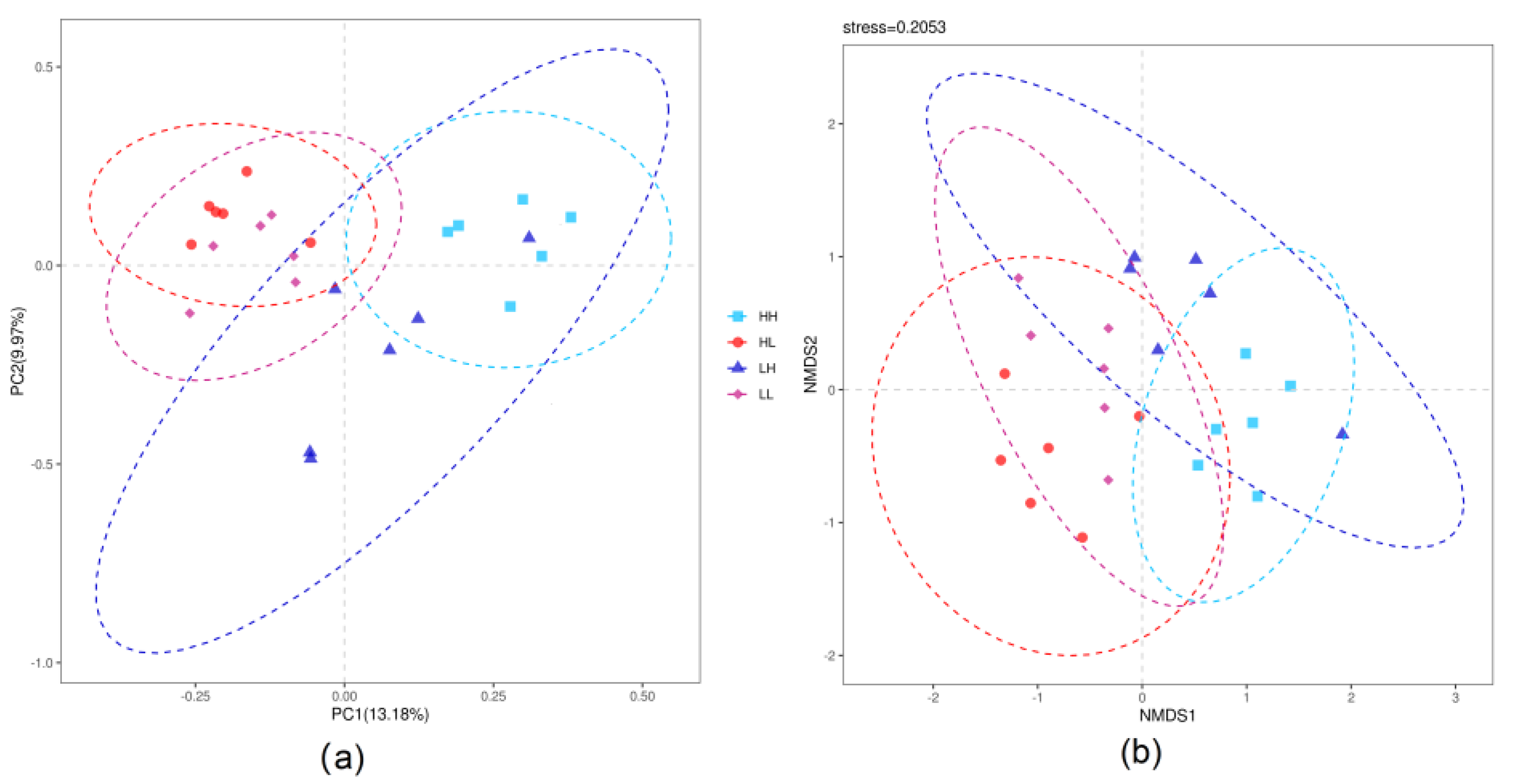
3.4. Beta-Diversity of Rumen Bacteria
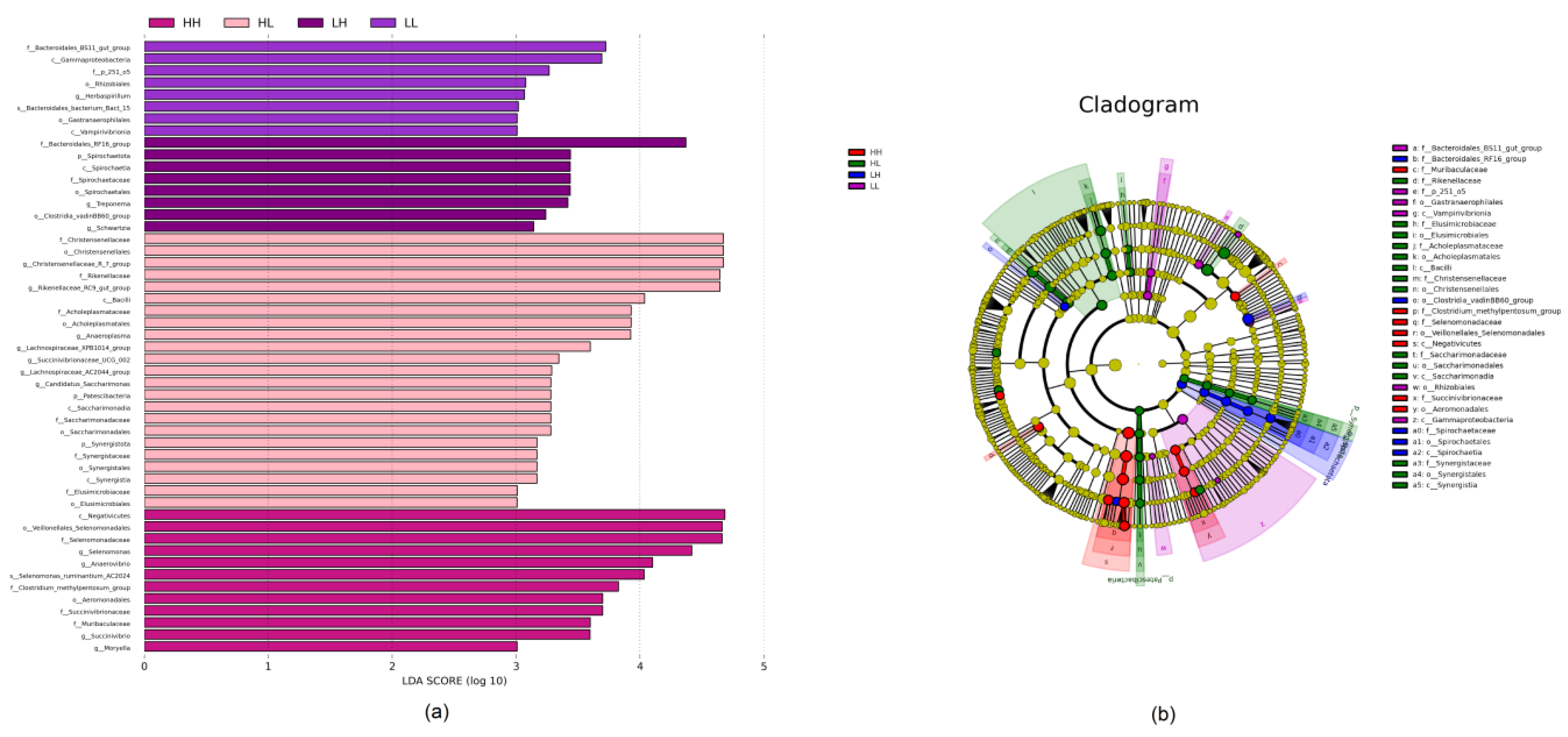
3.5. Biomarker Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Omontese, B.O.; Sharma, A.K.; Davison, S.; Jacobson, E.; DiConstanzo, A.; Webb, M.J.; Gomez, A. Microbiome network traits in the rumen predict average daily gain in beef cattle under different backgrounding systems. Anim. Microbiome 2022, 4, 25. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.Y.; Qi, W.P.; Zhang, T.; Zhang, J.Y.; Mei, S.J.; Mao, S.Y. Changes in rumen fermentation and bacterial community in lactating dairy cows with subacute rumen acidosis following rumen content transplantation. J. Dairy Sci. 2021, 104, 10780–10795. [Google Scholar] [CrossRef] [PubMed]
- Tapio, I.; Snelling, T.J.; Strozzi, F.; Wallace, R.J. The ruminal microbiome associated with methane emissions from ruminant livestock. J. Anim. Sci. Biotechnol. 2017, 8, 7. [Google Scholar] [CrossRef] [PubMed]
- Pitta, D.W.; Kumar, S.; Vecchiarelli, B.; Shirley, D.J.; Bittinger, K.; Baker, L.D.; Ferguson, J.D.; Thomsen, N. Temporal dynamics in the ruminal microbiome of dairy cows during the transition period. J. Anim. Sci. 2014, 92, 4014–4022. [Google Scholar] [CrossRef]
- Qiu, Q.; Gao, C.; Aziz ur Rahman, M.; Cao, B.; Su, H. Digestive ability, physiological characteristics, and rumen bacterial community of Holstein finishing steers in response to three nutrient density diets as fattening phases advanced. Microorganisms 2020, 8, 335. [Google Scholar] [CrossRef]
- Liu, H.J.; Xu, T.W.; Xu, S.X.; Ma, L.; Han, X.P.; Wang, X.G.; Zhang, X.L.; Hu, L.Y.; Zhao, N.; Chen, Y.W.; et al. Effect of dietary concentrate to forage ratio on growth performance, rumen fermentation and bacterial diversity of Tibetan sheep under barn feeding on the Qinghai-Tibetan plateau. PeerJ 2019, 7, e7462. [Google Scholar] [CrossRef]
- Pitta, D.W.; Indugu, N.; Baker, L.; Vecchiarelli, B.; Attwood, G. Symposium review: Understanding diet-microbe interactions to enhance productivity of dairy cows. J. Dairy Sci. 2018, 101, 7661–7679. [Google Scholar] [CrossRef]
- Ministry of Agriculture and Rural Affairs of the People’s Republic of China. Nutrient Requirements of Meat-Type Sheep and Goat; NY/T 816-2021; Ministry of Agriculture and Rural Affairs of the People’s Republic of China: Beijing, China, 2021.
- Qiu, Q.; Qiu, X.; Gao, C.; Muhammad, A.U.R.; Cao, B.; Su, H. High-density diet improves growth performance and beef yield but affects negatively on serum metabolism and visceral morphology of Holstein steers. J. Anim. Physiol. Anim. Nutr. 2020, 104, 1197–1208. [Google Scholar] [CrossRef]
- Rojen, B.A.; Theil, P.K.; Kristensen, N.B. Effects of nitrogen supply on inter-organ fluxes of urea-N and renal urea-N kinetics in lactating Holstein cows. J. Dairy Sci. 2011, 94, 2532–2544. [Google Scholar] [CrossRef]
- Wang, Q.; Zeng, Y.; Zeng, X.; Wang, X.; Wang, Y.; Dai, C.; Li, J.; Huang, P.; Huang, J.; Hussain, T.; et al. Effects of dietary energy levels on rumen fermentation, gastrointestinal tract histology, and bacterial community diversity in fattening male Hu lambs. Front. Microbiol. 2021, 12, 695445. [Google Scholar] [CrossRef]
- Wang, H.B.; Li, H.; Wu, F.; Qiu, X.J.; Yu, Z.T.; Niu, W.J.; He, Y.; Su, H.W.; Cao, B.H. Effects of dietary energy on growth performance, rumen fermentation and bacterial community, and meat quality of Holstein-Friesians bulls slaughtered at different ages. Animals 2019, 9, 1123. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Zeng, S.; Zhang, R.; Diao, Q.; Tu, Y. Effects of dietary energy levels on rumen bacterial community composition in Holstein heifers under the same forage to concentrate ratio condition. BMC Microbiol. 2018, 18, 69. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Cui, K.; Qi, M.; Wang, S.; Diao, Q.; Zhang, N. Ruminal microbiota and fermentation in response to dietary protein and energy levels in weaned lambs. Animals 2020, 10, 109. [Google Scholar] [CrossRef] [PubMed]
- Pang, K.; Dai, D.; Yang, Y.; Wang, X.; Liu, S.; Huang, W.; Xue, B.; Chai, S.; Wang, S. Effects of high concentrate rations on ruminal fermentation and microbiota of yaks. Front. Microbiol. 2022, 13, 957152. [Google Scholar] [CrossRef] [PubMed]
- Broderick, G.A.; Kang, J.H. Automated simultaneous determination of ammonia and total amino acids in ruminal fluid and in vitro media. J. Dairy Sci. 1980, 63, 64–75. [Google Scholar] [CrossRef]
- Makkar, H.P.; Sharma, O.P.; Dawra, R.K.; Negi, S.S. Simple determination of microbial protein in rumen liquor. J. Dairy Sci. 1982, 65, 2170–2173. [Google Scholar] [CrossRef]
- Wei, X.; Long, T.; Li, Y.; Ouyang, K.; Qiu, Q. Diet shift may trigger LuxS/AI-2 quorum sensing in rumen bacteria. Bioengineering 2022, 9, 379. [Google Scholar] [CrossRef]
- Wang, W.; Wu, Q.; Li, W.; Wang, Y.; Zhang, F.; Lv, L.; Li, S.; Yang, H. High-gossypol whole cottonseed exhibited mediocre rumen degradability and less microbial fermentation efficiency than cottonseed hull and cottonseed meal with an in vitro gas production technique. Fermentation 2022, 8, 103. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Q.; Wei, X.; Zhang, L.; Li, Y.; Qu, M.; Ouyang, K. Effect of dietary inclusion of tea residue and tea leaves on ruminal fermentation characteristics and methane production. Anim. Biotechnol. 2021, in press. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.H.; Zhao, Y.Y.; Xue, S.L.; Wang, W.; Wang, Y.J.; Cao, Z.J.; Yang, H.J.; Li, S.L. Feeding value assessment of substituting cassava (Manihot esculenta) residue for concentrate of dairy cows using an in vitro gas test. Animals 2021, 11, 307. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, J. Production and absorption of volatile fatty acids in the rumen. Livest. Prod. Sci. 1994, 39, 61–69. [Google Scholar] [CrossRef]
- Guan, L.L.; Nkrumah, J.D.; Basarab, J.A.; Moore, S.S. Linkage of microbial ecology to phenotype: Correlation of rumen microbial ecology to cattle’s feed efficiency. FEMS Microbiol. Lett. 2008, 288, 85–91. [Google Scholar] [CrossRef]
- Plaizier, J.C.; Li, S.; Danscher, A.M.; Derakshani, H.; Andersen, P.H.; Khafipour, E. Changes in microbiota in rumen digesta and feces due to a grain-based subacute ruminal acidosis (SARA) challenge. Microb. Ecol. 2017, 74, 485–495. [Google Scholar] [CrossRef]
- Hook, S.E.; Steele, M.A.; Northwood, K.S.; Dijkstra, J.; France, J.; Wright, A.D.G.; McBride, B.W. Impact of subacute ruminal acidosis (SARA) adaptation and recovery on the density and diversity of bacteria in the rumen of dairy cows. FEMS Microbiol. Ecol. 2011, 78, 275–284. [Google Scholar] [CrossRef]
- Zhou, S.; Luo, R.; Gong, G.; Wang, Y.; Gesang, Z.; Wang, K.; Xu, Z.; Suolang, S. Characterization of metagenome-assembled genomes and carbohydrate-degrading genes in the gut microbiota of Tibetan pig. Front. Microbiol. 2020, 11, 595066. [Google Scholar] [CrossRef]
- Herrmann, M.; Wegner, C.-E.; Taubert, M.; Geesink, P.; Lehmann, K.; Yan, L.; Lehmann, R.; Totsche, K.U.; Küsel, K. Predominance of Cand. Patescibacteria in groundwater is caused by their preferential mobilization from soils and flourishing under oligotrophic conditions. Front. Microbiol. 2019, 10, 1407. [Google Scholar] [CrossRef]
- Tian, R.; Ning, D.; He, Z.; Zhang, P.; Spencer, S.J.; Gao, S.; Shi, W.; Wu, L.; Zhang, Y.; Yang, Y.; et al. Small and mighty: Adaptation of superphylum Patescibacteria to groundwater environment drives their genome simplicity. Microbiome 2020, 8, 51. [Google Scholar] [CrossRef]
- Ribeiro, G.O.; Gruninger, R.J.; Jones, D.R.; Beauchemin, K.A.; Yang, W.Z.; Wang, Y.; Abbott, D.W.; Tsang, A.; McAllister, T.A. Effect of ammonia fiber expansion-treated wheat straw and a recombinant fibrolytic enzyme on rumen microbiota and fermentation parameters, total tract digestibility, and performance of lambs. J. Anim. Sci. 2020, 98, skaa116. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Guo, C.; Wang, L.; Chen, F.; Dong, X.; Li, X.; Ni, K.; Yang, F. Effects of paper mulberry silage on the growth performance, rumen microbiota and muscle fatty acid composition in Hu lambs. Fermentation 2021, 7, 286. [Google Scholar] [CrossRef]
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Sutter, J.L.; Koren, O.; Blekhman, R.; Beaumont, M.; Van Treuren, W.; Knight, R.; Bell, J.T.; et al. Human genetics shape the gut microbiome. Cell 2014, 159, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Wang, Y.; Zuo, S.; Peng, S.; Wang, Z.; Zhang, Y.; Luo, H. Untargeted and targeted metabolomics profiling of muscle reveals enhanced meat quality in artificial pasture grazing Tan lambs via rescheduling the rumen bacterial community. J. Agric. Food Chem. 2021, 69, 846–858. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Yao, R.; Liu, Q.; Deng, Y.; Fang, J. Effects of antibacterial peptides on rumen fermentation function and rumen microorganisms in goats. PLoS ONE 2019, 14, e0221815. [Google Scholar] [CrossRef]
- Kasperowicz, A.; Michałowski, T. Digestion and utilization of fructose polymers by the rumen bacterium Treponema sp. strain T. J. Anim. Feed Sci. 2007, 16, 577–589. [Google Scholar] [CrossRef]
- Hishinuma, F.; Kanegasaki, S.; Takahashi, H. Ruminal fermentation and sugar concentrations: A model experiment with Selenomonas ruminantium. Agri. Biol. Chem. 1968, 32, 1325–1330. [Google Scholar] [CrossRef]
- Florence, P.; Kaderbhai, N.N.; Susan, G.; Worgan, H.J.; Eric, P.; Scollan, N.D.; Huws, S.A.; Jamie, N.C.; Planet, P.J. Identification and characterization of three novel lipases belonging to families II and V from Anaerovibrio lipolyticus 5ST. PLoS ONE 2013, 8, e69076. [Google Scholar]
- Williams, A.G.; Withers, S.E. Bacillus spp. in the rumen ecosystem. Hemicellulose depolymerases and glycoside hydrolases of Bacillus spp. and rumen isolates grown under anaerobic conditions. J. Appl. Bacteriol. 1983, 55, 283–292. [Google Scholar] [CrossRef]
- Nardi, K.T.; Silva, K.G.S.; Favero, I.G.; Hinds, J.K.; Hoffmann, C.A.; Soto, S.M.; Hall, N.; Henry, D.D.; Sarturi, J.O. PSXII-3 Rumen microbiome from beef steers undergoing grain adaptation with steam-flaked corn varying in bulk density. J. Anim. Sci. 2020, 98, 446–447. [Google Scholar] [CrossRef]
- Choi, J.K.; Shah, M.; Yee, N. Anaerosporomusa subterranea gen. nov., sp. nov., a spore-forming anaerobe belonging to the class Negativicutes isolated from saprolite. Int. J. Syst. Evol. Microbiol. 2016, 66, 3848–3854. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.K.; Zhang, X.X.; Li, F.D.; Li, C.; Li, G.Z.; Zhang, D.Y.; Song, Q.Z.; Li, X.L.; Zhao, Y.; Wang, W.M. Characterization of the rumen microbiota and its relationship with residual feed intake in sheep. Animal 2021, 15, 100161. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zhang, T.; Yoon, I.; Khafipour, E.; Plaizier, J.C. 236 Effects of Saccharomyces cerevisiae fermentation products (SCFP) and subacute ruminal acidosis (SARA) on co-occurrence patterns and hub taxa of rumen liquid microbiota in lactating dairy cows. J. Anim. Sci. 2021, 99, 175–176. [Google Scholar] [CrossRef]
- He, J.; Zheng, W.; Tao, C.; Guo, H.; Xue, Y.; Zhao, R.; Yao, W. Heat stress during late gestation disrupts maternal microbial transmission with altered offspring’s gut microbial colonization and serum metabolites in a pig model. Environ. Pollut. 2020, 266, 115111. [Google Scholar] [CrossRef] [PubMed]
| Item | Low-Energy Diet | High-Energy Diet |
|---|---|---|
| Ingredient | ||
| Corn, g/kg | 111.4 | 490.9 |
| Wheat bran, g/kg | 46.2 | 18.1 |
| Soybean meal, g/kg | 110.9 | 143.1 |
| Peanut straw, g/kg | 687.9 | 304.1 |
| Calcium hydrophosphate, g/kg | 1.5 | 1.5 |
| Sodium bicarbonate, g/kg | 2.4 | 2.5 |
| Salt, g/kg | 4.9 | 5.0 |
| Premix 1, g/kg | 34.8 | 34.8 |
| Total | 1000 | 1000 |
| Chemical Composition | ||
| Metabolizable energy, Mcal/kg | 2.11 | 2.59 |
| Crude protein, g/kg | 157.6 | 158.3 |
| Ether extract, g/kg | 92.8 | 111.6 |
| Neutral detergent fiber, g/kg | 435.2 | 280.4 |
| Acid detergent fiber, g/kg | 322.8 | 158.6 |
| Item 1 | LL 2 | LH 3 | HL 4 | HH 5 | SEM 6 | p-Value |
|---|---|---|---|---|---|---|
| pH value | 7.04 | 7.36 | 7.24 | 6.80 | 0.244 | 0.415 |
| Ammoniacal nitrogen, mg/dL | 15.85 | 16.00 | 12.00 | 15.09 | 1.462 | 0.213 |
| Microbial crude protein, mg/L | 526.13 | 454.87 | 624.08 | 703.97 | 80.88 | 0.175 |
| Total volatile fatty acids, mM | 67.52 | 44.89 | 51.79 | 54.79 | 10.75 | 0.521 |
| Concentration, mM | ||||||
| Acetate | 46.42 | 30.69 | 37.08 | 33.61 | 6.994 | 0.433 |
| Propionate | 11.24 | 7.28 | 7.38 | 10.07 | 2.722 | 0.668 |
| Isobutyrate | 0.48 | 0.36 | 0.48 | 0.45 | 0.085 | 0.735 |
| Butyrate | 8.03 | 5.46 | 5.55 | 9.33 | 1.801 | 0.364 |
| Isovalerate | 0.60 | 0.51 | 0.67 | 0.58 | 0.089 | 0.640 |
| Valerate | 0.75 | 0.57 | 0.63 | 0.75 | 0.107 | 0.597 |
| BCVFAs | 1.82 | 1.45 | 1.79 | 1.77 | 0.244 | 0.679 |
| Proportion, % | ||||||
| Acetate | 69.82 ab | 68.68 ab | 71.16 a | 62.97 b | 1.878 | 0.029 |
| Propionate | 16.32 | 16.50 | 14.78 | 16.77 | 2.337 | 0.931 |
| Isobutyrate | 0.89 | 0.87 | 0.96 | 0.86 | 0.128 | 0.947 |
| Butyrate | 10.63 b | 11.44 b | 10.40 b | 16.93 a | 1.332 | 0.007 |
| Isovalerate | 1.10 | 1.17 | 1.41 | 1.11 | 0.181 | 0.597 |
| Valerate | 1.23 | 1.34 | 1.30 | 1.36 | 0.111 | 0.845 |
| BCVFAs | 3.22 | 3.38 | 3.66 | 3.32 | 0.382 | 0.865 |
| Rumen Fermentation Pattern | ||||||
| Acetate-to-propionate ratio | 4.48 | 4.21 | 4.10 | 4.41 | 0.430 | 0.919 |
| NGR | 5.52 | 5.29 | 4.90 | 6.30 | 0.630 | 0.484 |
| Fermentation efficiency | 0.72 | 0.73 | 0.73 | 0.74 | 0.008 | 0.579 |
| Item | LL | LH | HL | HH | SEM | p-Value |
|---|---|---|---|---|---|---|
| Chao 1 | 1060.80 a | 572.81 b | 567.39 b | 499.13 b | 74.15 | <0.001 |
| Observed species | 972.58 a | 557.00 b | 557.00 b | 481.15 b | 67.38 | <0.001 |
| PD whole tree | 79.27 a | 57.51 b | 60.02 b | 51.07 b | 4.050 | 0.001 |
| Shannon index | 7.44 a | 6.49 ab | 6.95 ab | 6.43 b | 0.249 | 0.033 |
| Simpson index | 0.97 | 0.95 | 0.98 | 0.97 | 0.008 | 0.198 |
| Phylum Name | LL | LH | HL | HH | SEM | p-Value |
|---|---|---|---|---|---|---|
| Bacteroidota | 69.93 | 62.95 | 58.87 | 60.91 | 3.645 | 0.191 |
| Firmicutes | 25.39 | 33.48 | 34.55 | 35.93 | 3.301 | 0.138 |
| Proteobacteria | 1.82 | 0.80 | 1.56 | 1.64 | 0.418 | 0.351 |
| Spirochaetota | 0.44 b | 0.86 a | 0.64 ab | 0.30 b | 0.104 | 0.006 |
| Verrucomicrobiota | 0.77 | 0.43 | 0.76 | 0.25 | 0.184 | 0.154 |
| Actinobacteriota | 0.25 | 0.11 | 1.42 | 0.18 | 0.661 | 0.465 |
| Desulfobacterota | 0.35 | 0.43 | 0.42 | 0.33 | 0.124 | 0.916 |
| Patescibacteria | 0.17 ab | 0.28 ab | 0.47 a | 0.08 b | 0.091 | 0.035 |
| Synergistota | 0.31 | 0.17 | 0.40 | 0.08 | 0.093 | 0.098 |
| Cyanobacteria | 0.21 | 0.10 | 0.15 | 0.06 | 0.039 | 0.066 |
| Genus Name | LL | LH | HL | HH | SEM | p-Value |
|---|---|---|---|---|---|---|
| Prevotella | 41.36 | 38.30 | 26.38 | 44.87 | 4.694 | 0.059 |
| Rikenellaceae RC9 gut group | 7.68 ab | 10.08 ab | 14.68 a | 4.83 b | 1.939 | 0.013 |
| Uncultured rumen bacterium | 6.48 | 4.79 | 6.20 | 4.78 | 1.066 | 0.553 |
| Christensenellaceae R-7 group | 3.55 b | 2.26 b | 11.38 a | 4.39 b | 1.513 | 0.002 |
| Uncultured bacterium | 5.46 | 5.87 | 6.80 | 2.59 | 1.257 | 0.134 |
| Prevotellaceae UCG-001 | 3.94 | 4.27 | 2.88 | 3.05 | 1.570 | 0.904 |
| Selenomonas | 2.67 ab | 3.57 ab | 1.08 b | 6.71 a | 1.188 | 0.022 |
| Succiniclasticum | 2.78 | 3.10 | 3.72 | 4.17 | 0.875 | 0.683 |
| Prevotellaceae UCG-003 | 5.76 | 1.64 | 3.43 | 2.55 | 1.211 | 0.128 |
| Veillonellaceae UCG-001 | 2.01 | 1.61 | 2.25 | 1.66 | 0.416 | 0.670 |
| NK4 A214 group | 1.27 | 1.26 | 1.16 | 0.51 | 0.263 | 0.165 |
| Anaerovibrio | 0.51 ab | 0.98 ab | 0.15 b | 2.49 a | 0.401 | 0.003 |
| Ruminococcus | 0.76 | 0.76 | 0.52 | 1.21 | 0.165 | 0.055 |
| Butyrivibrio | 0.77 | 1.20 | 0.62 | 0.55 | 0.364 | 0.603 |
| Lachnospiraceae NK3A20 group | 0.42 | 0.79 | 1.10 | 0.83 | 0.374 | 0.645 |
| Anaeroplasma | 0.52 b | 0.38 b | 1.85 a | 0.18 b | 0.310 | 0.004 |
| Lachnospiraceae ND3007 group | 0.33 | 0.93 | 0.77 | 0.91 | 0.363 | 0.629 |
| Succinivibrio | 0.89 | 0.36 | 0.31 | 1.08 | 0.253 | 0.108 |
| UCG-002 | 0.67 | 0.60 | 0.21 | 0.64 | 0.222 | 0.452 |
| Treponema | 0.39 b | 0.84 a | 0.57 ab | 0.30 b | 0.105 | 0.008 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiu, Q.; Zhang, J.; Qu, M.; Li, Y.; Zhao, X.; Ouyang, K. Effect of Energy Provision Strategy on Rumen Fermentation Characteristics, Bacterial Diversity and Community Composition. Bioengineering 2023, 10, 107. https://doi.org/10.3390/bioengineering10010107
Qiu Q, Zhang J, Qu M, Li Y, Zhao X, Ouyang K. Effect of Energy Provision Strategy on Rumen Fermentation Characteristics, Bacterial Diversity and Community Composition. Bioengineering. 2023; 10(1):107. https://doi.org/10.3390/bioengineering10010107
Chicago/Turabian StyleQiu, Qinghua, Jiantong Zhang, Mingren Qu, Yanjiao Li, Xianghui Zhao, and Kehui Ouyang. 2023. "Effect of Energy Provision Strategy on Rumen Fermentation Characteristics, Bacterial Diversity and Community Composition" Bioengineering 10, no. 1: 107. https://doi.org/10.3390/bioengineering10010107
APA StyleQiu, Q., Zhang, J., Qu, M., Li, Y., Zhao, X., & Ouyang, K. (2023). Effect of Energy Provision Strategy on Rumen Fermentation Characteristics, Bacterial Diversity and Community Composition. Bioengineering, 10(1), 107. https://doi.org/10.3390/bioengineering10010107