Direct Reprograming to Regenerate Myocardium and Repair Its Pacemaker and Conduction System
Abstract
1. Introduction
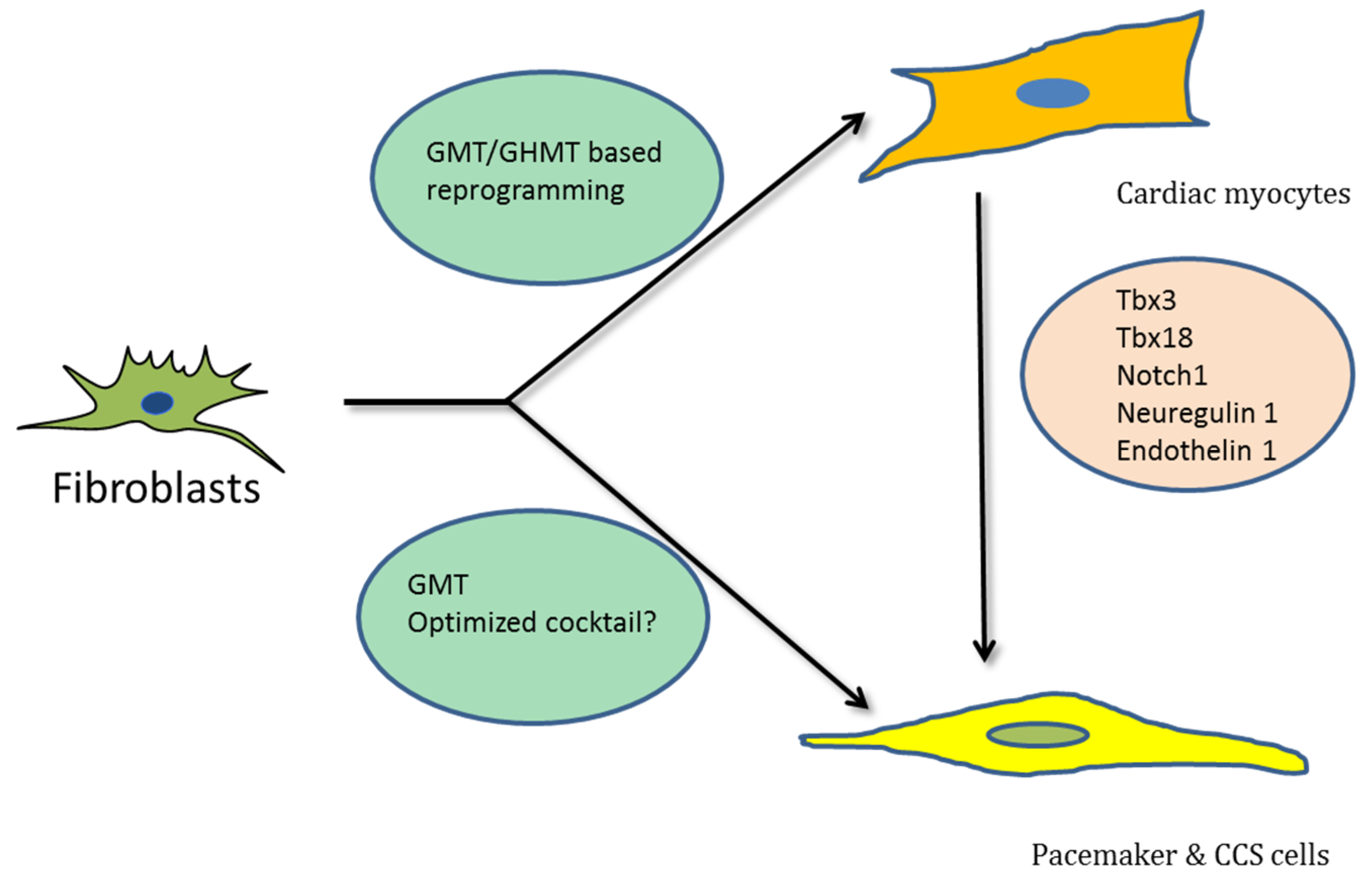
2. Direct Reprogramming for Cardiac Regeneration
2.1. Further Optimizing the TF Cocktail
2.2. Use of Small Molecules to Improve Reprogramming
2.3. Epigenetic Factors and RNA Splicing in iCM Generation
2.4. Direct Reprogramming of Human Fibroblasts
3. Reprogramming to Regenerate the CCS
4. Vector Systems Employed in Direct Reprogramming
5. Future Perspectives
6. Conclusions
Acknowledgments
Conflicts of Interest
Abbreviations
| TFs | Transcription factors |
| CMs | Cardiac myocytes |
| iCMs | induced cardiac myocytes |
| Micro RNAs | miRNAs |
| MCFs | Mouse cardiac fibroblasts |
| MEFs | Mouse embryonic fibroblasts |
| TTFs | Tail tip fibroblasts |
| GMT | Gata4, Mef2c, Tbx5 |
| iAMs | induced atrial myocytes |
| iVMs | induced ventricular myocytes |
| iPMs | induced pacemaker myocytes |
| HCFs | Human cardiac fibroblasts |
| CCS | cardiac conduction system |
References
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Devalla, H.D.; Schwach, V.; Ford, J.W.; Milnes, J.T.; El-Haou, S.; Jackson, C.; Gkatzis, K.; Elliott, D.A.; Chuva de Sousa Lopes, S.M.; Mummery, C.L.; et al. Atrial-like cardiomyocytes from human pluripotent stem cells are a robust preclinical model for assessing atrial-selective pharmacology. EMBO Mol. Med. 2015, 7, 394–410. [Google Scholar] [CrossRef] [PubMed]
- Karakikes, I.; Senyei, G.D.; Hansen, J.; Kong, C.W.; Azeloglu, E.U.; Stillitano, F.; Lieu, D.K.; Wang, J.; Ren, L.; Hulot, J.S.; et al. Small molecule-mediated directed differentiation of human embryonic stem cells toward ventricular cardiomyocytes. Stem Cells Transl. Med. 2014, 3, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Protze, S.I.; Liu, J.; Nussinovitch, U.; Ohana, L.; Backx, P.H.; Gepstein, L.; Keller, G.M. Sinoatrial node cardiomyocytes derived from human pluripotent cells function as a biological pacemaker. Nat. Biotechnol. 2017, 35, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Yamakawa, H.; Ieda, M. Strategies for heart regeneration: Approaches ranging from induced pluripotent stem cells to direct cardiac reprogramming. Int. Heart J. 2015, 56, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Brown, J.; Kanarek, A.; Rajagopal, J.; Melton, D.A. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature 2008, 455, 627–632. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, S.; Suzuki, A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 2011, 475, 390–393. [Google Scholar] [CrossRef] [PubMed]
- Vierbuchen, T.; Ostermeier, A.; Pang, Z.P.; Kokubu, Y.; Sudhof, T.C.; Wernig, M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 2010, 463, 1035–1041. [Google Scholar] [CrossRef] [PubMed]
- Ieda, M.; Fu, J.D.; Delgado-Olguin, P.; Vedantham, V.; Hayashi, Y.; Bruneau, B.G.; Srivastava, D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 2010, 142, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Boyett, M.R.; Inada, S.; Yoo, S.; Li, J.; Liu, J.; Tellez, J.; Greener, I.D.; Honjo, H.; Billeter, R.; Lei, M.; et al. Connexins in the sinoatrial and atrioventricular nodes. Adv. Cardiol. 2006, 42, 175–197. [Google Scholar] [PubMed]
- Efimov, I.R.; Fedorov, V.V.; Joung, B.; Lin, S.F. Mapping cardiac pacemaker circuits: methodological puzzles of the sinoatrial node optical mapping. Circ. Res. 2010, 106, 255–271. [Google Scholar] [CrossRef] [PubMed]
- Yaniv, Y.; Lakatta, E.G.; Maltsev, V.A. From two competing oscillators to one coupled-clock pacemaker cell system. Front. Physiol. 2015, 6, 28. [Google Scholar] [CrossRef] [PubMed]
- Sneddon, J.F.; Camm, A.J. Sinus node disease. Current concepts in diagnosis and therapy. Drugs 1992, 44, 728–737. [Google Scholar] [CrossRef] [PubMed]
- Vardas, P.E.; Auricchio, A.; Blanc, J.J.; Daubert, J.C.; Drexler, H.; Ector, H.; Gasparini, M.; Linde, C.; Morgado, F.B.; Oto, A.; et al. Guidelines for cardiac pacing and cardiac resynchronization therapy. The Task Force for Cardiac Pacing and Cardiac Resynchronization Therapy of the European Society of Cardiology. Developed in collaboration with the European Heart Rhythm Association. Europace 2007, 9, 959–998. [Google Scholar] [PubMed]
- Tellez, J.O.; McZewski, M.; Yanni, J.; Sutyagin, P.; Mackiewicz, U.; Atkinson, A.; Inada, S.; Beresewicz, A.; Billeter, R.; Dobrzynski, H.; et al. Ageing-dependent remodelling of ion channel and Ca2+ clock genes underlying sino-atrial node pacemaking. Exp. Physiol. 2011, 96, 1163–1178. [Google Scholar] [CrossRef] [PubMed]
- Csepe, T.A.; Kalyanasundaram, A.; Hansen, B.J.; Zhao, J.; Fedorov, V.V. Fibrosis: A structural modulator of sinoatrial node physiology and dysfunction. Front. Physiol. 2015, 6, 37. [Google Scholar] [CrossRef] [PubMed]
- Boink, G.J.; Christoffels, V.M.; Robinson, R.B.; Tan, H.L. The past, present, and future of pacemaker therapies. Trends Cardiovasc. Med. 2015, 25, 661–673. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, J.K.; Bruneau, B.G. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature 2009, 459, 708–711. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Huang, Y.; Spencer, C.I.; Foley, A.; Vedantham, V.; Liu, L.; Conway, S.J.; Fu, J.D.; Srivastava, D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 2012, 485, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Wang, L.; Yin, C.; Liu, J.; Qian, L. In vivo cardiac reprogramming using an optimal single polycistronic construct. Cardiovasc. Res. 2015, 108, 217–219. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; Nam, Y.J.; Luo, X.; Qi, X.; Tan, W.; Huang, G.N.; Acharya, A.; Smith, C.L.; Tallquist, M.D.; Neilson, E.G.; et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors1. Nature 2012, 485, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.X.; Krane, M.; Deutsch, M.A.; Wang, L.; Rav-Acha, M.; Gregoire, S.; Engels, M.C.; Rajarajan, K.; Karra, R.; Abel, E.D.; et al. Inefficient reprogramming of fibroblasts into cardiomyocytes using Gata4, Mef2c, and Tbx5. Circ. Res. 2012, 111, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Protze, S.; Khattak, S.; Poulet, C.; Lindemann, D.; Tanaka, E.M.; Ravens, U. A new approach to transcription factor screening for reprogramming of fibroblasts to cardiomyocyte-like cells. J. Mol. Cell. Cardiol. 2012, 53, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Addis, R.C.; Ifkovits, J.L.; Pinto, F.; Kellam, L.D.; Esteso, P.; Rentschler, S.; Christoforou, N.; Epstein, J.A.; Gearhart, J.D. Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. J. Mol. Cell. Cardiol. 2013, 60, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Dickson, M.E.; Kim, M.S.; Bassel-Duby, R.; Olson, E.N. Akt1/protein kinase B enhances transcriptional reprogramming of fibroblasts to functional cardiomyocytes. Proc. Natl. Acad. Sci. USA 2015, 112, 11864–11869. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Morales, M.G.; Hashimoto, H.; Dickson, M.E.; Song, K.; Ye, W.; Kim, M.S.; Niederstrasser, H.; Wang, Z.; Chen, B.; et al. ZNF281 enhances cardiac reprogramming by modulating cardiac and inflammatory gene expression. Genes Dev. 2017, 31, 1770–1783. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Ambasudhan, R.; Yuan, X.; Li, W.; Hilcove, S.; Abujarour, R.; Lin, X.; Hahm, H.S.; Hao, E.; Hayek, A.; et al. A chemical platform for improved induction of human iPSCs. Nat. Methods 2009, 6, 805–808. [Google Scholar] [CrossRef] [PubMed]
- Maherali, N.; Hochedlinger, K. Tgfbeta signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Current Biol. 2009, 19, 1718–1723. [Google Scholar] [CrossRef] [PubMed]
- Lian, X.; Hsiao, C.; Wilson, G.; Zhu, K.; Hazeltine, L.B.; Azarin, S.M.; Raval, K.K.; Zhang, J.; Kamp, T.J.; Palecek, S.P. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. USA 2012, 109, E1848–E1857. [Google Scholar] [CrossRef] [PubMed]
- Ifkovits, J.L.; Addis, R.C.; Epstein, J.A.; Gearhart, J.D. Inhibition of TGFbeta signaling increases direct conversion of fibroblasts to induced cardiomyocytes. PLoS ONE 2014, 9, e89678. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Londono, P.; Cao, Y.; Sharpe, E.J.; Proenza, C.; O’Rourke, R.; Jones, K.L.; Jeong, M.Y.; Walker, L.A.; Buttrick, P.M.; et al. High-efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro-fibrotic signalling. Nat. Commun. 2015, 6, 8243. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, T.M.; Stone, N.R.; Berry, E.C.; Radzinsky, E.; Huang, Y.; Pratt, K.; Ang, Y.S.; Yu, P.; Wang, H.; Tang, S.; et al. Chemical Enhancement of In Vitro and In Vivo Direct Cardiac Reprogramming. Circulation 2017, 135, 978–995. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, O.; Zheng, M.; Wang, L.; Zhou, Y.; Yin, C.; Liu, J.; Qian, L. Re-patterning of H3K27me3, H3K4me3 and DNA methylation during fibroblast conversion into induced cardiomyocytes. Stem Cell Res. 2016, 16, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, J.J.; Kieboom, K.; Marino, S.; DePinho, R.A.; van Lohuizen, M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 1999, 397, 164–168. [Google Scholar] [CrossRef] [PubMed]
- Park, I.K.; Morrison, S.J.; Clarke, M.F. Bmi1, stem cells, and senescence regulation. J. Clin. Investig. 2004, 113, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, L.; Vaseghi, H.R.; Liu, Z.; Lu, R.; Alimohamadi, S.; Yin, C.; Fu, J.D.; Wang, G.G.; Liu, J.; et al. Bmi1 Is a Key Epigenetic Barrier to Direct Cardiac Reprogramming. Cell Stem Cell 2016, 18, 382–395. [Google Scholar] [CrossRef] [PubMed]
- Wamstad, J.A.; Alexander, J.M.; Truty, R.M.; Shrikumar, A.; Li, F.; Eilertson, K.E.; Ding, H.; Wylie, J.N.; Pico, A.R.; Capra, J.A.; et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell 2012, 151, 206–220. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Lei, I.; Karatas, H.; Li, Y.; Wang, L.; Gnatovskiy, L.; Dou, Y.; Wang, S.; Qian, L.; Wang, Z. Targeting Mll1 H3K4 methyltransferase activity to guide cardiac lineage specific reprogramming of fibroblasts. Cell Discov. 2016, 2, 16036. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, Z.; Yin, C.; Zhou, Y.; Liu, J.; Qian, L. Improved Generation of Induced Cardiomyocytes Using a Polycistronic Construct Expressing Optimal Ratio of Gata4, Mef2c and Tbx5. J. Vis. Exp. 2015, 105. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, L.; Welch, J.D.; Ma, H.; Zhou, Y.; Vaseghi, H.R.; Yu, S.; Wall, J.B.; Alimohamadi, S.; Zheng, M.; et al. Single-cell transcriptomics reconstructs fate conversion from fibroblast to cardiomyocyte. Nature 2017, 551, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Alimohamadi, S.; Wang, L.; Liu, Z.; Wall, J.B.; Yin, C.; Liu, J.; Qian, L. A Loss of Function Screen of Epigenetic Modifiers and Splicing Factors during Early Stage of Cardiac Reprogramming. Stem Cells Int. 2018, 2018, 3814747. [Google Scholar] [CrossRef] [PubMed]
- Purvis, N.; Bahn, A.; Katare, R. The Role of MicroRNAs in Cardiac Stem Cells. Stem Cells Int. 2015, 2015, 194894. [Google Scholar] [CrossRef] [PubMed]
- Gangaraju, V.K.; Lin, H. MicroRNAs: Key regulators of stem cells. Nat. Rev. Mol. Cell. Biol. 2009, 10, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Jayawardena, T.M.; Egemnazarov, B.; Finch, E.A.; Zhang, L.; Payne, J.A.; Pandya, K.; Zhang, Z.; Rosenberg, P.; Mirotsou, M.; Dzau, V.J. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ. Res. 2012, 110, 1465–1473. [Google Scholar] [CrossRef] [PubMed]
- Nam, Y.J.; Song, K.; Luo, X.; Daniel, E.; Lambeth, K.; West, K.; Hill, J.A.; DiMaio, J.M.; Baker, L.A.; Bassel-Duby, R.; et al. Reprogramming of human fibroblasts toward a cardiac fate. Proc. Natl. Acad. Sci. USA 2013, 110, 5588–5593. [Google Scholar] [CrossRef] [PubMed]
- Wada, R.; Muraoka, N.; Inagawa, K.; Yamakawa, H.; Miyamoto, K.; Sadahiro, T.; Umei, T.; Kaneda, R.; Suzuki, T.; Kamiya, K.; et al. Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc. Natl. Acad. Sci. USA 2013, 110, 12667–12672. [Google Scholar] [CrossRef] [PubMed]
- Muraoka, N.; Yamakawa, H.; Miyamoto, K.; Sadahiro, T.; Umei, T.; Isomi, M.; Nakashima, H.; Akiyama, M.; Wada, R.; Inagawa, K.; et al. MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J. 2014, 33, 1565–1581. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.P.; Mathison, M.; Patel, V.; Sanagasetti, D.; Gibson, B.W.; Yang, J.; Rosengart, T.K. MiR-590 Promotes Transdifferentiation of Porcine and Human Fibroblasts Toward a Cardiomyocyte-Like Fate by Directly Repressing Specificity Protein 1. J. Am. Heart Assoc. 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, K.; Akiyama, M.; Tamura, F.; Isomi, M.; Yamakawa, H.; Sadahiro, T.; Muraoka, N.; Kojima, H.; Haginiwa, S.; Kurotsu, S.; et al. Direct In Vivo Reprogramming with Sendai Virus Vectors Improves Cardiac Function after Myocardial Infarction. Cell Stem Cell 2018, 22, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.D.; Stone, N.R.; Liu, L.; Spencer, C.I.; Qian, L.; Hayashi, Y.; Delgado-Olguin, P.; Ding, S.; Bruneau, B.G.; Srivastava, D. Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem Cell Rep. 2013, 1, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Nam, Y.J.; Lubczyk, C.; Bhakta, M.; Zang, T.; Fernandez-Perez, A.; McAnally, J.; Bassel-Duby, R.; Olson, E.N.; Munshi, N.V. Induction of diverse cardiac cell types by reprogramming fibroblasts with cardiac transcription factors. Development 2014, 141, 4267–4278. [Google Scholar] [CrossRef] [PubMed]
- Van Weerd, J.H.; Christoffels, V.M. The formation and function of the cardiac conduction system. Development 2016, 143, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Hoogaars, W.M.; Tessari, A.; Moorman, A.F.; de Boer, P.A.; Hagoort, J.; Soufan, A.T.; Campione, M.; Christoffels, V.M. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovasc. Res. 2004, 62, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Hoogaars, W.M.; Engel, A.; Brons, J.F.; Verkerk, A.O.; de Lange, F.J.; Wong, L.Y.; Bakker, M.L.; Clout, D.E.; Wakker, V.; Barnett, P.; et al. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 2007, 21, 1098–1112. [Google Scholar] [CrossRef] [PubMed]
- Bakker, M.L.; Boink, G.J.; Boukens, B.J.; Verkerk, A.O.; van den Boogaard, M.; den Haan, A.D.; Hoogaars, W.M.; Buermans, H.P.; de Bakker, J.M.; Seppen, J.; et al. T-box transcription factor TBX3 reprograms mature cardiac myocytes into pacemaker-like cells. Cardiovasc. Res. 2012, 94, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, N.; Liang, W.; Marban, E.; Cho, H.C. Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat. Biotechnol. 2013, 31, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.F.; Dawkins, J.F.; Cho, H.C.; Marban, E.; Cingolani, E. Biological pacemaker created by minimally invasive somatic reprogramming in pigs with complete heart block. Sci. Transl. Med. 2014, 6, 245ra94. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Gonzalez, S.; Lehman, J.; Cho, H.C. Stable and synchronous pacing generated from the Tbx18-induced pacemaker cells by epithelial-to-mesenchymal transition inhibition. Circulation 2016, 134, A18724. [Google Scholar]
- Kim, N.K.; Li, J.; Wolfson, D.; Fernandez, N.; Gu, J.; Han, P.; Grijalva, S.; Cho, H.C. Stable in vivo ventricular pacing created by Tbx18-induced pacemaker cells upon inhibition of epithelial-to-mesenchymal transformation in an ambulatory rat model of complete atrioventricular block. Circulation 2017, 136, A17025. [Google Scholar]
- Rentschler, S.; Harris, B.S.; Kuznekoff, L.; Jain, R.; Manderfield, L.; Lu, M.M.; Morley, G.E.; Patel, V.V.; Epstein, J.A. Notch signaling regulates murine atrioventricular conduction and the formation of accessory pathways. J. Clin. Investig. 2011, 121, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Rentschler, S.; Yen, A.H.; Lu, J.; Petrenko, N.B.; Lu, M.M.; Manderfield, L.J.; Patel, V.V.; Fishman, G.I.; Epstein, J.A. Myocardial Notch signaling reprograms cardiomyocytes to a conduction-like phenotype. Circulation 2012, 126, 1058–1066. [Google Scholar] [CrossRef] [PubMed]
- Gillers, B.S.; Chiplunkar, A.; Aly, H.; Valenta, T.; Basler, K.; Christoffels, V.M.; Efimov, I.R.; Boukens, B.J.; Rentschler, S. Canonical wnt signaling regulates atrioventricular junction programming and electrophysiological properties. Circ. Res. 2015, 116, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Rentschler, S.; Zander, J.; Meyers, K.; France, D.; Levine, R.; Porter, G.; Rivkees, S.A.; Morley, G.E.; Fishman, G.I. Neuregulin-1 promotes formation of the murine cardiac conduction system. Proc. Natl. Acad. Sci. USA 2002, 99, 10464–10469. [Google Scholar] [CrossRef] [PubMed]
- Shekhar, A.; Lin, X.; Liu, F.Y.; Zhang, J.; Mo, H.; Bastarache, L.; Denny, J.C.; Cox, N.J.; Delmar, M.; Roden, D.M.; et al. Transcription factor ETV1 is essential for rapid conduction in the heart. J. Clin. Investig. 2016, 126, 4444–4459. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Freed, C.R. Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem Cells 2009, 27, 2667–2674. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Jayandharan, G.R.; Li, B.; Ling, C.; Ma, W.; Srivastava, A.; Zhong, L. High-efficiency transduction of fibroblasts and mesenchymal stem cells by tyrosine-mutant AAV2 vectors for their potential use in cellular therapy. Hum. Gene Ther. 2010, 21, 1527–1543. [Google Scholar] [CrossRef] [PubMed]
- Umei, T.C.; Yamakawa, H.; Muraoka, N.; Sadahiro, T.; Isomi, M.; Haginiwa, S.; Kojima, H.; Kurotsu, S.; Tamura, F.; Osakabe, R.; et al. Single-Construct Polycistronic Doxycycline-Inducible Vectors Improve Direct Cardiac Reprogramming and Can Be Used to Identify the Critical Timing of Transgene Expression. Int. J. Mol. Sci. 2017, 18, 1805. [Google Scholar] [CrossRef] [PubMed]
- Merentie, M.; Rissanen, R.; Lottonen-Raikaslehto, L.; Huusko, J.; Gurzeler, E.; Turunen, M.P.; Holappa, L.; Makinen, P.; Yla-Herttuala, S. Doxycycline modulates VEGF-A expression: Failure of doxycycline-inducible lentivirus shRNA vector to knockdown VEGF-A expression in transgenic mice. PLoS ONE 2018, 13, e0190981. [Google Scholar] [CrossRef] [PubMed]
- Tse, H.F.; Xue, T.; Lau, C.P.; Siu, C.W.; Wang, K.; Zhang, Q.Y.; Tomaselli, G.F.; Akar, F.G.; Li, R.A. Bioartificial sinus node constructed via in vivo gene transfer of an engineered pacemaker HCN Channel reduces the dependence on electronic pacemaker in a sick-sinus syndrome model. Circulation 2006, 114, 1000–1011. [Google Scholar] [CrossRef] [PubMed]
- Donahue, J.K.; Heldman, A.W.; Fraser, H.; McDonald, A.D.; Miller, J.M.; Rade, J.J.; Eschenhagen, T.; Marban, E. Focal modification of electrical conduction in the heart by viral gene transfer. Nat. Med. 2000, 6, 1395–1398. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, E.; Yee, K.; Shehata, M.; Chugh, S.S.; Marban, E.; Cho, H.C. Biological pacemaker created by percutaneous gene delivery via venous catheters in a porcine model of complete heart block. Heart Rhythm 2012, 9, 1310–1318. [Google Scholar] [CrossRef] [PubMed]
- Bucchi, A.; Plotnikov, A.N.; Shlapakova, I.; Danilo, P., Jr.; Kryukova, Y.; Qu, J.; Lu, Z.; Liu, H.; Pan, Z.; Potapova, I.; et al. Wild-type and mutant HCN channels in a tandem biological-electronic cardiac pacemaker. Circulation 2006, 114, 992–999. [Google Scholar] [CrossRef] [PubMed]
- Boink, G.J.; Nearing, B.D.; Shlapakova, I.N.; Duan, L.; Kryukova, Y.; Bobkov, Y.; Tan, H.L.; Cohen, I.S.; Danilo, P., Jr.; Robinson, R.B.; et al. Ca(2+)-stimulated adenylyl cyclase AC1 generates efficient biological pacing as single gene therapy and in combination with HCN2. Circulation 2012, 126, 528–536. [Google Scholar] [CrossRef] [PubMed]
- Boink, G.J.; Duan, L.; Nearing, B.D.; Shlapakova, I.N.; Sosunov, E.A.; Anyukhovsky, E.P.; Bobkov, E.; Kryukova, Y.; Ozgen, N.; Danilo, P., Jr.; et al. HCN2/SkM1 gene transfer into canine left bundle branch induces stable, autonomically responsive biological pacing at physiological heart rates. J. Am. Coll. Cardiol. 2013, 61, 1192–1201. [Google Scholar] [CrossRef] [PubMed]
| Reprogramming Factors | Weeks Post MI | Ejection Fraction | Stroke Volume | Scar Area (%) | References |
|---|---|---|---|---|---|
| G/M/T vs. dsRed group | 12 w | ~32% vs. 22% | ~42 mL/min vs. 30 mL/min | ~18% vs. 40% | [19] |
| G/M/T/H vs. GFP group | 12 w | ~58% vs. 30% | ~55 µL vs. 40 µL | ~18% vs. 40% | [21] |
| MGT vs. G/M/T | 8 w | ~38% vs. 24% | ND | ~18% vs. 28% * | [20] |
| Reprogramming Factors | Cell Type | Reprogramming Read-Out | Reprogramming Efficiency | Beating Cells | References |
|---|---|---|---|---|---|
| Gata4, Hand2, Tbx5, Myocardin, miR-1, miR-133 | Human neonatal FFs, adult CFs and DFs | cTNT | 13% | + (Rare) | [45] |
| GMT, Mesp1, Myocardin | Human CFs and DFs | Multiple cardiac gene expression, sarcomeric organization structure and calcium oscillations | 6% | - | [46] |
| GMT-Mesp1 or GMT, miR-133a | Mouse & Human CFs | a-actinin protein, c-TnT calcium oscillations | 10% (mouse) & 8% (human) | + | [47] |
| GMT, Hand2, Myocardin or GMT, miR-590 | Porcine & Human CFs | cTNT | 5% | + | [48] |
| SeV-GMT/H | Human CFs | cTNT | 15% | + | [49] |
| GMT, ESRRG, MESP1, Myocardin, ZFPM2 | Human ESC derived fibroblasts, neonatal and skin fibroblasts | cTNT | 5% | - | [50] |
| GMT, ESRRG, MESP1, Myocardin, ZFPM2, TGFβ inhibitor, Wnt inhibitor | Adult HCF cell line | cTNT | 12% | + | [51] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adepu, S.; Oosterwerff, E.F.J.; Christoffels, V.M.; Boink, G.J.J. Direct Reprograming to Regenerate Myocardium and Repair Its Pacemaker and Conduction System. Medicines 2018, 5, 48. https://doi.org/10.3390/medicines5020048
Adepu S, Oosterwerff EFJ, Christoffels VM, Boink GJJ. Direct Reprograming to Regenerate Myocardium and Repair Its Pacemaker and Conduction System. Medicines. 2018; 5(2):48. https://doi.org/10.3390/medicines5020048
Chicago/Turabian StyleAdepu, Saritha, Erik F. J. Oosterwerff, Vincent M. Christoffels, and Gerard J. J. Boink. 2018. "Direct Reprograming to Regenerate Myocardium and Repair Its Pacemaker and Conduction System" Medicines 5, no. 2: 48. https://doi.org/10.3390/medicines5020048
APA StyleAdepu, S., Oosterwerff, E. F. J., Christoffels, V. M., & Boink, G. J. J. (2018). Direct Reprograming to Regenerate Myocardium and Repair Its Pacemaker and Conduction System. Medicines, 5(2), 48. https://doi.org/10.3390/medicines5020048




