Optimizing the Seeding Density of Human Mononuclear Cells to Improve the Purity of Highly Proliferative Mesenchymal Stem Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Isolation and Culture of Human BM-MSCs
2.3. Time-Lapse Imaging
2.4. Senescence-Associated β-Galactosidase Staining
2.5. Flow Cytometry
2.6. Single-Cell Cloning Assay
2.7. Colony-Forming Assay
2.8. Quantitative Real-Time PCR
2.9. Serial Analysis of Gene Expression (SAGE)
2.10. Differentiation Assay
2.11. Statistical Analysis
3. Results
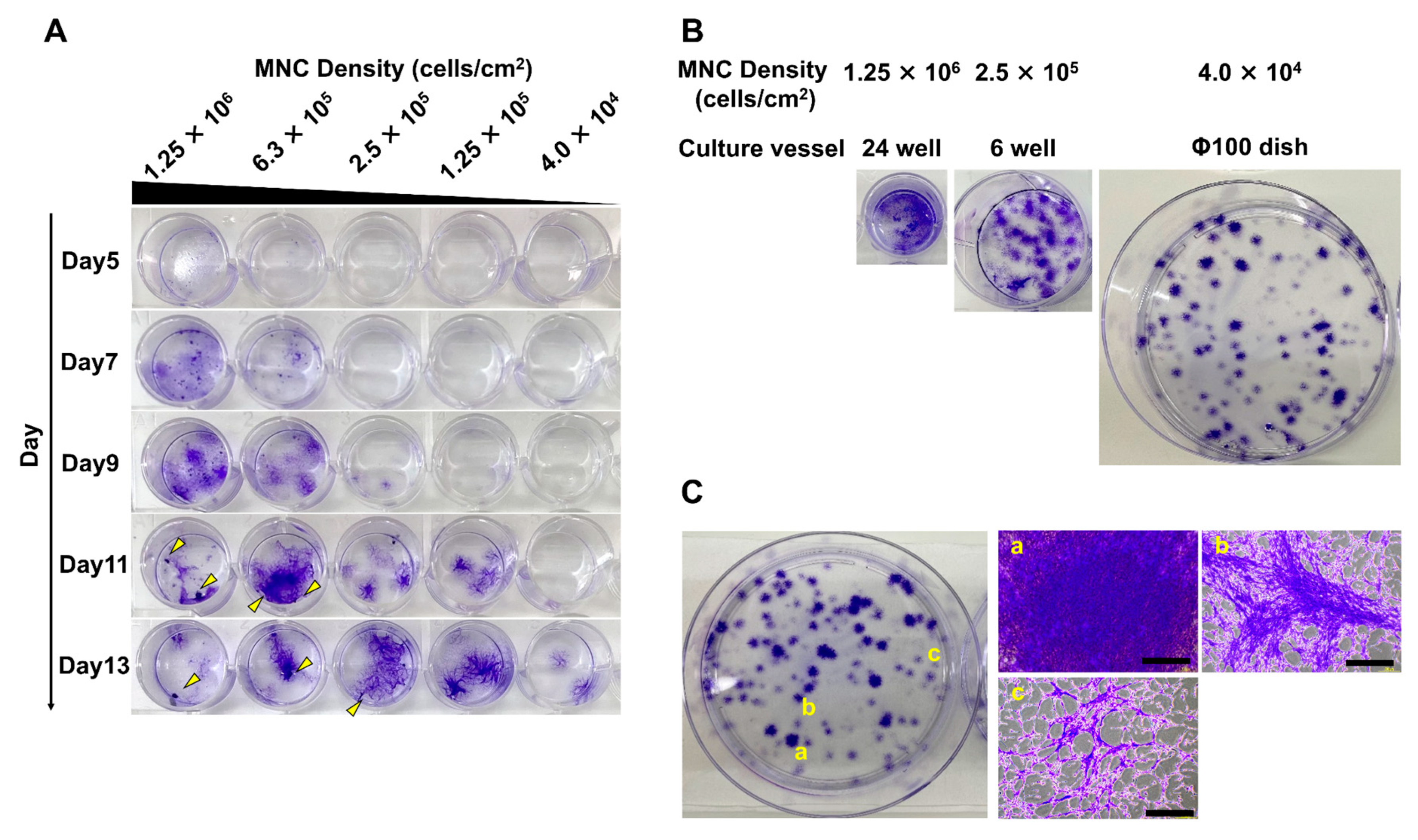
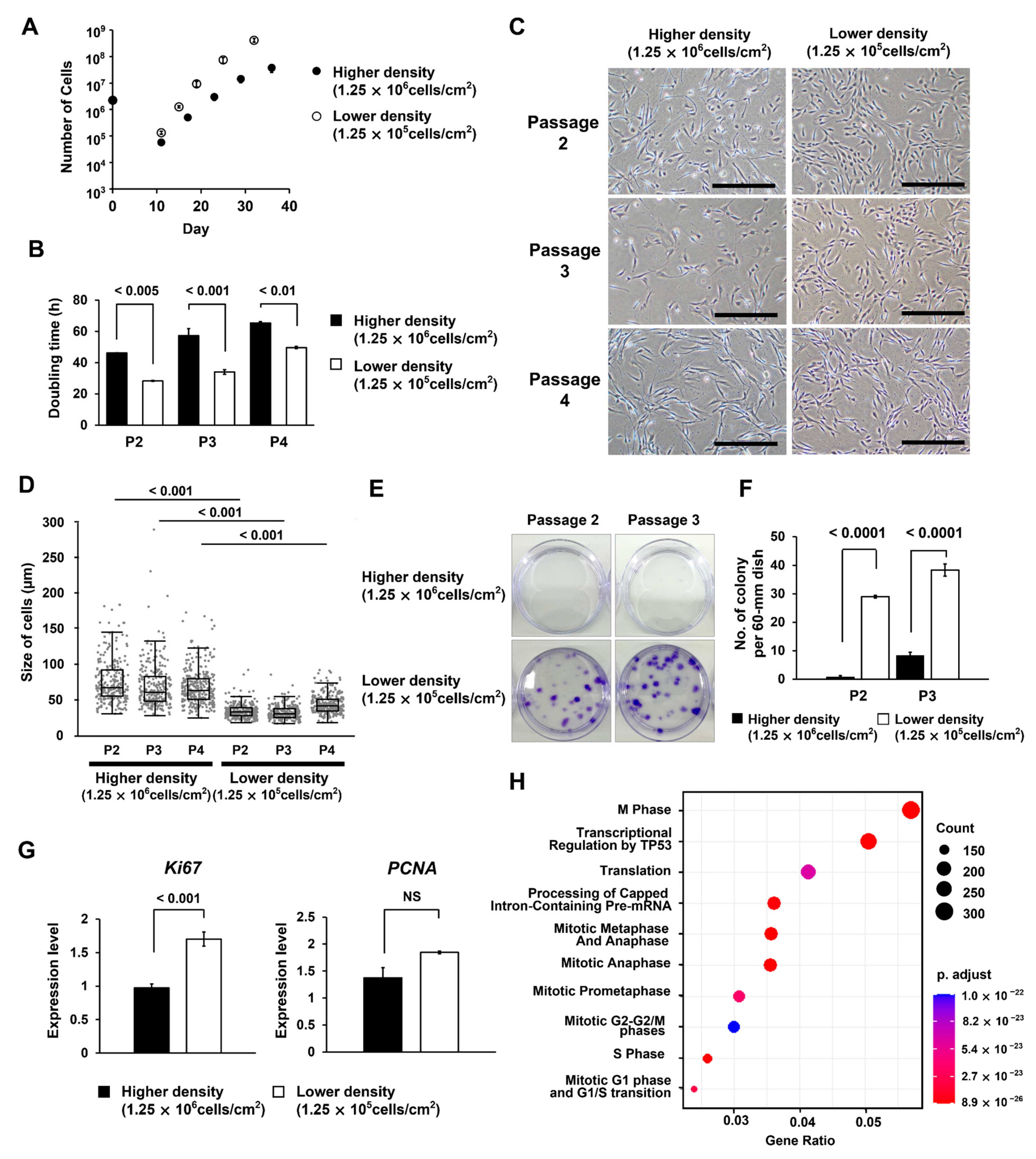
3.1. Different Seeding Densities of MNCs Cause Differences in the Timing of MSC Colony Formation
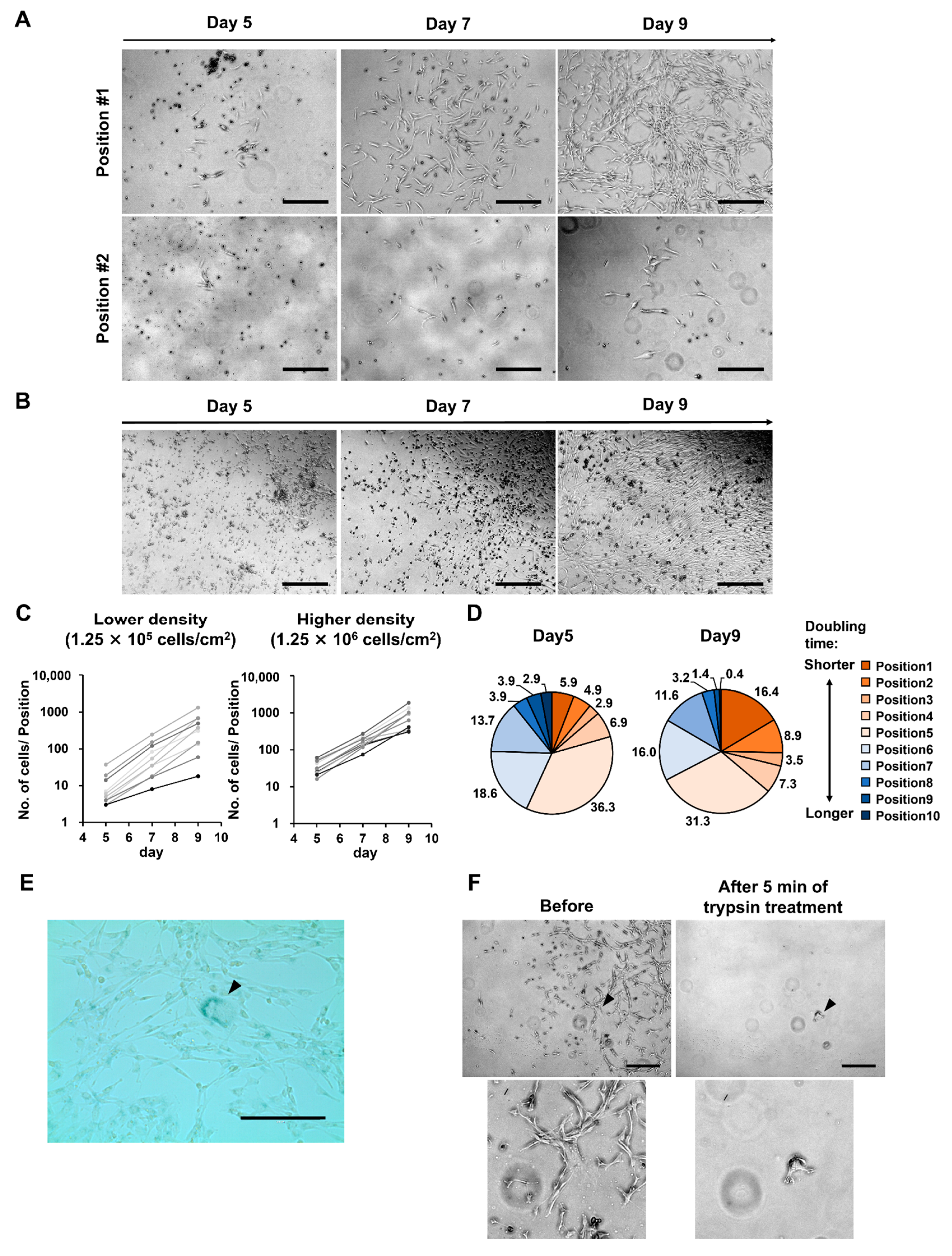
3.2. At Lower MNC Seeding Density, the Superiority of Proliferative Ability Results in Significant Differences in Numbers between Cells of Different Quality
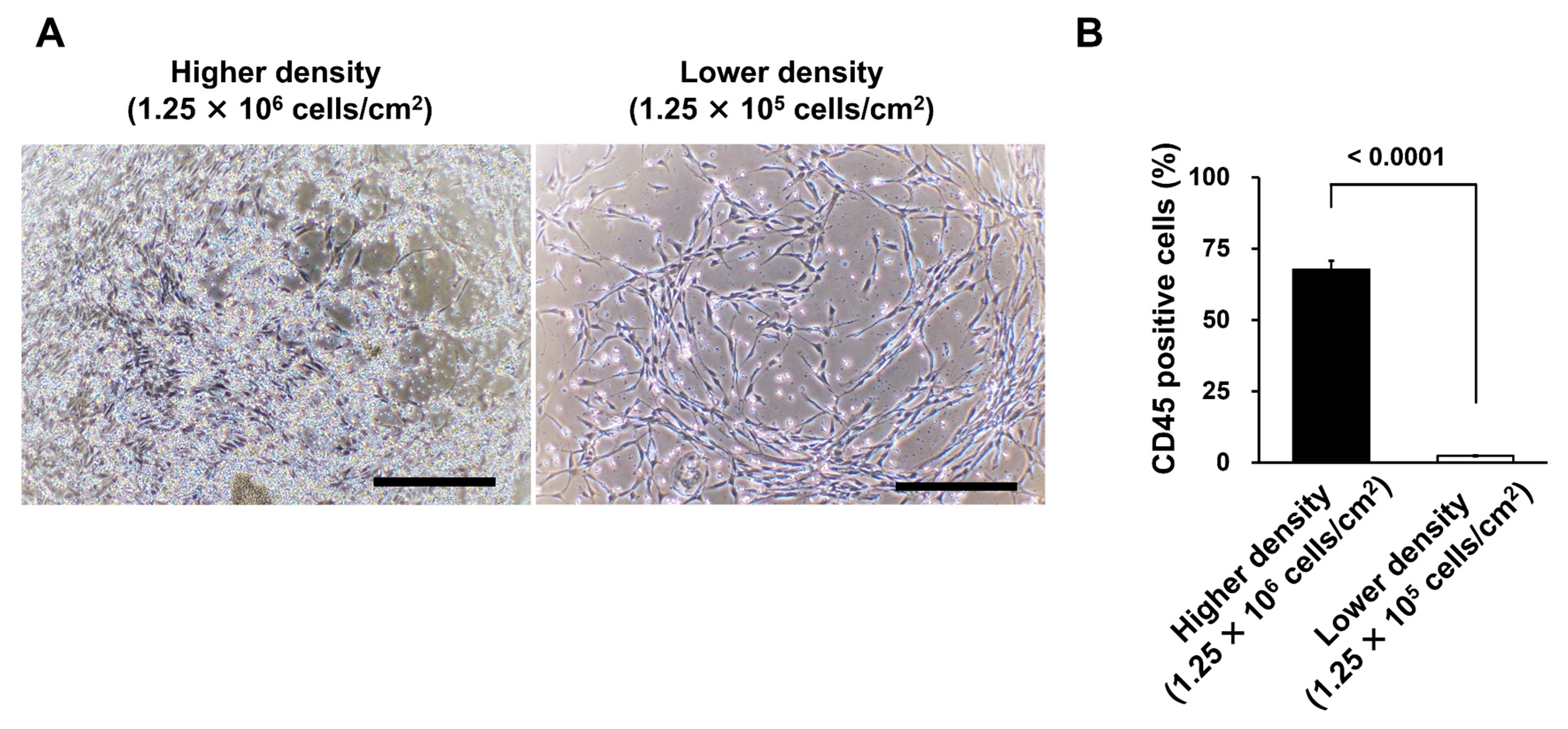
3.3. Different Seeding Densities of MNCs Affect the Number of Remaining CD45-Positive Cells
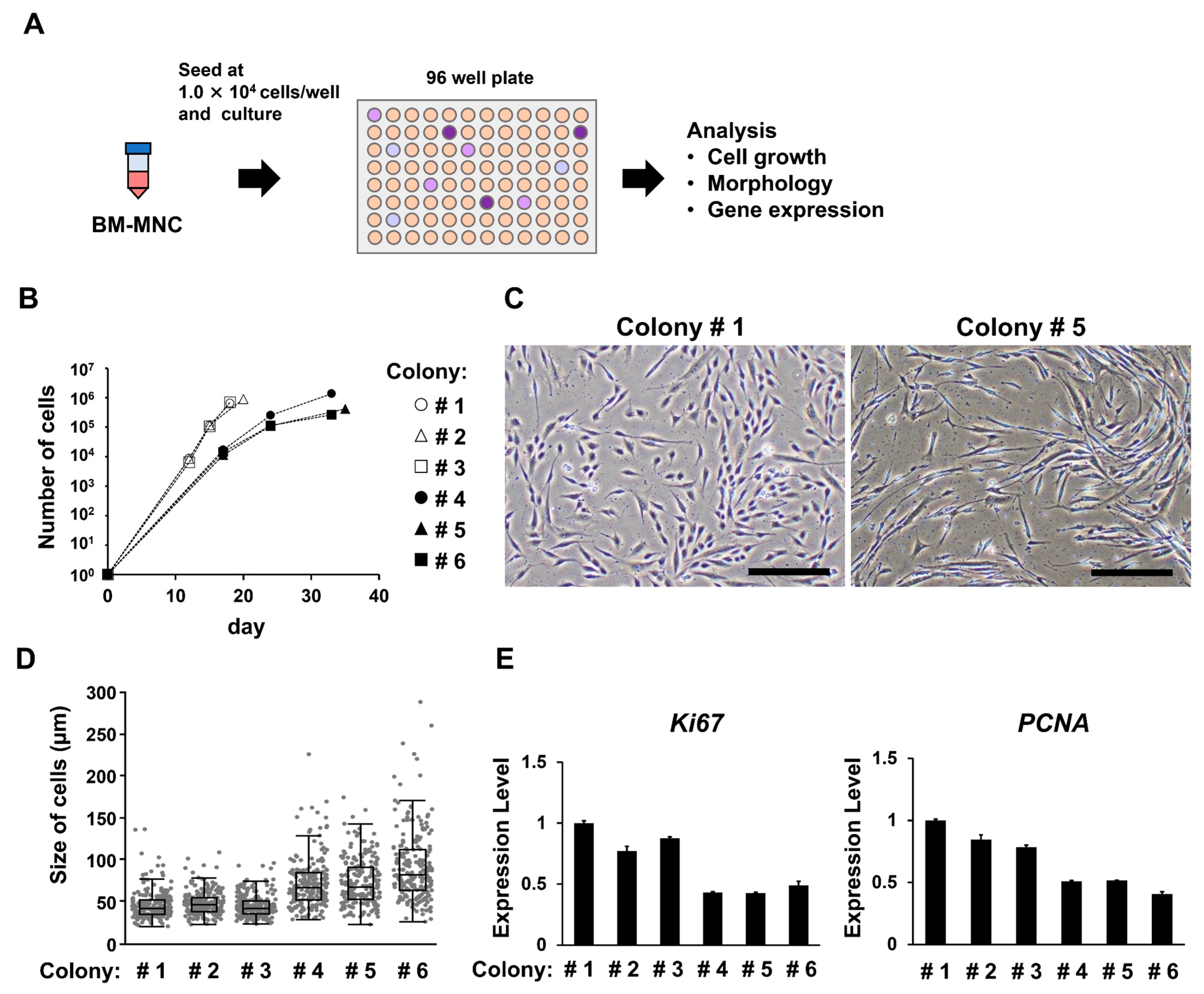
3.4. Groups of Cells That Form Colonies Quickly Show High Proliferative Ability
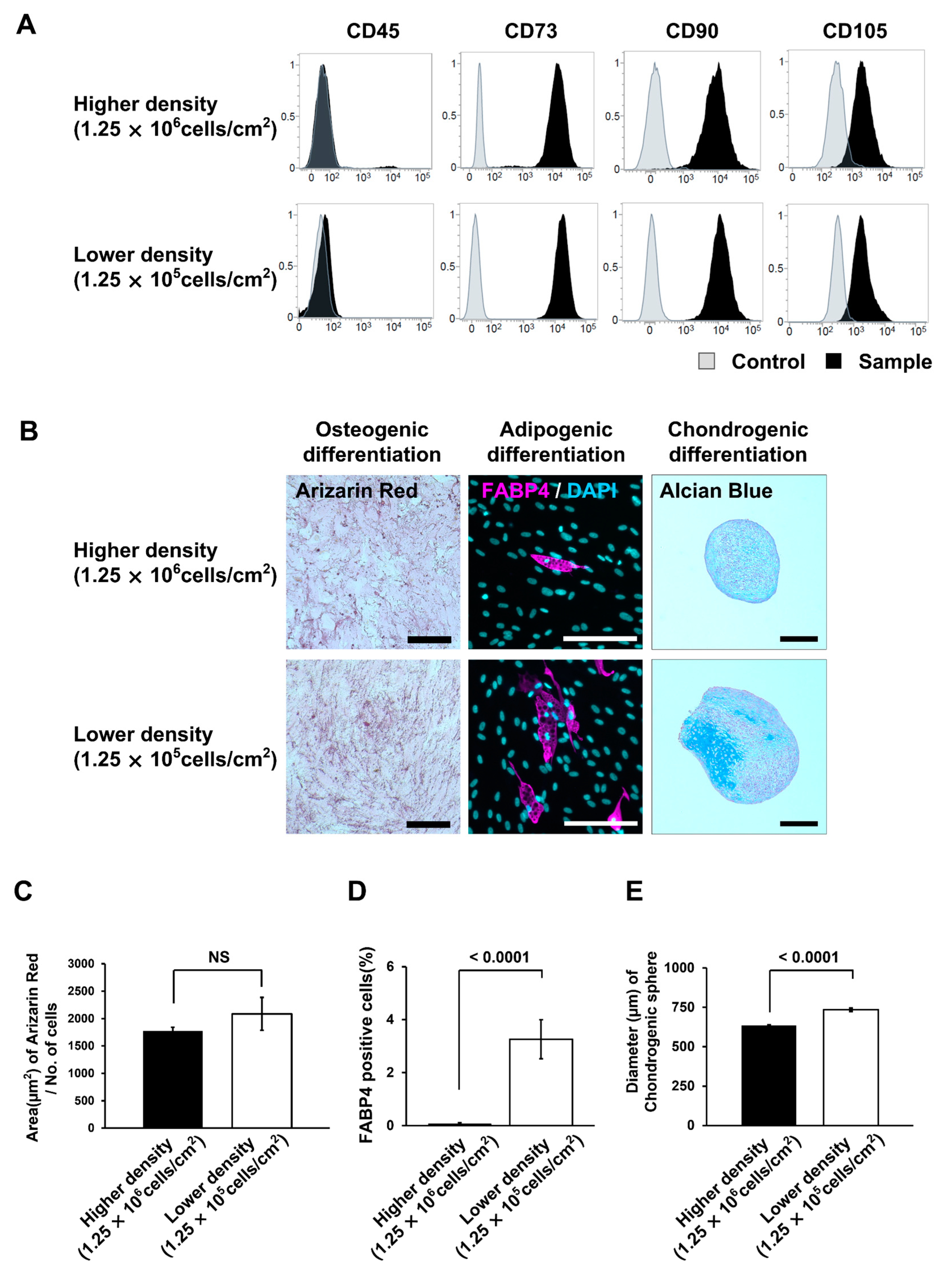
3.5. MSCs Isolated under the Low MNC Seeding Density Condition Have Higher Proliferative Ability and Improved Differentiation Potential
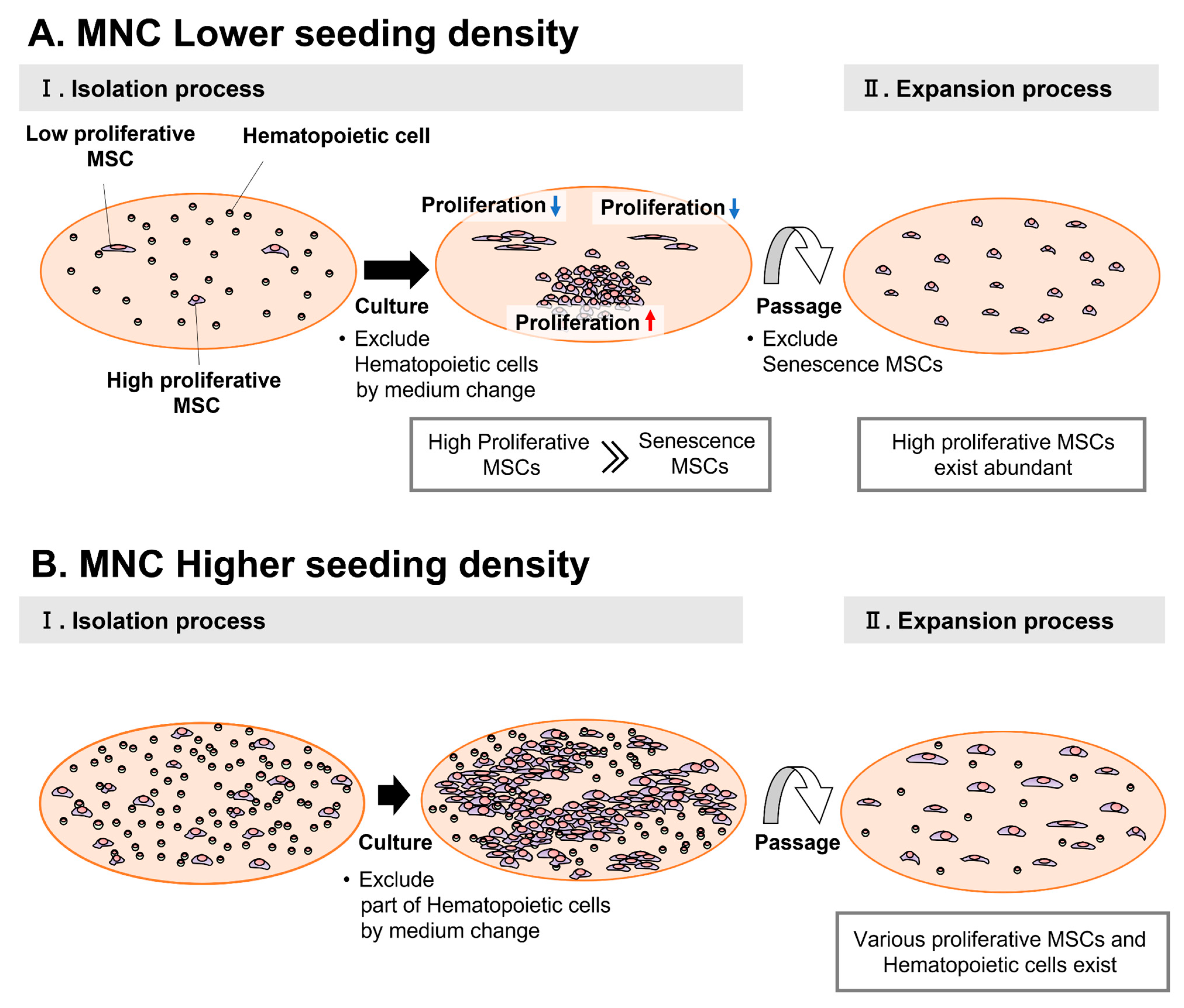
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Squillaro, T.; Peluso, G.; Galderisi, U. Clinical trials with mesenchymal stem cells: An update. Cell Transplant. 2016, 25, 829–848. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.R.; Pollock, K.; Hubel, A.; McKenna, D. Mesenchymal stem or stromal cells: A review of clinical applications and manufacturing practices. Transfusion 2014, 54, 1418–1437. [Google Scholar] [CrossRef] [PubMed]
- Seong, J.M.; Kim, B.-C.; Park, J.-H.; Kwon, K.I.; Mantalaris, A.; Hwang, Y.-S. Stem cells in bone tissue engineering. Biomed. Mater. 2010, 5, 062001. [Google Scholar] [CrossRef] [PubMed]
- Stoltz, J.-F.; Huselstein, C.; Schiavi, J.; Li, Y.; Bensoussan, D.; Decot, V.; De Isla, N. Human stem cells and articular cartilage tissue engineering. Curr. Pharm. Biotechnol. 2012, 13, 2682–2691. [Google Scholar] [CrossRef] [PubMed]
- Namioka, T.; Namioka, A.; Sasaki, M.; Kataoka-Sasaki, Y.; Oka, S.; Nakazaki, M.; Onodera, R.; Suzuki, J.; Sasaki, Y.; Nagahama, H.; et al. Intravenous infusion of mesenchymal stem cells promotes functional recovery in a rat model of chronic cerebral infarction. J. Neurosurg. 2019, 131, 1289–1296. [Google Scholar] [CrossRef]
- Lee, R.H.; Pulin, A.A.; Seo, M.J.; Kota, D.J.; Ylostalo, J.; Larson, B.L.; Semprun-Prieto, L.; Delafontaine, P.; Prockop, D.J. Intravenous hmscs improve myocardial infarction in mice because cells embolized in lung are activated to secrete the anti-inflammatory protein TSG-6 Cell. Stem Cell 2009, 5, 54–63. [Google Scholar] [CrossRef]
- Chen, Y.-S.; Lin, E.-Y.; Chiou, T.-W.; Harn, H.-J. Exosomes in clinical trial and their production in compliance with good manufacturing practice. Tzu. Chi. Med. J. 2020, 32, 113–120. [Google Scholar]
- Terai, S.; Ishikawa, T.; Omori, K.; Aoyama, K.; Marumoto, Y.; Urata, Y.; Yokoyama, Y.; Uchida, K.; Yamasaki, T.; Fujii, Y.; et al. Improved liver function in patients with liver cirrhosis after autologous bone marrow cell infusion therapy. STEM CELLS 2006, 24, 2292–2298. [Google Scholar] [CrossRef]
- Tanimoto, H.; Terai, S.; Taro, T.; Murata, Y.; Fujisawa, K.; Yamamoto, N.; Sakaida, I. Improvement of liver fibrosis by infusion of cultured cells derived from human bone marrow. Cell Tissue Res. 2013, 354, 717–728. [Google Scholar] [CrossRef]
- Sakaida, I.; Terai, S.; Yamamoto, N.; Aoyama, K.; Ishikawa, T.; Nishina, H.; Okita, K. Transplantation of bone marrow cells reduces CCl4-induced liver fibrosis in mice. Hepatology 2004, 40, 1304–1311. [Google Scholar] [CrossRef]
- Matsuda, T.; Takami, T.; Sasaki, R.; Nishimura, T.; Aibe, Y.; Paredes, B.D.; Quintanilha, L.F.; Matsumoto, T.; Ishikawa, T.; Yamamoto, N.; et al. A canine liver fibrosis model to develop a therapy for liver cirrhosis using cultured bone marrow-derived cells. Hepatol. Commun. 2017, 1, 691–703. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, T.; Takami, T.; Sasaki, R.; Aibe, Y.; Matsuda, T.; Fujisawa, K.; Matsumoto, T.; Yamamoto, N.; Tani, K.; Taura, Y.; et al. Liver regeneration therapy through the hepatic artery-infusion of cultured bone marrow cells in a canine liver fibrosis model. PLoS ONE 2019, 14, e0210588. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ding, Y.; Liu, Z.; Liang, X. Senescence in mesenchymal stem cells: Functional alterations, molecular mechanisms, and rejuvenation strategies. Front. Cell Dev. Biol. 2020, 8, 258. [Google Scholar] [CrossRef]
- Sidler, C.; Kovalchuk, O.; Kovalchuk, I. Epigenetic regulation of cellular senescence and aging. Front. Genet. 2017, 8, 138. [Google Scholar] [CrossRef]
- Hussain, A.; Tebyaniyan, H.; Khayatan, D. The role of epigenetic in dental and oral regenerative medicine by different types of dental stem cells: A comprehensive overview. Stem Cells Int. 2022, 2022, 5304860. [Google Scholar] [CrossRef]
- Wang, R.; Wang, Y.; Zhu, L.; Liu, Y.; Li, W. Epigenetic regulation in mesenchymal stem cell aging and differentiation and osteoporosis. Stem Cells Int. 2020, 2020, 8836258. [Google Scholar] [CrossRef]
- Bieback, K.; Schallmoser, K.; Klüter, H.; Strunk, D. Clinical protocols for the isolation and expansion of mesenchymal stromal cells. Transfus. Med. Hemotherapy 2008, 35, 4. [Google Scholar] [CrossRef] [PubMed]
- Zahedi, M.; Parham, A.; Dehghani, H.; Mehrjerdi, H.K. Equine bone marrow-derived mesenchymal stem cells: Optimization of cell density in primary culture. Stem Cell Investig. 2018, 5, 31. [Google Scholar] [CrossRef]
- Sotiropoulou, P.A.; Perez, S.A.; Salagianni, M.; Baxevanis, C.N.; Papamichail, M. Characterization of the optimal culture conditions for clinical scale production of human mesenchymal stem cells. Stem Cells 2005, 24, 462–471. [Google Scholar] [CrossRef]
- Pittenger, M.F.; Mackay, A.M.; Beck, S.C.; Jaiswal, R.K.; Douglas, R.; Mosca, J.D.; Moorman, M.A.; Simonetti, D.W.; Craig, S.; Marshak, D.R. Multilineage potential of adult human mesenchymal stem cells. Science 1999, 284, 143–147. [Google Scholar] [CrossRef]
- Fuhrmann, A.; Engler, A.J. The cytoskeleton regulates cell attachment strength. Biophys. J. 2015, 109, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.A.; Sung, J.R.; Yoon, S.O.; Ji, H.P.; Jung, W.L.; Kim, H.P.; Kyung, T.K.; Ik, S.J.; Sang, C.P. Morphological adjustment of senescent cells by modulating caveolin-1 status. J. Biol. Chem. 2004, 279, 42270–42278. [Google Scholar] [CrossRef] [PubMed]
- Sarugaser, R.; Hanoun, L.; Keating, A.; Stanford, W.L.; Davies, J.E. Human mesenchymal stem cells self-renew and differentiate according to a deterministic hierarchy. PLoS ONE 2009, 4, e6498. [Google Scholar] [CrossRef] [PubMed]
- Marceau, K.; Ruttle, P.L.; Shirtcliff, E.A.; Essex, M.J.; Susman, E.J.; Studies, A.; Hospital, R.I.; Studies, F.; Orleans, N. Human bone marrow stromal cell confluence: Effects on cell characteristics and methods of assessment. Cytotherapy 2015, 57, 742–768. [Google Scholar]
- Nakamura, K.; Tsuji, K.; Mizuno, M.; Koga, H.; Muneta, T.; Sekiya, I. Initial cell plating density affects properties of human primary synovial mesenchymal stem cells. J. Orthop. Res. 2019, 37, 1358–1367. [Google Scholar] [CrossRef]
- Mabuchi, Y.; Morikawa, S.; Harada, S.; Niibe, K.; Suzuki, S.; Renault-Mihara, F.; Houlihan, D.D.; Akazawa, C.; Okano, H.; Matsuzaki, Y. LNGFR+THY-1 + VCAM-1hi + cells reveal functionally distinct subpopulations in mesenchymal stem cells. Stem Cell Rep. 2013, 1, 152–165. [Google Scholar] [CrossRef]
- Mizuno, M.; Katano, H.; Shimozaki, Y.; Sanami, S.; Ozeki, N.; Koga, H.; Sekiya, I. Time-lapse image analysis for whole colony growth curves and daily distribution of the cell number per colony during the expansion of mesenchymal stem cells. Sci. Rep. 2019, 9, 16835. [Google Scholar] [CrossRef]
- Owens, R.B.; Smith, H.S.; Hackett, A.J. Epithelial cell cultures from normal glandular tissue of mice. J. Natl. Cancer Inst. 1974, 53, 261–269. [Google Scholar] [CrossRef]
- Tarifa, C.M.; Jiménez, G.; Garcia, M.A.; Entrena, J.M.; Griñán-Lisón, C.; Aguilera, M.; Picon-Ruiz, M.; Marchal, J.A. Low adherent cancer cell subpopulations are enriched in tumorigenic and metastatic epithelial-to-mesenchymal transition-induced cancer stem-like cells. Sci. Rep. 2016, 6, srep18772. [Google Scholar] [CrossRef]
- Segawa, Y.; Muneta, T.; Makino, H.; Nimura, A.; Mochizuki, T.; Ju, Y.-J.; Ezura, Y.; Umezawa, A.; Sekiya, I. Mesenchymal stem cells derived from synovium, meniscus, anterior cruciate ligament, and articular chondrocytes share similar gene expression profiles. J. Orthop. Res. 2009, 27, 435–441. [Google Scholar] [CrossRef]
- Rennerfeldt, D.A.; Van Vliet, K.J. Concise review: When colonies are not clones: Evidence and implications of intracolony heterogeneity in mesenchymal stem cells. Stem Cells 2016, 34, 1135–1141. [Google Scholar] [CrossRef]
- Bruedigam, C.; Eijken, M.; Koedam, M.; Peppel, J.; Drabek, K.; Chiba, H.; Leeuwen, J.P.T.M. A new concept underlying stem cell lineage skewing that explains the detrimental effects of thiazolidinediones on bone. Stem Cells 2010, 28, 916–927. [Google Scholar] [CrossRef]
- Gnani, D.; Crippa, S.; Volpe, L.D.; Rossella, V.; Conti, A.; Lettera, E.; Rivis, S.; Ometti, M.; Fraschini, G.; Bernardo, M.E.; et al. An early-senescence state in aged mesenchymal stromal cells contributes to hematopoietic stem and progenitor cell clonogenic impairment through the activation of a pro-inflammatory program. Aging Cell 2019, 18, e12933. [Google Scholar] [CrossRef] [PubMed]
- Andrä, I.; Ulrich, H.; Dürr, S.; Soll, D.; Henkel, L.; Angerpointner, C.; Ritter, J.; Przibilla, S.; Stadler, H.; Effenberger, M.; et al. An evaluation of T-cell functionality after flow cytometry sorting revealed p38 MAPK activation. Cytom. Part A 2020, 97, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Ryan, K.; Rose, R.E.; Jones, D.R.; Lopez, P.A. Sheath fluid impacts the depletion of cellular metabolites in cells afflicted by sorting induced cellular stress (SICS). Cytom. Part A 2021, 99, 921–929. [Google Scholar] [CrossRef]
- Pfister, G.; Toor, S.M.; Nair, V.S.; Elkord, E. An evaluation of sorter induced cell stress (SICS) on peripheral blood mononuclear cells (PBMCs) after different sort conditions—Are your sorted cells getting SICS? J. Immunol. Methods 2020, 487, 112902. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-N.; Choi, B.; Lee, C.-J.; Moon, J.H.; Kim, M.K.; Chung, E.; Song, S.U. Culturing at low cell density delays cellular senescence of human bone marrow-derived mesenchymal stem cells in long-term cultures. Int. J. Stem Cells 2020, 14, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Harada, S.; Mabuchi, Y.; Kohyama, J.; Shimojo, D.; Suzuki, S.; Kawamura, Y.; Araki, D.; Suyama, T.; Kajikawa, M.; Akazawa, C.; et al. FZD5 regulates cellular senescence in human mesenchymal stem/stromal cells. Stem Cells 2020, 39, 318–330. [Google Scholar] [CrossRef]
- Tushinski, R.J.; Oliver, I.T.; Guilbert, L.J.; Tynan, P.; Warner, J.R.; Stanley, E. Survival of mononuclear phagocytes depends on a lineage-specific growth factor that the differentiated cells selectively destroy. Cell 1982, 28, 71–81. [Google Scholar] [CrossRef]
- Takizawa, N.; Okubo, N.; Kamo, M.; Chosa, N.; Mikami, T.; Suzuki, K.; Yokota, S.; Ibi, M.; Ohtsuka, M.; Taira, M.; et al. Bone marrow-derived mesenchymal stem cells propagate immunosuppressive/anti-inflammatory macrophages in cell-to-cell contact-independent and dependent manners under hypoxic culture. Exp. Cell Res. 2017, 358, 411–420. [Google Scholar] [CrossRef]
| Gene Name | Primer Sequence (5′–3′) | Product Size (bp) |
|---|---|---|
| Ki67 | Forward: GGAACAGCCTCAACCATCAG | 210 |
| Reverse: CCACTCTTTCTCCCTCCTCTC | ||
| PCNA | Forward: TGGAGAACTTGGAAATGGAAA | 95 |
| Reverse: GAACTGGTTCATTCATCTCTATGG | ||
| β-actin | Forward: CGGGACCTGACTGACTACCT | 96 |
| Reverse: CTCCTTAATGTCACGCACGA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nagai, H.; Miwa, A.; Yoneda, K.; Fujisawa, K.; Takami, T. Optimizing the Seeding Density of Human Mononuclear Cells to Improve the Purity of Highly Proliferative Mesenchymal Stem Cells. Bioengineering 2023, 10, 102. https://doi.org/10.3390/bioengineering10010102
Nagai H, Miwa A, Yoneda K, Fujisawa K, Takami T. Optimizing the Seeding Density of Human Mononuclear Cells to Improve the Purity of Highly Proliferative Mesenchymal Stem Cells. Bioengineering. 2023; 10(1):102. https://doi.org/10.3390/bioengineering10010102
Chicago/Turabian StyleNagai, Hiroyuki, Akihiro Miwa, Kenji Yoneda, Koichi Fujisawa, and Taro Takami. 2023. "Optimizing the Seeding Density of Human Mononuclear Cells to Improve the Purity of Highly Proliferative Mesenchymal Stem Cells" Bioengineering 10, no. 1: 102. https://doi.org/10.3390/bioengineering10010102
APA StyleNagai, H., Miwa, A., Yoneda, K., Fujisawa, K., & Takami, T. (2023). Optimizing the Seeding Density of Human Mononuclear Cells to Improve the Purity of Highly Proliferative Mesenchymal Stem Cells. Bioengineering, 10(1), 102. https://doi.org/10.3390/bioengineering10010102







