1. Introduction
National statutory bodies responsible for monitoring water quality within water bodies must fulfil many, often competing and changing, policy objectives. To fulfil their remit, national monitoring programmes must be designed to have adequate spatial and temporal coverage for reliable assessment of water quality standards, while keeping costs of sample collection and analysis within reasonable limits. Primarily (though not exclusively), the aims of such programmes are to protect human health and prevent environmental degradation. In certain cases, water quality may be monitored more frequently, for example, to assess the cause of suspected deterioration or contamination events, or to examine the effectiveness of long-term environmental improvement measures. However, in the absence of specific needs, financial and human resource constraints limit the capacity for high-resolution monitoring throughout the national waterbody network; therefore, data are rarely available at a sufficiently high spatial resolution or temporal frequency to inform the implementation of site-specific measures to support a healthy aquatic environment. Additionally, there is no spatial or temporal uniformity to current routine monitoring at the river catchment level, and it is common to find large gaps in spatial and temporal records. Often, this leads to mitigative measures having to be put in place once negative impacts (e.g., disease outbreaks in human or animal populations; environmental degradation) are observed [
1,
2].
To assist with estimation of water quality in data-limited areas, models at the scale of individual river catchments have the potential to be used in combination with in situ monitoring data to assess the environmental characteristics and human activities that are likely to have the greatest influence on water quality. Such tools can help environmental managers understand both spatial and temporal variations in the sources and loadings of various substances entering watercourses, including nutrients, sediments, chemical compounds, and pathogenic organisms.
In England and Wales, there is a strong focus on hydrological and physico-chemical monitoring of water bodies. River flow gauges capture hydrological data, which are examined against historical records to identify and interpret emerging hydrological trends over time. The findings are used to improve water management strategies, increase awareness of water resources issues, and assess and report on flood and drought risk. Although the Environment Agency routinely monitors physico-chemical parameters (e.g., temperature, turbidity, pH, dissolved oxygen, and nitrates), monitoring of individual parameters can be sporadic and limited in certain locations. Additionally, few measurements relate to bacterial content, and monitoring of the faecal indicator Escherichia coli (E. coli) tends to be restricted to bathing waters, most of which are in coastal marine, rather than inland freshwater, locations: this is carried out only between March and October, during the UK ‘bathing season’, to protect human health. Other bacterial monitoring tends to be on a case basis, e.g., to support classification of shellfish harvesting areas for food safety purposes.
Faecal indicator bacteria, such as
E. coli, faecal coliforms, and enterococci, are used globally as proxies for the presence of faecally derived pathogens in freshwater bodies [
3]. Consequently, the use of high-quality monitoring data for these indicators as inputs to high-resolution catchment models can assist decision-making around effective mitigation measures for environmental improvement and protection of human health. Catchment models have become increasingly advanced and more widely available, resulting in a wealth of studies on estimating changes in surface run-off and water yield for flood mitigation and soil erosion purposes [
4,
5] and estimating chemical loadings, including nutrients, to riverine environments [
6,
7]. Incorporation of the fate of microbial or faecal indicator organisms is often less well advanced and, therefore, there are typically fewer studies on the assessment of the fate of bacterial loadings to waterbodies [
8,
9]. Faecal contamination of freshwater, estuarine and coastal waters can have various causes, but principal sources are faecal matter from humans (e.g., via point source wastewater discharges), agricultural livestock (e.g., via diffuse run-off from agricultural land), and wildlife (via direct deposition into waterbodies or diffuse terrestrial run-off). The magnitude and extent of both point source discharges and diffuse inputs can be influenced by environmental factors, such as topography and rainfall. For example, it is well known that episodic bacterial loads occur, often because of high flow conditions due to storm rainfall events [
10]. As a result, the tracing of sources and subsequent transmission pathways of microbiological pathogens is complex, not least due to their spatial and temporal variability. Therefore, successful reduction and mitigation of the negative impacts of such pathogens require routine detailed observation and analysis to enable predictive and mitigative measures to be focused effectively.
The work presented here formed one element of a wider project under the Pathogen Surveillance in Agriculture, Food and Environment (PATH-SAFE) programme [
11], which examined potential sources and pathways of pathogens implicated in foodborne disease. Although the programme focused on foodborne disease risk, the work has wider application to situations where microbial contamination of waterbodies poses a risk to human, animal or ecosystem health [
12,
13]. Through a case study at the scale of a river catchment in southwest England [
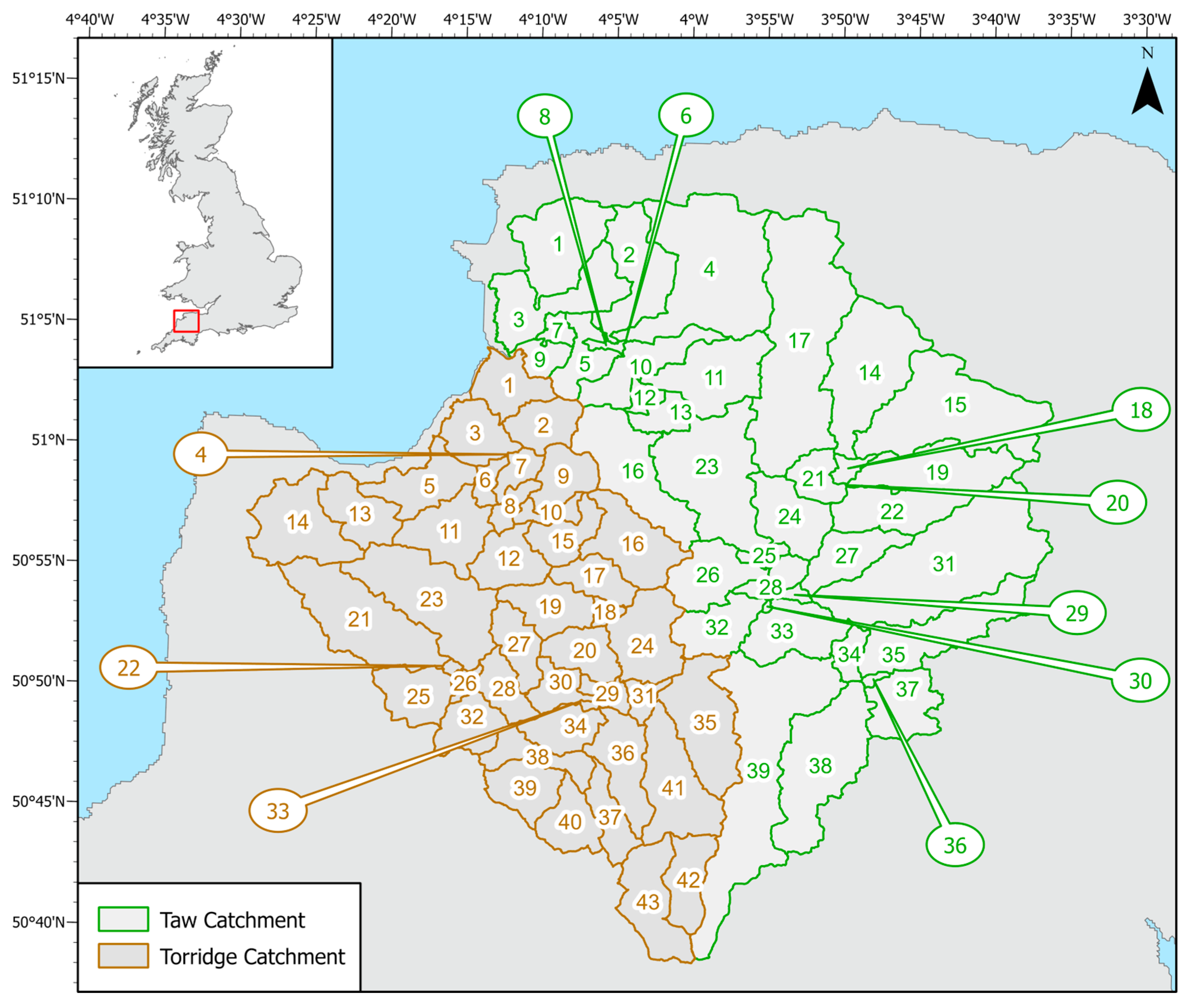
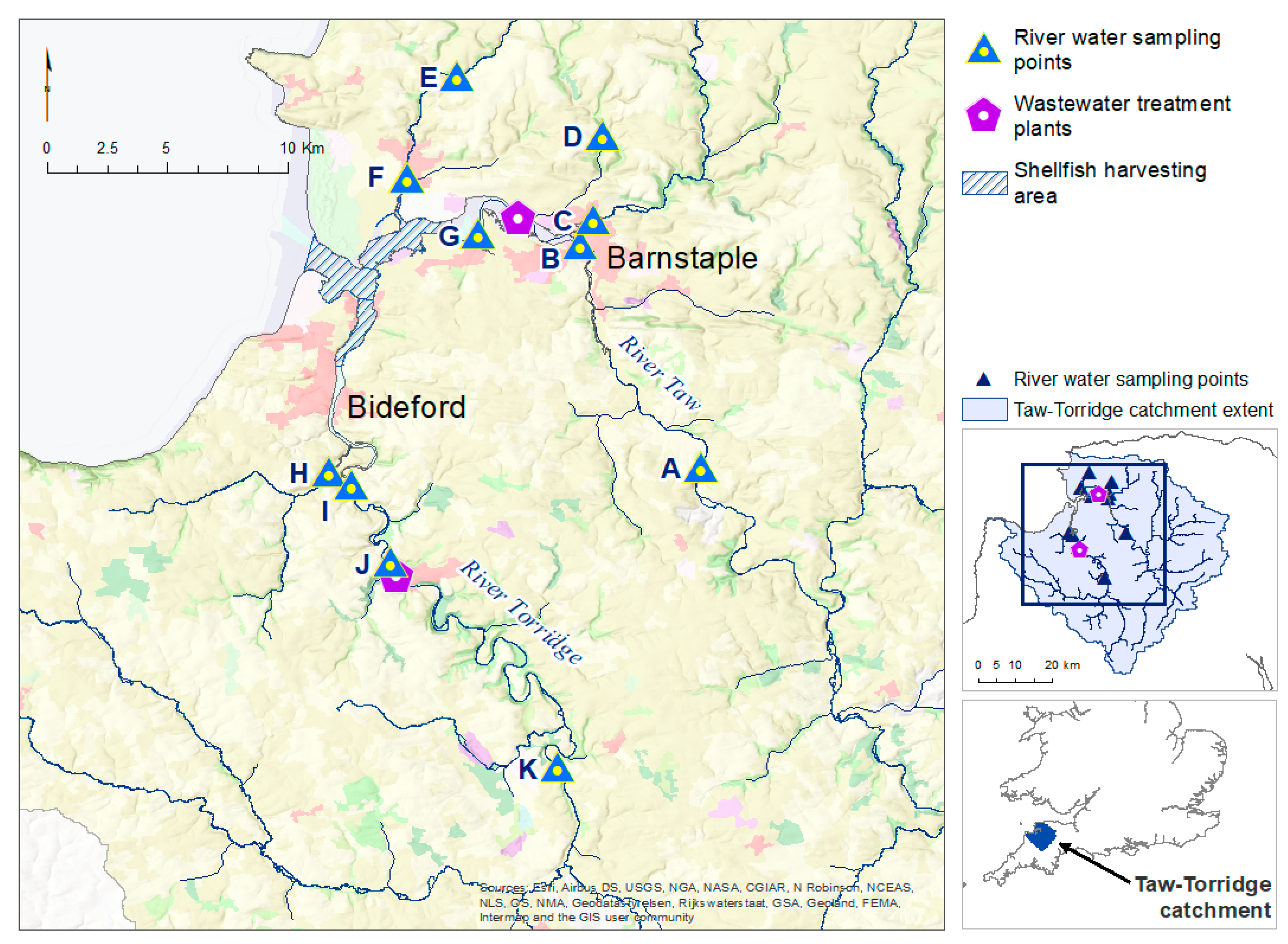
14], the work investigated how microbiological contaminants, from both diffuse and point sources, could be transported via overland run-off and watercourses to adjacent coastal waters, where high levels of microbiological contamination could pose a potential human health risk. The aim was to better understand pathogen transport and loadings in various parts of the river network, under different seasonal conditions, and to identify possible gaps and limitations in the existing monitoring framework, to provide recommendations for application to the national surveillance network.
SWAT (Soil and Water Assessment Tool) is a river basin hydrologic model used to simulate water flow, and nutrient and sediment mass transport, within a defined watershed [
15]. The model has been used extensively in studies worldwide to assess the influence of land use change [
16,
17], farm management practices [
18], and climate change [
17,
19] on water availability and nutrient contamination. More recently, the model has been used to model salinity changes [
20,
21] and microbial loads within river catchments [
22,
23]. Conceptually, the SWAT model is a continuous model, typically operating on a daily time scale, simulating water, transport processes of nutrients and other components (such as pesticides and bacteria) in surface run-off, soil percolation, soil lateral flow, groundwater flow with discharge to streams, and stream flow. Its approach is to divide a watershed into subbasins connected via a stream network. These subbasins are then divided into hydrological response units (HRUs), each representing a unique combination of land use, soil type, and topographic slope. Vegetative growth, management practices, and hydrology are simulated at the level of the HRU, driven by input data on climatic conditions (precipitation, temperature, humidity, solar radiation and wind), land use operations (for example, the application of fertilizer), and regional soil properties. At the level of the HRU, water, nutrients, sediment and other constituents (such as pesticides) are summarized and routed through the watershed via the stream network, eventually reaching the watershed outlet.
In addition, SWAT 2012 integrates a bacterial module that enables the fate and transport of two types of bacterial populations to be modelled concurrently, namely persistent and non-persistent bacteria. This allows bacterial (or viral) populations with distinct growth and die-off patterns to be incorporated, enabling separate consideration of bacteria that persist in the soil and those that do not. The bacterial module of SWAT includes the ability to incorporate loadings from a variety of sources, including livestock, wildlife, point sources (such as water company sewage discharges), and septic systems of private dwellings. Equations are employed to control the movement of bacteria from land to the stream network, and to model bacterial die-off and regrowth within the watercourse (reach) of each subbasin. Bacteria can enter the watercourse through surface run-off, especially important during rainfall events, or via sediment transport and/or resuspension from streambed sediments. The SWAT model assumes that bacteria present in the top 10 mm of the soil are available for transport under run-off conditions, and transport of bacteria via groundwater to streams is not considered, with bacteria in groundwater assumed to be lost to the system.
Using this case study as an example, our paper examines the challenge of combining diverse environmental data within a river catchment model to estimate diffuse and point source bacterial inputs into, and transport through, the system, with the aim of obtaining a close approximation to observed microbiological loadings. The following challenges are investigated and explored in this paper:
Challenge 1: Monitoring data on bacterial concentrations in riverine water are typically sparse both temporally and spatially. For the Taw and Torridge catchments, is seasonal variation evident within the monitored data, and how does it respond to rainfall events within the catchment?
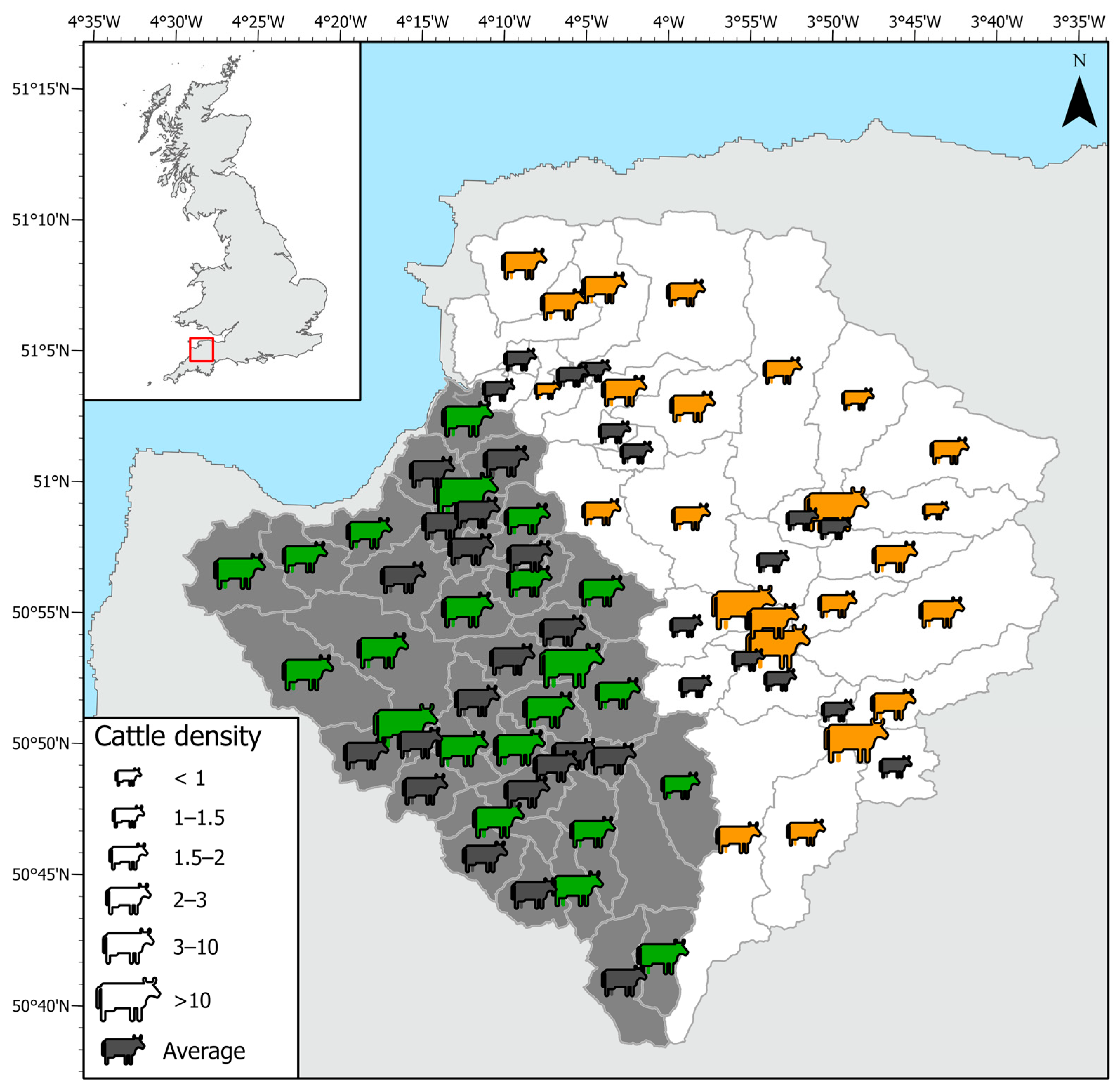
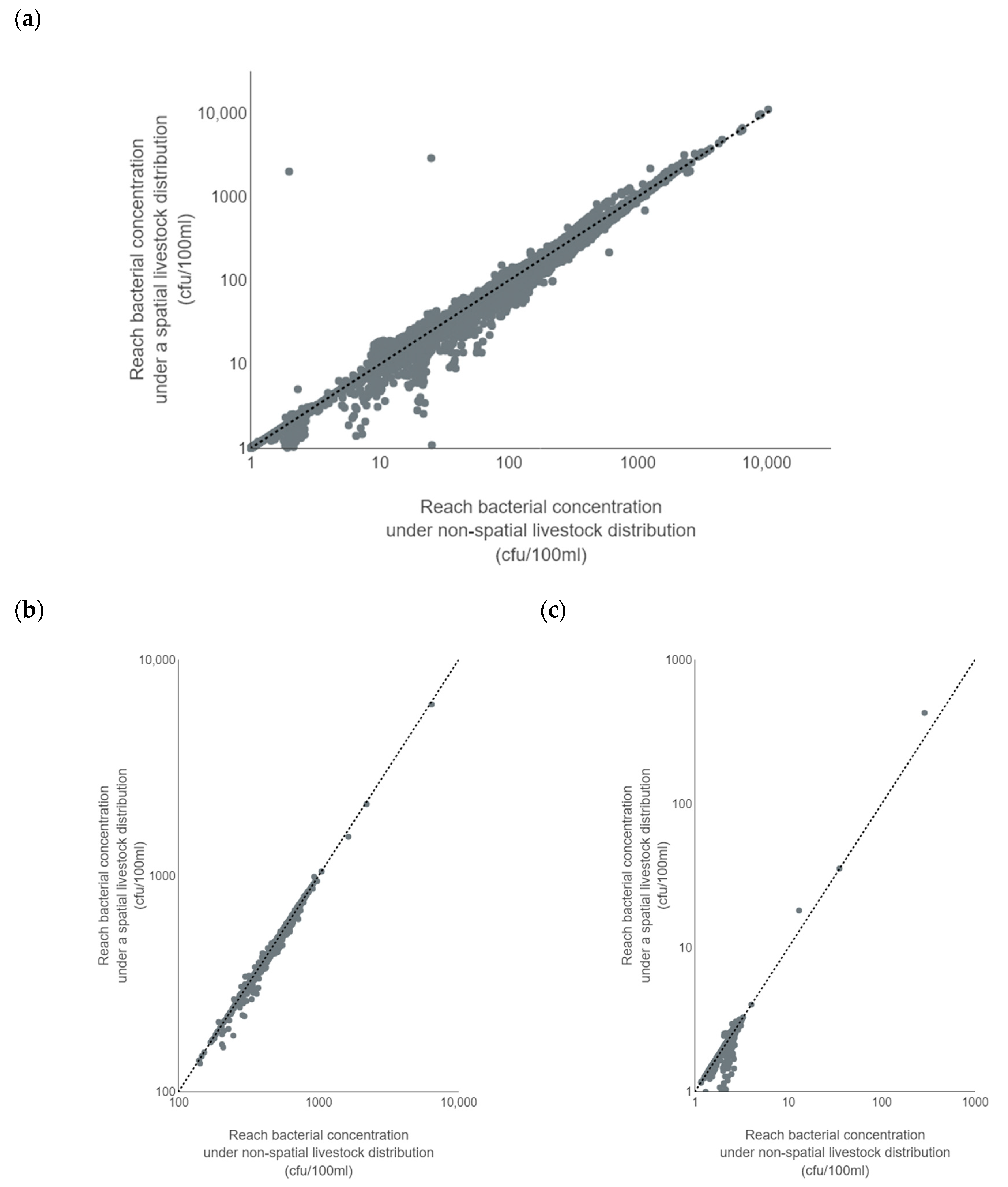
Challenge 2: Often, livestock numbers for cattle and sheep are apportioned equally across a catchment by land use. Does the spatial apportionment of livestock to specific areas within the catchment result in a significant improvement in modelling of bacteria?
Challenge 3: An effective catchment model should respond in a similar manner to observed data measured in the catchment. Does the bacterial modelling within SWAT respond in a similar way to rainfall events as seen in the monitoring data?
4. Discussion
Ideally, a national water quality monitoring programme should adopt a mix of monitoring approaches underpinned by scientific reasoning, to ensure that all objectives are met for all necessary water quality metrics and to allow for readjustment in response to policy or other changes. Breaking down the various national monitoring programmes at the catchment level offers an approach to answering questions about best practice for water quality monitoring within riverine environments and providing actionable information for policy standards. Not only does this provide the opportunity to ensure the best monitoring approach for the resources available, but it also presents the opportunity to fill in monitoring gaps by using output from a validated model.
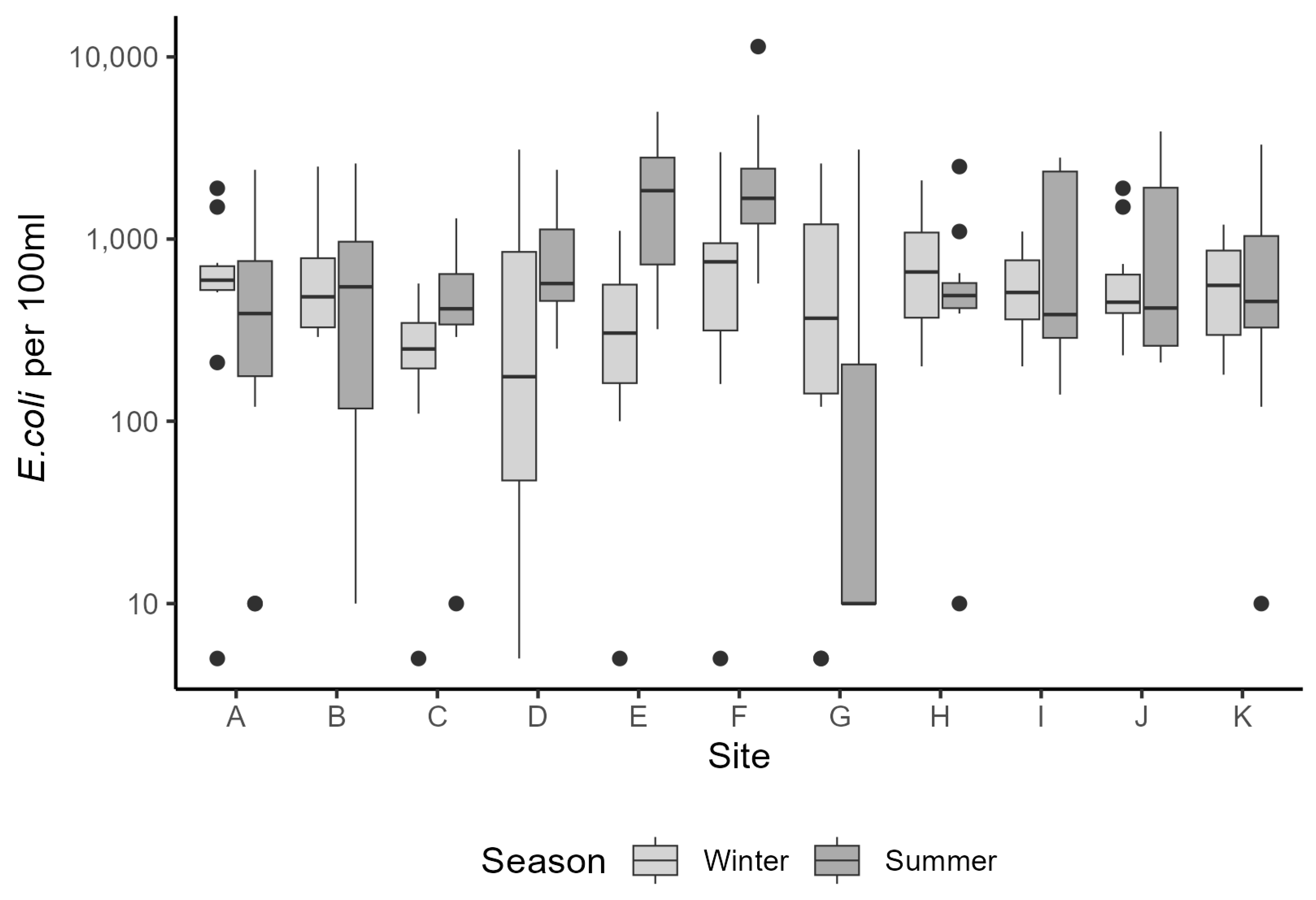
In our pilot study, Faecal Indicator Organism (FIO) data from the river water sampling were broadly in line with measured data from the national monitoring programme, albeit with higher
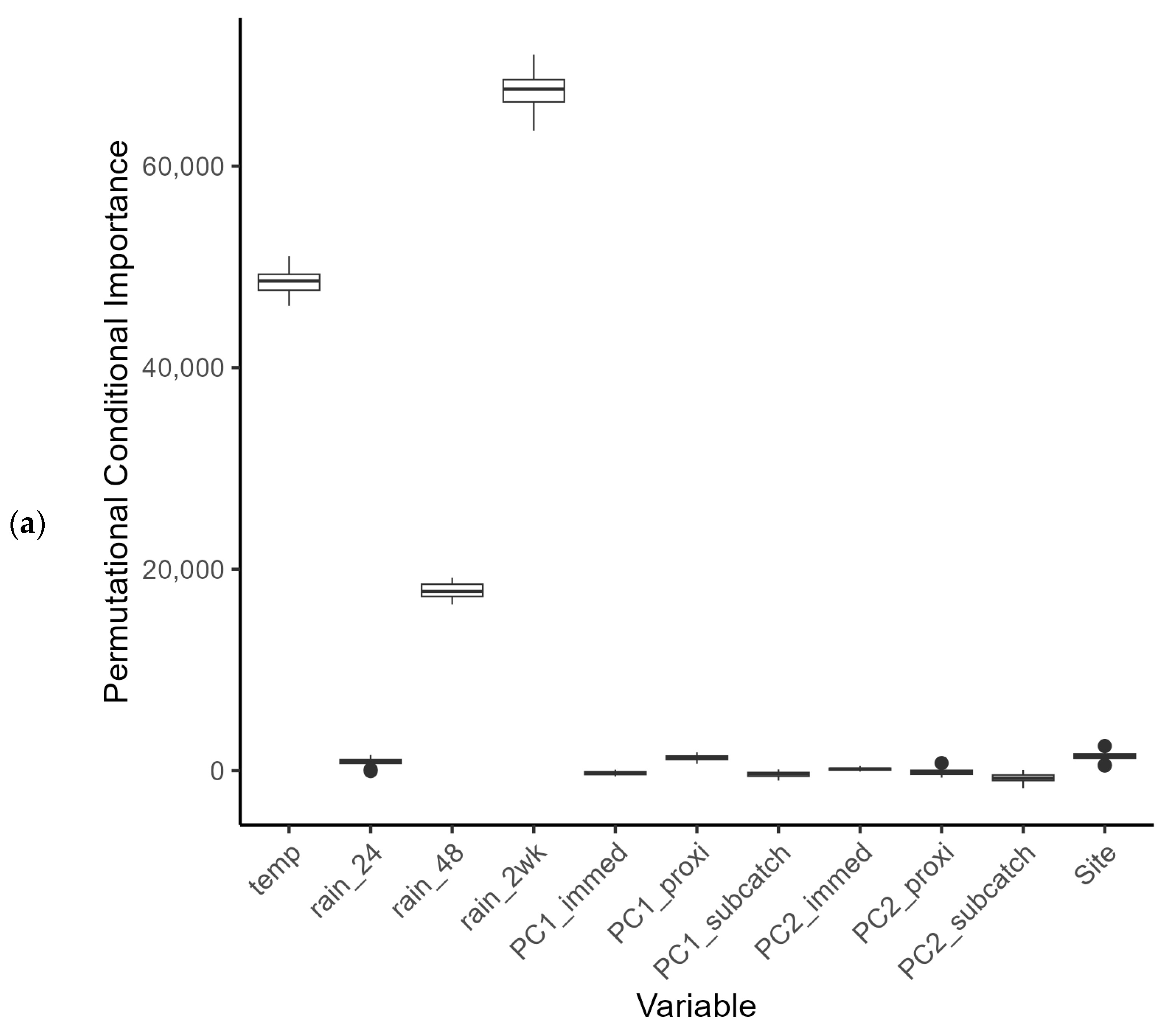
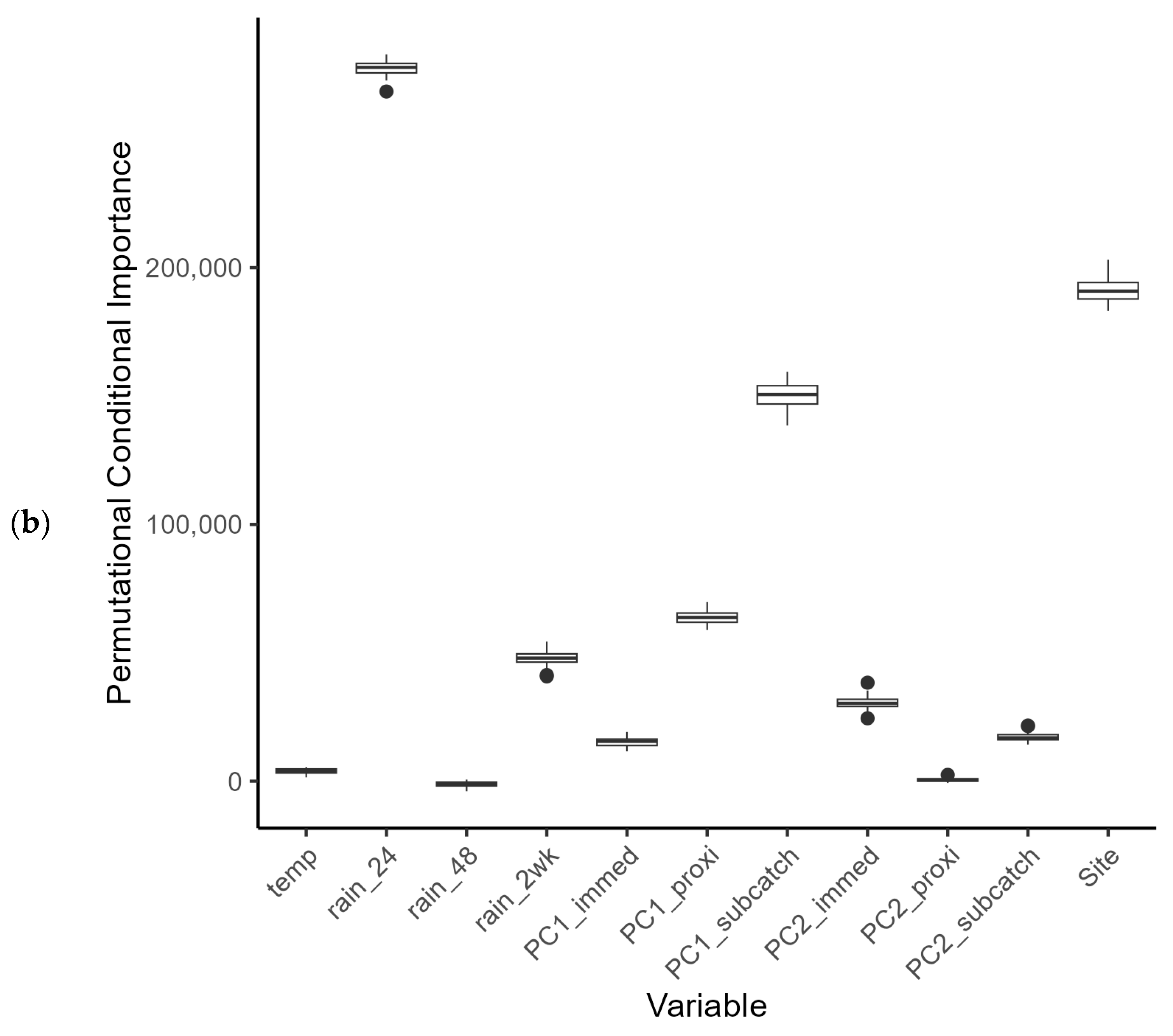
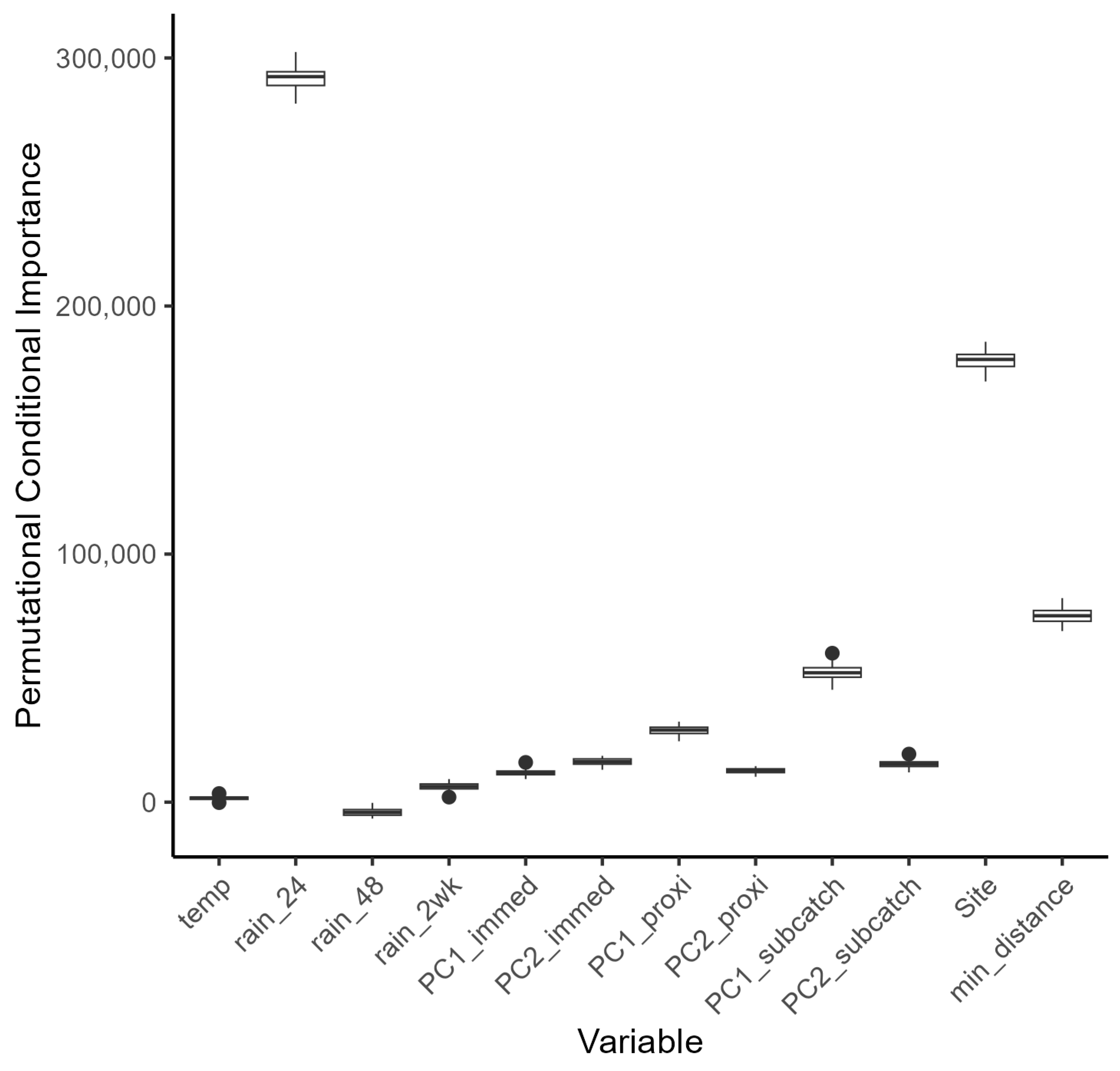
E. coli counts overall. Both data sets were subject to large variation, with several very high readings above 13,000 counts (with a mean count of 3202 cfu per 100 mL). At present it is difficult to know whether these fluctuations are genuine spikes in bacterial load, or whether they represent a confounding effect during the sampling or measurement process (although the latter is unlikely, since standard protocols were used for sampling and analysis). Riverine bacterial loads did show a partial dependence on rainfall events within the catchments that was subtly different between the summer and winter periods (Challenge 1—see Introduction). Observational data for bacterial loads in watercourses can be highly variable (for example, see [
22,
56,
57]) and in our study, variability was seen under the low flow regimen in summer and during peak flows in winter. This variation hampered statistical modelling based on catchment characteristics, but land use, more strictly land use within hydrological response units, was identified as a predictor variable for the summer period. Export of FIOs under dry and wet weather conditions has been shown to be associated with varying land use conditions [
10], with improved pastureland identified as a primary control factor in catchments in Scotland [
56]. During the UK summer, livestock are more likely to be outdoors, and those grazing in fields adjacent to watercourses are likely to add to the reservoir of faecal bacteria available to enter the river network [
58]. According to UK government statistics, in 2023 69% of farm holdings
always prevented livestock from entering watercourses, a percentage that has been rising since 2020 [
37]. However, around 8% of farm holdings
never prevented access, and this has remained relatively constant since 2020. It is well documented that quantities of faecal bacteria on fields, accumulated during long periods of dry weather, can be washed directly into watercourses by intense rainfall during subsequent summer storms, thereby causing large spikes in bacterial loadings [
59]. This may be a contributing factor to the variation in bacterial loads during the summer in our catchments.
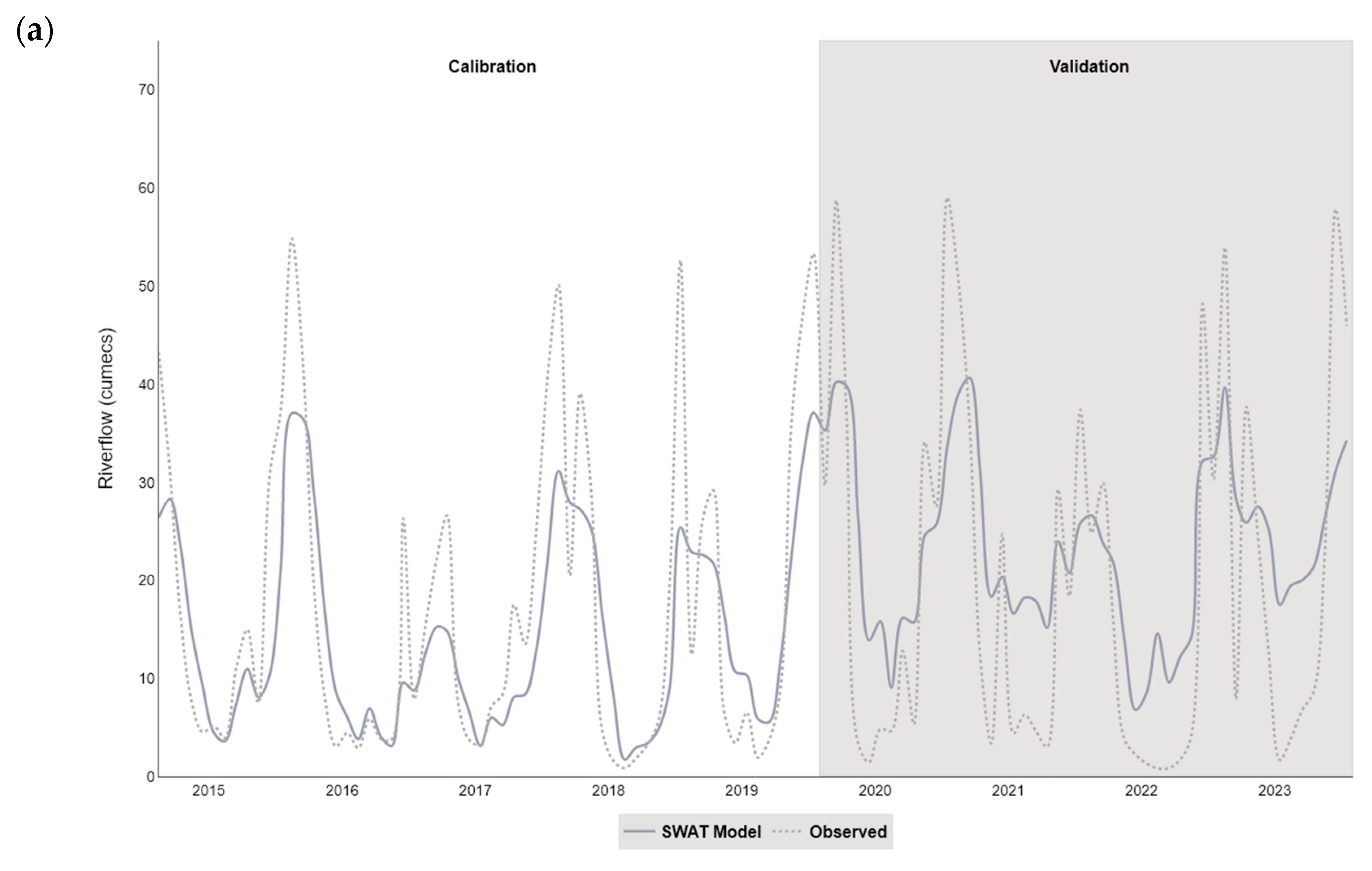
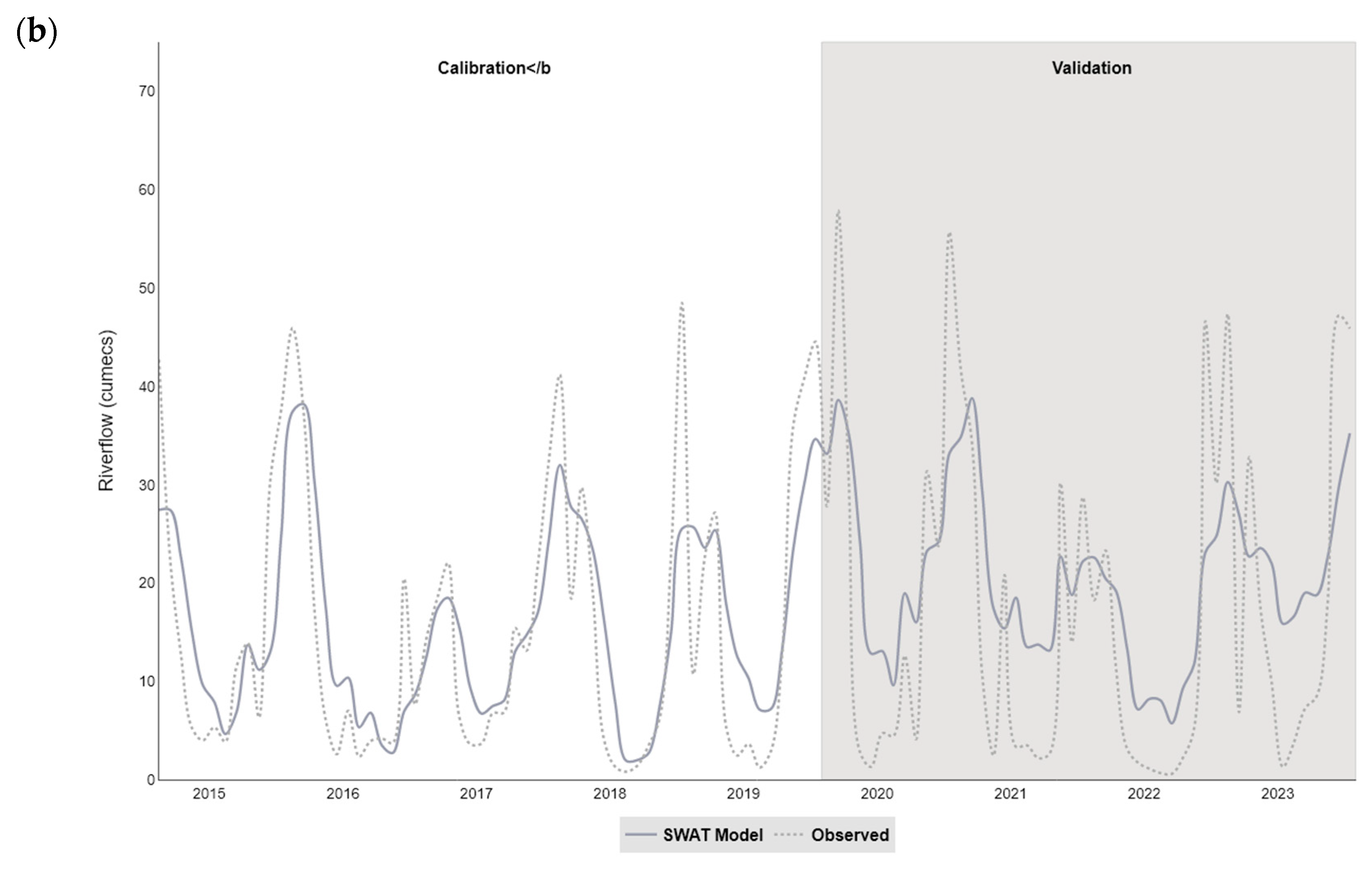
The catchment (SWAT) models developed in our study simulated the streamflow satisfactorily within the two catchments (NSE values of 0.62 for Taw and 0.65 for Torridge), and were more successful at modelling the monthly, rather than daily, time step. In particular, the models tended to overestimate flow during the summer months and underestimate flow in winter, a pattern that has been observed in studies using SWAT to model similar catchments (for example, see [
16,
22]). The SWAT 2012 architecture was superseded by SWAT+ [
60], with more emphasis on spatial connectivity between features within the watershed. However, both models are limited in groundwater-driven watersheds, and a groundwater flow module is available for SWAT+ [
61], which offers improvements to groundwater-surface water locations and rates, and nutrient loading via subsurface flow. Furthermore, the empirical curve number approach used in SWAT for rainfall-runoff simulation has recently come under scrutiny, and was outperformed by other methods for high-flow events [
62]. The inability to faithfully simulate high rainfall-runoff events represents a major limitation for modelling faecal bacterial fate and transport from livestock in SWAT. Accounting for measured flow, even in a relatively small catchment, can be problematic due to uncertainties surrounding the contributions from field drains and other anthropogenic sources [
57]. Whilst operation of SWAT at a sub-daily timestep appears to improve model efficiency for peak flows, this comes at a cost for medium flows, where the efficiency drops [
63].
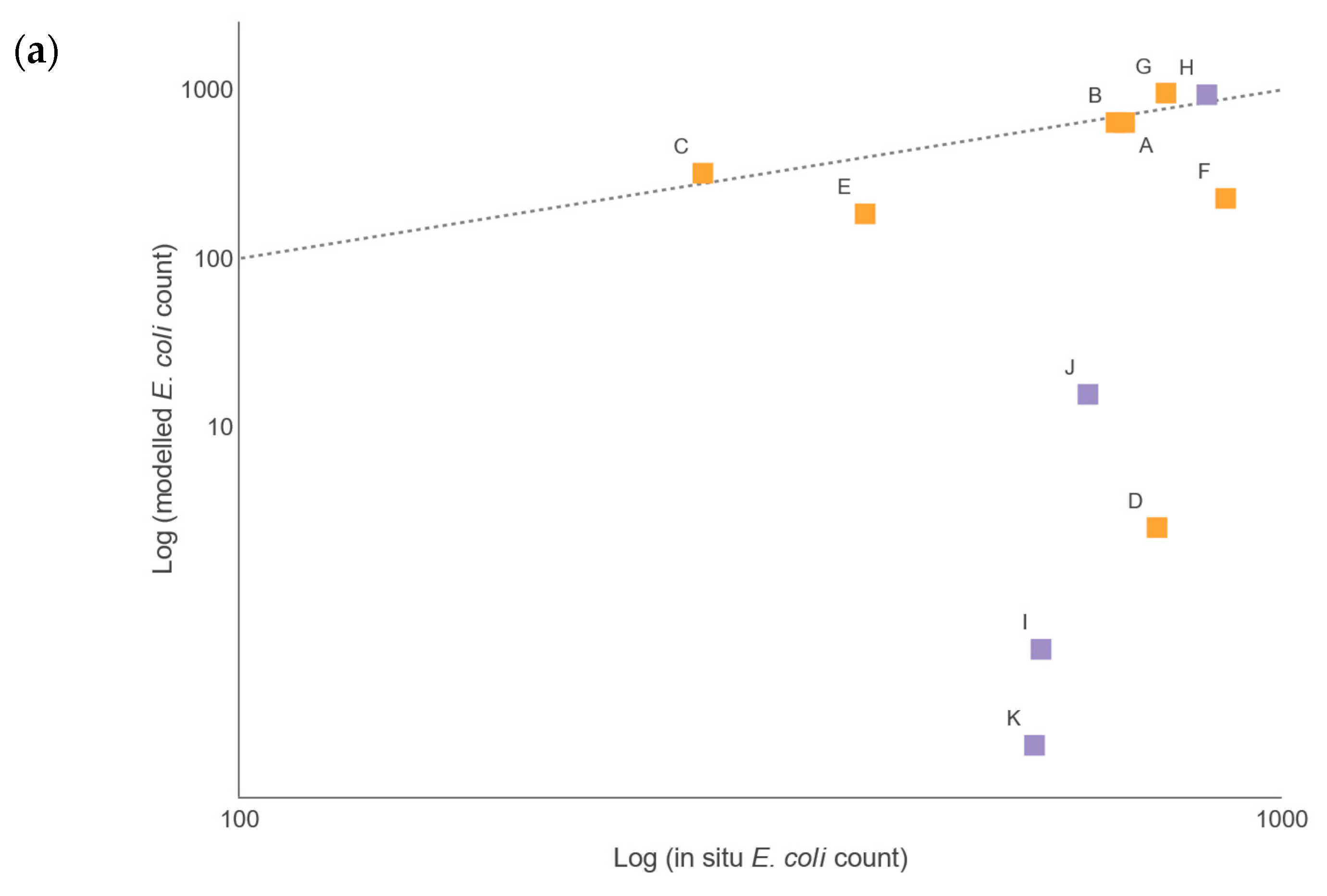
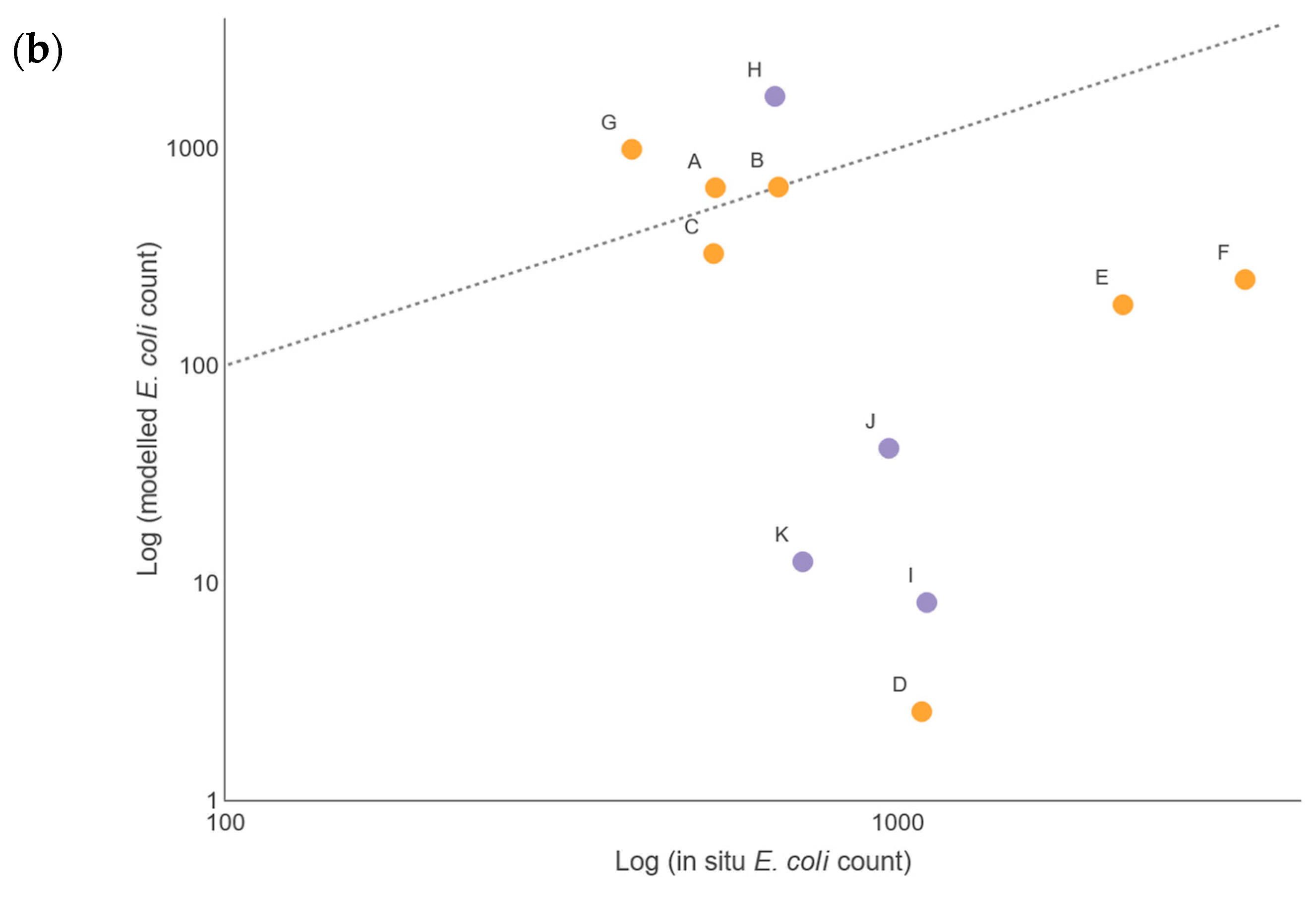
There were insufficient in situ observational data to validate our models for water quality assessment. In England, nutrient data (including nitrate, nitrite, ammonium, and ortho-phosphate) are collected monthly for many of the major rivers, but the collection points are rarely close to a river gauge, leading to greater inaccuracies in estimation of daily nutrient levels using flow rates. This hampers the creation and validation of accurate catchment models operating at a daily, or even sub-daily, time scale, a factor that is especially important with the increasing intensity of rainfall events in a changing climate. This has implications for modelling episodic bacterial contamination events that can occur under intense, short-duration rainfall events [
10,
56,
64] affected by antecedent wetness [
65], and was a severe limitation of the models developed in our study. Simulated bacterial counts at locations corresponding with the sampling sites showed reasonable levels of agreement for those rivers with good flow rates during the summer and winter periods. For example, the R
2 (Pearson) value for Sites A, B, C, and H was 0.64 and 0.95 for the respective summer and winter periods, when comparing the mean monthly simulated and observed bacterial loads. This comparison is often used to assess performance of SWAT models for bacterial load simulation (see [
22,
23,
64]). However, when comparing the daily values directly, the correlation coefficient dropped to 0.03 for summer and 0.34 for winter, revealing the limitations of the model to operate faithfully at the daily level. Nevertheless, the modelled data responded to rainfall in the catchments in a similar manner, such that for the winter period, the 2-week rainfall pattern could be used in part as a predictor of the bacterial loads, and for the summer period the 24-h and, to some extent, the 2-week rainfall pattern, were significant predictors. This partially satisfies Challenge 3 (see Introduction), as it suggests that catchment modelling using SWAT has many of the routines needed to simulate the fate of bacteria in the watershed. Future work will consider the use of sub-daily rainfall in the SWAT models to improve the resolution; however, rainfall patterns alone did not produce a satisfactory model of bacterial output in the watercourses, and it was clear that rainfall was not the only predictor. For sites in low-flow regimens, the agreement between simulated and observed data was less good, with the model underestimating the bacterial load. Coffey et al. [
22] observed a similar pattern for predictions of
E. coli in Irish catchments, with best agreement between predicted and observed data found for moderate flow rates. Therefore, when modelling it is critically important to understand the flow rates and bacterial loads across many different parts of a catchment [
57,
66]. Often, calibration and validation of catchment models focuses on the watershed outlet, and this has come under criticism recently [
62]. While this may seem intuitive, it exposes a major data gap for many regions and, while the model may show good agreement at the outlet, the upstream components may not reflect actual processes within the catchment. Recognition of this is important for policy advice and scenario testing and could be used to suggest sampling strategies to help fill the void.
Successful modelling of faecal coliforms within a catchment requires reliable information on the locations and loads for point source and diffuse inputs within a watershed. Implementation of diffuse source input from livestock can be problematic in the UK due to difficulties such as (1) obtaining precise numbers of animals for different livestock groups; (2) temporal and locational changes associated with the management of grazing livestock; (3) statistics only available for commercial farms; and (4) differences in livestock access to rivers, affecting the level of risk of faecal contamination. Livestock inputs are often incorporated into models by apportioning them evenly across a catchment as ‘average number of animals per hectare’ according to land use type (see, for example [
22,
23]). In this study, to investigate Challenge 2 (see Introduction), we explored spatial apportionment of livestock in the model that is more representative of their actual distribution within the catchment. A barrier to universal application of this approach is the limited availability of livestock data for small subsections of catchments (subbasins), or restrictions in areas where only a few farms are present for data protection and privacy concerns. This issue may become more pronounced with an increase in larger holdings in UK farming. In 2023, the average farm size was 69 ha for the southwest region of England, which includes the Taw and Torridge catchments [
67]. This was the smallest average farm size for all regions in England, and 19 ha smaller than the average English farm. Despite this our study indicates that, for the smaller upstream subbasins within a catchment, better spatial apportionment of livestock can have a significant effect on bacterial input to the model.
In theory, inclusion of point sources in the catchment model should be relatively easy as their locations are often well known, as is the case for the UK, where point source locations of licenced discharges to watercourses (consented discharges) and SOs are readily available [
39]. The major deficiency, however, is precise detail on what is being discharged, and when. For the consented continuous-treated effluent discharges, we used the dry weather flow as an estimate of daily discharge, but this assumes a steady discharge rate and does not account for discharges during wet flow conditions. Moreover, our model assumes that the spill pattern from SOs is related to rainfall within the catchment, although studies have suggested that SOs appear to spill more than the design criteria [
68,
69,
70]. Furthermore, there are notable uncertainties around the actual effluent volumes and concentration of faecal bacteria within a given volume of discharge, and these are exacerbated for SO spills during heavy rainfall events, where the discharge may range from raw sewage to highly diluted effluent [
71].
This study has highlighted some of the issues associated with setting up and refining models in data-limited situations. While modelling can assist with the estimation of water quality in unmonitored areas, the very existence of spatial and temporal gaps in established monitoring programmes creates challenges for model calibration and validation [
72,
73,
74]. A potential solution to this problem is the deployment of remote sampling devices (sondes) to increase spatiotemporal density of sampling while minimizing manpower costs of sample collection and processing. However, to date, such devices have had issues regarding their reliability, calibration, and biofouling (see [
75] for a review); therefore, siting them in optimal locations (for example, for ease of access, but without risk of vandalization) is key. Alternatively, a growing number of new initiatives involving citizen scientists (i.e., members of the public who volunteer to monitor and capture data in their locality) could help fill the gaps in formal monitoring programmes [
76].
With increased complexities surrounding the nature and scale of inputs into UK rivers from land use and other activities, there is a need for further development of catchment models to assess and predict the likely impacts of future scenarios, not only on riverine water quality, but also on that of estuarine and coastal waters [
64]. This paper has highlighted the challenges of creating a model representing the sources and pathways of faecal bacteria in a UK river catchment. In any such model there is considerable uncertainty in quantifying the bacterial input loads and spatial distribution of those inputs. This is especially pertinent when trying to assess the relationships between riverine loadings and intense rainfall events, for instance, which may produce wastewater spills or high volumes of run-off from the land, leading to potential shellfish hygiene issues and eutrophication of estuarine waters [
64]. This type of assessment requires analysis at a daily, if not higher, temporal resolution, with the uncertainty of outputs increasing at finer spatial resolutions of the sub-catchment. While the model inputs could be refined with better data, such as improved spatial apportionment of livestock, due to current data limitations, validation of UK catchment models is almost always restricted to the drainage outlet. As well as preventing fine tuning of the model upstream and failing to properly account for processes in the upstream reaches, this could be a significant issue when considering the likely impacts of different scenarios, including the influence of climate change, on faecal contamination downstream.