Insights into the Toxicity and Degradation Mechanisms of Imidacloprid Via Physicochemical and Microbial Approaches
Abstract
1. Introduction
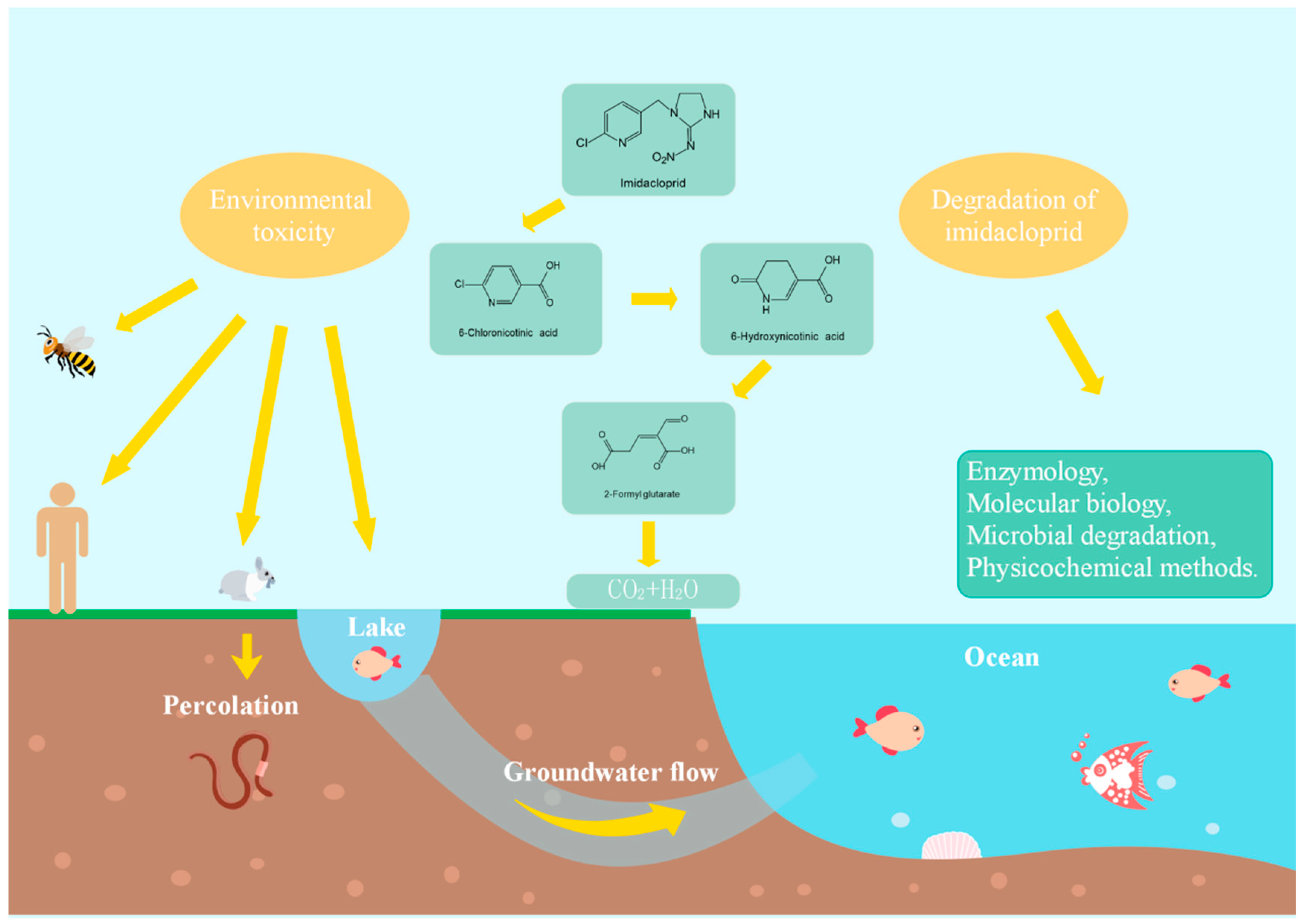
2. Toxicity of Imidacloprid
2.1. Toxicity to Humans and Terrestrial Animals
2.2. Toxicity to Aquatic Organisms
2.3. Toxicity to Insects
2.4. Synergistic Effects of Pesticides
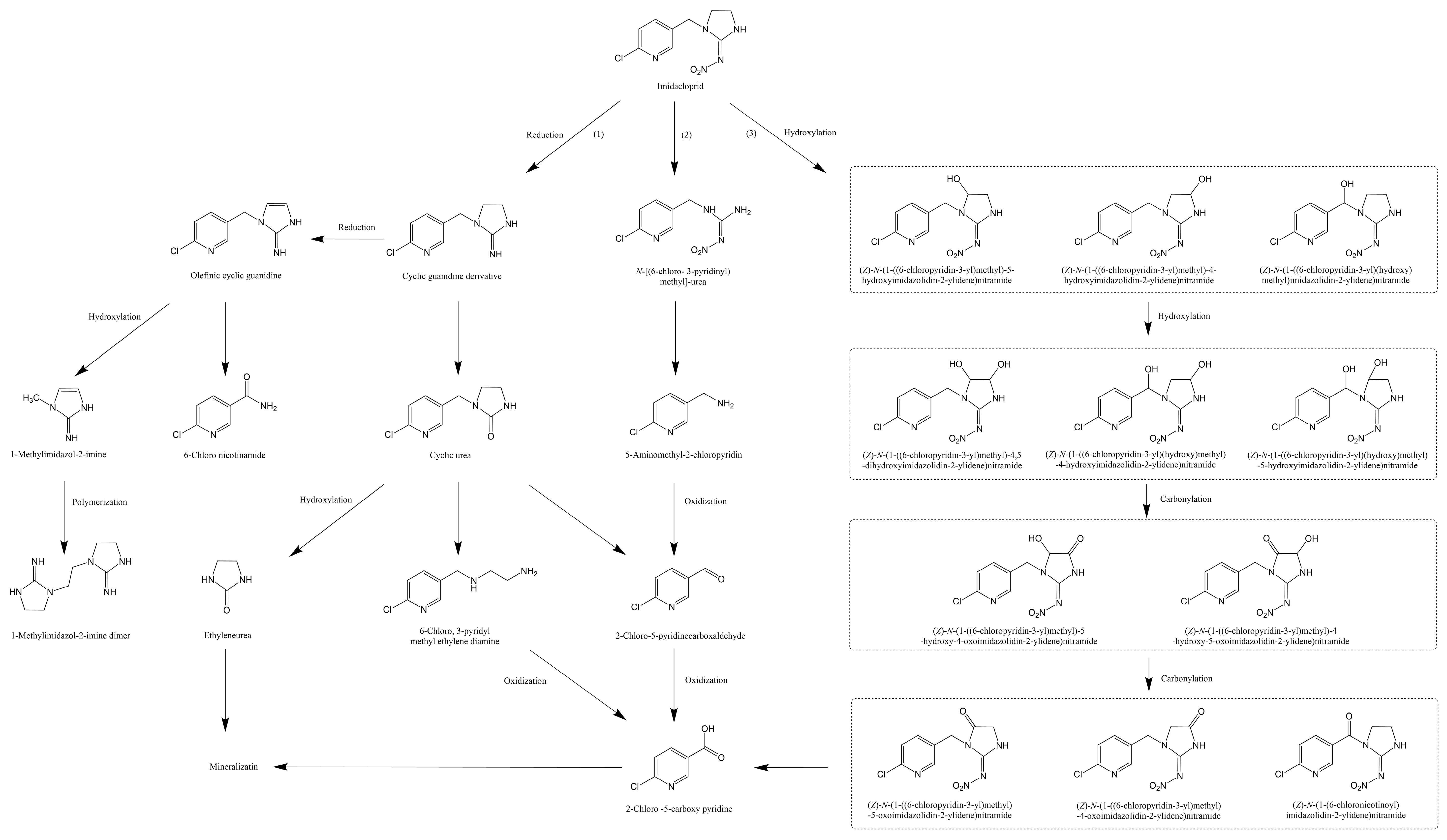
3. Degradation of Imidacloprid
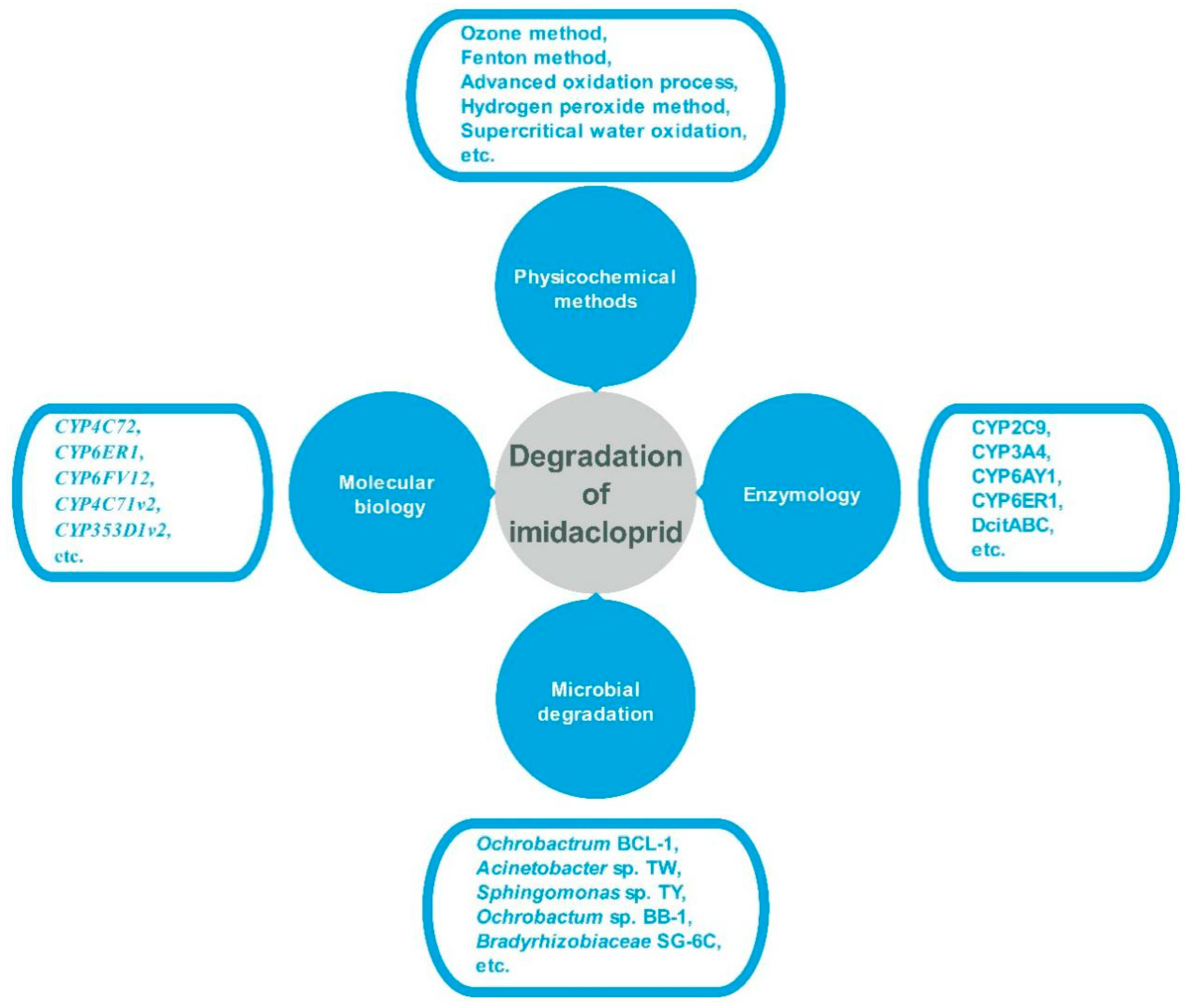
3.1. Degradation of Imidacloprid by Physicochemical Methods
3.2. Degradation of Imidacloprid by Microbial Strains
3.3. Molecular Biology of Imidacloprid Degradation
3.4. Enzymology of Imidacloprid Degradation
4. Conclusions and Future Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Chen, S.; Deng, J.; Deng, Y.; Gao, N. Influencing factors and kinetic studies of imidacloprid degradation by ozonation. Environ. Technol. 2019, 40, 2127–2134. [Google Scholar] [CrossRef] [PubMed]
- Diakou, A.; Dimzas, D.; Astaras, C.; Savvas, I.; Di Cesare, A.; Morelli, S.; Migli, D.; Traversa, D. Clinical investigations and treatment outcome in a European wildcat (Felis silvestris silvestris) infected by cardio-pulmonary nematodes. Vet. Parasitol. 2019, 19, 100357. [Google Scholar] [CrossRef] [PubMed]
- Sgolastra, F.; Medrzycki, P.; Bortolotti, L.; Maini, S.; Porrini, C.; Simon-Delso, N.; Bosch, J. Bees and pesticide regulation: Lessons from the neonicotinoid experience. Biol. Conserv. 2020, 241, 108356. [Google Scholar] [CrossRef]
- Motaung, T.E. Chloronicotinyl insecticide imidacloprid: Agricultural relevance, pitfalls and emerging opportunities. Crop. Prot. 2020, 131, 105097. [Google Scholar] [CrossRef]
- DiBartolomeis, M.; Kegley, S.; Mineau, P.; Radford, R.; Klein, K. An assessment of acute insecticide toxicity loading (AITL) of chemical pesticides used on agricultural land in the United States. PLoS ONE 2019, 14, e0220029. [Google Scholar] [CrossRef]
- Wei, F.; Wang, D.; Li, H.; Xia, P.; Ran, Y.; You, J. Toxicogenomics provides insights to toxicity pathways of neonicotinoids to aquatic insect, Chironomus dilutus. Environ. Pollut. 2020, 260, 114011. [Google Scholar] [CrossRef]
- Huang, Y.; Zhan, H.; Bhatt, P.; Chen, S. Paraquat degradation from contaminated environments: Current achievements and perspectives. Front. Microbiol. 2019, 10, 1754. [Google Scholar] [CrossRef]
- Bhatt, P.; Huang, Y.; Rene, E.R.; Kumar, A.J.; Chen, S. Mechanism of allethrin biodegradation by a newly isolated Sphingomonas trueperi strain CW3 from wastewater sludge. Bioresour. Technol. 2020, 305, 123074. [Google Scholar] [CrossRef]
- Mishra, S.; Zhang, W.; Lin, Z.; Pang, S.; Huang, Y.; Bhatt, P.; Chen, S. Carbofuran toxicity and its microbial degradation in contaminated environments. Chemosphere 2020, 259, 127419. [Google Scholar] [CrossRef]
- Pang, S.; Lin, Z.; Zhang, W.; Mishra, S.; Bhatt, P.; Chen, S. Insights into the Microbial Degradation and Biochemical Mechanisms of Neonicotinoids. Front. Microbiol. 2020, 11, 868. [Google Scholar] [CrossRef]
- Castillo Diaz, J.M.; Martin-Laurent, F.; Beguet, J.; Nogales, R.; Romero, E. Fate and effect of imidacloprid on vermicompost-amended soils under dissimilar conditions: Risk for soil functions, structure, and bacterial abundance. Sci. Total Environ. 2017, 579, 1111–1119. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Chang, C.; Deng, Y.; An, S.; Dong, Y.H.; Zhou, J.; Zhong, G.; Hu, M.; Zhang, L.H. Fenpropathrin biodegradation pathway in Bacillus sp. DG-02 and its potential for bioremediation of pyrethroid-contaminated soils. J. Agric. Food Chem. 2014, 62, 2147–2157. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Feng, Y.; Zhan, H.; Liu, J.; Yang, F.; Zhang, K.; Zhang, L.; Chen, S. Characterization of a pyrethroid-degrading Pseudomonas fulva strain P31 and biochemical degradation pathway of D-phenothrin. Front. Microbiol. 2018, 9, 1003. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, P.; Huang, Y.; Zhan, H.; Chen, S. Insight into microbial applications for the biodegradation of pyrethroid insecticides. Front. Microbiol. 2019, 10, 1778. [Google Scholar] [CrossRef]
- Huang, Y.; Lin, Z.; Zhang, W.; Pang, S.; Bhatt, P.; Rene, E.R.; Kumar, A.J.; Chen, S. New insights into the microbial degradation of D-cyphenothrin in contaminated water/soil environments. Microorganisms 2020, 8, 473. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, Y.M.M.; Badawy, M.E.I. Biodegradation of imidacloprid in liquid media by an isolated wastewater fungus Aspergillus terreus YESM3. J. Environ. Sci. Health B. 2017, 52, 752–761. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Tian, Y.; Shen, X. Human exposure to neonicotinoid insecticides and the evaluation of their potential toxicity: An overview. Chem. Eng. J. 2018, 192, 59–65. [Google Scholar] [CrossRef]
- Khalil, S.R.; Awad, A.; Mohammed, H.H.; Nassan, M.A. Imidacloprid insecticide exposure induces stress and disrupts glucose homeostasis in male rats. Environ. Toxic Pharm. 2017, 55, 165–174. [Google Scholar] [CrossRef]
- Wu, C.H.; Lin, C.L.; Wang, S.E.; Lu, C.W. Effects of imidacloprid, a neonicotinoid insecticide, on the echolocation system of insectivorous bats. Pestic. Biochem. Phys. 2020, 163, 94–101. [Google Scholar] [CrossRef]
- Vignet, C.; Cappello, T.; Fu, Q.; Lajoie, K.; De Marco, G.; Clérandeau, C.; Mottaz, H.; Maisano, M.; Hollender, J.; Schirmer, K.; et al. Imidacloprid induces adverse effects on fish early life stages that are more severe in Japanese medaka (Oryzias latipes) than in zebrafish (Danio rerio). Chemosphere 2019, 225, 470–478. [Google Scholar] [CrossRef]
- Wang, K.; Pang, S.; Mu, X.; Qi, S.; Li, D.; Cui, F.; Wang, C. Biological response of earthworm, Eisenia fetida, to five neonicotinoid insecticides. Chemosphere 2015, 132, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Martínez, L.C.; Plata-Rueda, A.; Gonçalves, W.G.; Freire, A.F.P.A.; Zanuncio, J.C.; Bozdoğan, H.; Serrão, J.E. Toxicity and cytotoxicity of the insecticide imidacloprid in the midgut of the predatory bug, Podisus nigrispinus. Ecotoxicol. Environ. Saf. 2019, 167, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Phung, D.; Dong, F.; Xu, J.; Liu, X.; Wu, X.; Liu, Q.; He, M.; Pan, X.; Li, R.; et al. Urinary monitoring of neonicotinoid imidacloprid exposure to pesticide applicators. Sci. Total Environ. 2019, 669, 721–728. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Dong, F.; Xu, J.; Phung, D.; Liu, Q.; Li, R.; Liu, X.; Wu, X.; He, M.; Zheng, Y. Characteristics of neonicotinoid imidacloprid in urine following exposure of humans to orchards in China. Environ. Int. 2019, 132, 105079. [Google Scholar] [CrossRef]
- Želježić, D.; Mladinić, M.; Žunec, S.; Lucić Vrdoljak, A.; Kašuba, V.; Tariba, B.; Živković, T.; Marjanović, A.M.; Pavičić, I.; Milić, M.; et al. Cytotoxic, genotoxic and biochemical markers of insecticide toxicity evaluated in human peripheral blood lymphocytes and an HepG2 cell line. Food Chem. Toxicol. 2016, 96, 90–106. [Google Scholar] [CrossRef]
- Kumar, A.; Verma, A.; Kumar, A. Accidental human poisoning with a neonicotinoid insecticide, imidacloprid: A rare case report from rural India with a brief review of literature. Egypt. J. Forensic Sci. 2013, 3, 123–126. [Google Scholar] [CrossRef]
- Lv, Y.; Bing, Q.; Lv, Z.; Xue, J.; Li, S.; Han, B.; Yang, Q.; Wang, X.; Zhang, Z. Imidacloprid-induced liver fibrosis in quails via activation of the TGF-β1/Smad pathway. Sci. Total Environ. 2020, 705, 135915. [Google Scholar] [CrossRef]
- Wang, X.; Ji, R.; Zhang, Y.; Yang, Y.; Fu, C.; Yang, D. Research on characterization and modeling for ultraviolet degradation of imidacloprid based on absorbance change. Optik 2018, 154, 315–319. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, P.; Chang, J.; Li, W.; Yang, L.; Tian, H. Unraveling the toxic effects of neonicotinoid insecticides on the thyroid endocrine system of lizards. Environ. Pollut. 2020, 258, 113731. [Google Scholar] [CrossRef]
- Yang, L.; Shen, Q.; Zeng, T.; Li, J.; Li, W.; Wang, Y. Enrichment of imidacloprid and its metabolites in lizards and its toxic effects on gonads. Environ. Pollut. 2020, 258, 113748. [Google Scholar] [CrossRef]
- Al-Badran, A.A.; Fujiwara, M.; Mora, M.A. Effects of insecticides, fipronil and imidacloprid, on the growth, survival, and behavior of brown shrimp Farfantepenaeus aztecus. PLoS ONE 2019, 14, e0223641. [Google Scholar] [CrossRef] [PubMed]
- Ewere, E.E.; Reichelt-Brushett, A.; Benkendorff, K. Imidacloprid and formulated product impacts the fatty acids and enzymatic activities in tissues of Sydney rock oysters, Saccostrea glomerata. Mar. Environ. Res. 2019, 151, 104765. [Google Scholar] [CrossRef] [PubMed]
- Shan, Y.; Yan, S.; Hong, X.; Zha, J.; Qin, J. Effect of imidacloprid on the behavior, antioxidant system, multixenobiotic resistance, and histopathology of Asian freshwater clams (Corbicula fluminea). Aquat. Toxicol. 2020, 218, 105333. [Google Scholar] [CrossRef] [PubMed]
- Vieira, C.E.D.; Pérez, M.R.; Acayaba, R.D.A.; Raimundo, C.C.M.; dos Reis Martinez, C.B. DNA damage and oxidative stress induced by imidacloprid exposure in different tissues of the Neotropical fish Prochilodus lineatus. Chemosphere 2018, 195, 125–134. [Google Scholar] [CrossRef]
- Lukaszewicz, G.; Iturburu, F.G.; Garanzini, D.S.; Menone, M.L.; Pflugmacher, S. Imidacloprid modifies the mitotic kinetics and causes both aneugenic and clastogenic effects in the macrophyte Bidens laevis L. Heliyon 2019, 5, e02118. [Google Scholar] [CrossRef]
- Njattuvetty Chandran, N.; Fojtova, D.; Blahova, L.; Rozmankova, E.; Blaha, L. Acute and (sub)chronic toxicity of the neonicotinoid imidacloprid on Chironomus riparius. Chemosphere 2018, 209, 568–577. [Google Scholar] [CrossRef]
- Oppold, A.; Kreß, A.; Vanden Bussche, J.; Diogo, J.B.; Kuch, U.; Oehlmann, J.; Vandegehuchte, M.B.; Müller, R. Epigenetic alterations and decreasing insecticide sensitivity of the Asian tiger mosquito Aedes albopictus. Ecotox. Environ. Saf. 2015, 122, 45–53. [Google Scholar] [CrossRef]
- Morales, S.I.; Martínez, A.M.; Figueroa, J.I.; Campos-García, J.; Gómez-Tagle, A.; Lobit, P.; Smagghe, G.; Pineda, S. Foliar persistence and residual activity of four insecticides of different mode of action on the predator Engytatus varians (Hemiptera: Miridae). Chemosphere 2019, 235, 76–83. [Google Scholar] [CrossRef]
- Ricupero, M.; Desneux, N.; Zappalà, L.; Biondi, A. Target and non-target impact of systemic insecticides on a polyphagous aphid pest and its parasitoid. Chemosphere 2020, 247, 125728. [Google Scholar] [CrossRef]
- Skouras, P.J.; Brokaki, M.; Stathas, G.J.; Demopoulos, V.; Louloudakis, G.; Margaritopoulos, J.T. Lethal and sub-lethal effects of imidacloprid on the aphidophagous coccinellid Hippodamia variegata. Chemosphere 2019, 229, 392–400. [Google Scholar] [CrossRef]
- Reid, R.J.; Troczka, B.J.; Kor, L.; Randall, E.; Williamson, M.S.; Field, L.M.; Nauen, R.; Bass, C.; Emyr Davies, T.G. Assessing the acute toxicity of insecticides to the buff-tailed bumblebee (Bombus terrestris audax). Pestic. Biochem. Phys. 2020, 166, 104562. [Google Scholar] [CrossRef] [PubMed]
- Bernardes, R.C.; Barbosa, W.F.; Martins, G.F.; Lima, M.A.P. The reduced-risk insecticide azadirachtin poses a toxicological hazard to stingless bee Partamona helleri (Friese, 1900) queens. Chemosphere 2018, 201, 550–556. [Google Scholar] [CrossRef] [PubMed]
- Brandt, A.; Gorenflo, A.; Siede, R.; Meixner, M.; Büchler, R. The neonicotinoids thiacloprid, imidacloprid, and clothianidin affect the immunocompetence of honey bees (Apis mellifera L.). J. Insect Physiol. 2016, 86, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Ospina, M.; Jayatilaka, N.K.; Wong, L.Y.; Restrepo, P.; Calafat, A.M. Exposure to organophosphate flame retardant chemicals in the U.S. general population: Data from the 2013–2014 National Health and Nutrition Examination Survey. Environ. Int. 2018, 110, 32–41. [Google Scholar] [CrossRef] [PubMed]
- López-García, M.; Romero-González, R.; Lacasaña, M.; Garrido Frenich, A. Semiautomated determination of neonicotinoids and characteristic metabolite in urine samples using TurboFlow™ coupled to ultra high performance liquid chromatography coupled to Orbitrap analyzer. J. Pharmaceut. Biomed. 2017, 146, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Vardavas, A.I.; Ozcagli, E.; Fragkiadaki, P.; Stivaktakis, P.D.; Tzatzarakis, M.N.; Alegakis, A.K.; Vasilaki, F.; Kaloudis, K.; Tsiaoussis, J.; Kouretas, D.; et al. The metabolism of imidacloprid by aldehyde oxidase contributes to its clastogenic effect in New Zealand rabbits. Mutat. Res. Gen. Tox. Environ. Mutagen. 2018, 829, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Bonmatin, J.-M.; Noome, D.A.; Moreno, H.; Mitchell, E.A.D.; Glauser, G.; Soumana, O.S.; van Lexmond, M.B.; Sánchez-Bayo, F. A survey and risk assessment of neonicotinoids in water, soil and sediments of Belize. Environ. Pollut. 2019, 249, 949–958. [Google Scholar] [CrossRef]
- Butcherine, P.; Benkendorff, K.; Kelaher, B.; Barkla, B.J. The risk of neonicotinoid exposure to shrimp aquaculture. Chemosphere 2019, 217, 329–348. [Google Scholar] [CrossRef]
- Paquet-Walsh, A.; Bertolo, A.; Landry, C.; Deschamps, L.; Boily, M. Interactive effects of neonicotinoids and natural ultraviolet radiation on yellow perch (Perca flavescens) larvae. Sci. Total Environ. 2019, 685, 690–701. [Google Scholar] [CrossRef]
- Ewere, E.E.; Powell, D.; Rudd, D.; Reichelt-Brushett, A.; Mouatt, P.; Voelcker, N.H.; Benkendorff, K. Uptake, depuration and sublethal effects of the neonicotinoid, imidacloprid, exposure in Sydney rock oysters. Chemosphere 2019, 230, 1–13. [Google Scholar] [CrossRef]
- Raby, M.; Zhao, X.; Hao, C.; Poirier, D.G.; Sibley, P.K. Chronic effects of an environmentally-relevant, short-term neonicotinoid insecticide pulse on four aquatic invertebrates. Sci. Total Environ. 2018, 639, 1543–1552. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, R.A.; Sutherland, A.M.; Hengel, M.J.; Parrella, M.P.; Gubler, W.D. Imidacloprid movement into fungal conidia is lethal to mycophagous beetles. bioRxiv 2020. [Google Scholar] [CrossRef]
- Prosser, R.S.; de Solla, S.R.; Holman, E.A.M.; Osborne, R.; Robinson, S.A.; Bartlett, A.J.; Maisonneuve, F.J.; Gillis, P.L. Sensitivity of the early-life stages of freshwater mollusks to neonicotinoid and butenolide insecticides. Environ. Pollut. 2016, 218, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Shigetou, S.; Shimada, S.; Makoto, I.; Matsuda, K. Modulation by neonicotinoids of honeybee α1/chicken β2 hybrid nicotinic acetylcholine receptors expressed in Xenopus laevis oocytes. Pestic. Biochem. Phys. 2020, 166, 104545. [Google Scholar] [CrossRef]
- Almasi, A.; Rasekh, A.; Esfandiari, M.; Askari Seyahooei, M.; Ziaee, M. The prospect of using sub-lethal imidacloprid or pirimicarb and a parasitoid wasp, Lysiphlebus fabarum, simultaneously, to control Aphis gossypii on cucumber plants. J. Asia Pac. Entomol. 2018, 21, 161–167. [Google Scholar] [CrossRef]
- Calatayud-Vernich, P.; Calatayud, F.; Simó, E.; Pascual Aguilar, J.A.; Picó, Y. A two-year monitoring of pesticide hazard in-hive: High honey bee mortality rates during insecticide poisoning episodes in apiaries located near agricultural settings. Chemosphere 2019, 232, 471–480. [Google Scholar] [CrossRef]
- Crall, J.D.; Switzer, C.M.; Myers, S.S.; Combes, S.A.; de Bivort, B.L. Pesticides and pollinators, an automated platform to assess the effects of neonicotinoid exposure and other environmental stressors on bee colonies: A computational, ethological study. Lancet 2017, 389, S4. [Google Scholar] [CrossRef]
- Gross, M. Bee worries beyond neonicotinoids. Curr. Biol. 2018, 28, R1121–R1123. [Google Scholar] [CrossRef]
- Chang, Y.; Mao, L.; Zhang, L.; Zhang, Y.; Jiang, H. Combined toxicity of imidacloprid, acetochlor, and tebuconazole to zebrafish (Danio rerio): Acute toxicity and hepatotoxicity assessment. Environ. Sci. Pollut. Res. Int. 2020, 27, 10286–10295. [Google Scholar] [CrossRef]
- Graily Moradi, F.; Hejazi, M.J.; Hamishehkar, H.; Enayati, A.A. Co-encapsulation of imidacloprid and lambda-cyhalothrin using biocompatible nanocarriers: Characterization and application. Ecotox. Environ. Saf. 2019, 175, 155–163. [Google Scholar] [CrossRef]
- Alvim, T.T.; Martinez, C.B.d.R. Genotoxic and oxidative damage in the freshwater teleost Prochilodus lineatus exposed to the insecticides lambda-cyhalothrin and imidacloprid alone and in combination. Mutat. Res. Gen. Tox. Environ. Mutagen. 2019, 842, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.; Jhamtani, R.C.; Dahiya, M.S.; Agarwal, R. Oxidative injury caused by individual and combined exposure of neonicotinoid, organophosphate and herbicide in zebrafish. Toxicol. Rep. 2017, 4, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Li, X.; Liu, X.; Yang, G.; An, X.; Wang, Q.; Wang, Y. Joint toxic effects of triazophos and imidacloprid on zebrafish (Danio rerio). Environ. Pollut. 2018, 235, 470–481. [Google Scholar] [CrossRef] [PubMed]
- Ishtiaq, F.; Bhatti, H.N.; Khan, A.; Iqbal, M.; Kausar, A. Polypyrole, polyaniline and sodium alginate biocomposites and adsorption-desorption efficiency for imidacloprid insecticide. Int. J. Biol. Macromol. 2020, 147, 217–232. [Google Scholar] [CrossRef]
- Liu, W.; Zheng, W.; Ma, Y.; Liu, K.K. Sorption and degradation of imidacloprid in soil and water. J. Environ. Sci. Health B 2006, 41, 623–634. [Google Scholar] [CrossRef]
- Joice, J.A.I.; Aishwarya, S.; Sivakumar, T. Nano structured Ni and Ru impregnated TiO2 photocatalysts: Synthesis, characterization and photocatalytic degradation of neonicotinoid insecticides. J. Nanosci. Nanotechno. 2019, 19, 2575–2589. [Google Scholar] [CrossRef]
- Patil, P.N.; Bote, S.D.; Gogate, P.R. Degradation of imidacloprid using combined advanced oxidation processes based on hydrodynamic cavitation. Ultrason. Sonochem. 2014, 21, 1770–1777. [Google Scholar] [CrossRef]
- Lacson, C.F.Z.; de Luna, M.D.G.; Dong, C.; Garcia-Segura, S.; Lu, M.-C. Fluidized-bed Fenton treatment of imidacloprid: Optimization and degradation pathway. Sustain. Environ. Res. 2018, 28, 309–314. [Google Scholar] [CrossRef]
- De Luna, M.D.G.; Sablas, M.M.; Hung, C.M.; Chen, C.W.; Garcia-Segura, S.; Dong, C.D. Modeling and optimization of imidacloprid degradation by catalytic percarbonate oxidation using artificial neural network and Box-Behnken experimental design. Chemosphere 2020, 251, 126254. [Google Scholar] [CrossRef]
- Zhang, Y.; He, P.; Zhou, L.; Dong, F.; Yang, D.; Lei, H.; Du, L.; Jia, L.; Zhou, S. Optimized terbium doped Ti/PbO2 dimensional stable anode as a strong tool for electrocatalytic degradation of imidacloprid waste water. Ecotoxicol. Environ. Saf. 2020, 188, 109921. [Google Scholar] [CrossRef]
- Jiang, X.; Song, D.; Wang, D.; Zhang, R.; Fang, Q.; Sun, H.; Kong, F. Eliminating imidacloprid and its toxicity by permanganate via highly selective partial oxidation. Ecotoxicol. Environ. Saf. 2020, 191, 110234. [Google Scholar] [CrossRef] [PubMed]
- Bogatu, C.; Covei, M.; Tismanar, I.; Perniu, D.; Duta, A. 10—Composite nanostructures as potential materials for water and air cleaning with enhanced efficiency. In Advanced Nanostructures for Environmental Health; Baia, L., Pap, Z., Hernadi, K., Baia, M., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 431–463. [Google Scholar]
- Covei, M.; Bogatu, C.; Perniu, D.; Tismanar, I.; Duta, A. Comparative study on the photodegradation efficiency of organic pollutants using n-p multi-junction thin films. Catal. Today 2019, 328, 57–64. [Google Scholar] [CrossRef]
- Naghizadeh, M.; Taher, M.A.; Tamaddon, A.-M. Facile synthesis and characterization of magnetic nanocomposite ZnO/CoFe2O4 hetero-structure for rapid photocatalytic degradation of imidacloprid. Heliyon 2019, 5, e02870. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Li, L.; Xu, D.; Huang, X.; Xu, X.; Zheng, S.; Zhang, Y.; Lin, H. Metal–organic framework preparation using magnetic graphene oxide–β-cyclodextrin for neonicotinoid pesticide adsorption and removal. Carbohyd. Polym. 2017, 175, 584–591. [Google Scholar] [CrossRef]
- Acero, J.L.; Real, F.J.; Javier Benitez, F.; Matamoros, E. Degradation of neonicotinoids by UV irradiation: Kinetics and effect of real water constituents. Sep. Purif. Technol. 2019, 211, 218–226. [Google Scholar] [CrossRef]
- Wang, Y.; Han, Y.; Xu, P.; Guo, B.; Li, W.; Wang, X. The metabolism distribution and effect of imidacloprid in chinese lizards (Eremias argus) following oral exposure. Ecotox. Environ. Saf. 2018, 165, 476–483. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, X. Efficient visible-light photocatalytic degradation of imidacloprid and acetamiprid using a modified carbon nitride/tungstophosphoric acid composite induced by a nucleophilic addition reaction. Appl. Surf. Sci. 2019, 485, 423–431. [Google Scholar] [CrossRef]
- Banić, N.D.; Abramović, B.F.; Šojić, D.V.; Krstić, J.B.; Finčur, N.L.; Bočković, I.P. Efficiency of neonicotinoids photocatalytic degradation by using annular slurry reactor. Che. Eng. J. 2016, 286, 184–190. [Google Scholar] [CrossRef]
- Wang, Q.; Rao, P.; Li, G.; Dong, L.; Zhang, X.; Shao, Y.; Gao, N.; Chu, W.; Xu, B.; An, N.; et al. Degradation of imidacloprid by UV-activated persulfate and peroxymonosulfate processes: Kinetics, impact of key factors and degradation pathway. Ecotoxicol. Environ. Saf. 2020, 187, 109779. [Google Scholar] [CrossRef]
- Heng, H.; Gan, Q.; Meng, P.; Liu, X. The visible-light-driven type III heterojunction H3PW12O40/TiO2-In2S3: A photocatalysis composite with enhanced photocatalytic activity. J. Alloy. Comp. 2017, 696, 51–59. [Google Scholar] [CrossRef]
- Andronic, L.; Isac, L.; Miralles-Cuevas, S.; Visa, M.; Oller, I.; Duta, A.; Malato, S. Pilot-plant evaluation of TiO2 and TiO2-based hybrid photocatalysts for solar treatment of polluted water. J. Hazard. Mater. 2016, 320, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.; Tang, F.; Wang, J.; Yue, Y. Photo-degradation dynamics of five neonicotinoids: Bamboo vinegar as a synergistic agent for improved functional duration. PLoS ONE 2019, 14, e0223708. [Google Scholar] [CrossRef] [PubMed]
- Fiorenza, R.; Di Mauro, A.; Cantarella, M.; Iaria, C.; Scalisi, E.M.; Brundo, M.V.; Gulino, A.; Spitaleri, L.; Nicotra, G.; Dattilo, S.; et al. Preferential removal of pesticides from water by molecular imprinting on TiO2 photocatalysts. Chem. Eng. J. 2020, 379, 122309. [Google Scholar] [CrossRef]
- González, T.; Dominguez, J.R.; Correia, S. Neonicotinoids removal by associated binary, tertiary and quaternary advanced oxidation processes: Synergistic effects, kinetics and mineralization. J. Environ. Manage. 2020, 261, 110156. [Google Scholar] [CrossRef] [PubMed]
- Mandal, A.; Singh, N. Optimization of atrazine and imidacloprid removal from water using biochars: Designing single or multi-staged batch adsorption systems. Int. J. Hyg. Environ. Health 2017, 220, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Kalhor, M.M.; Rafati, A.A.; Rafati, L.; Rafati, A.A. Synthesis, characterization and adsorption studies of amino functionalized silica nano hollow sphere as an efficient adsorbent for removal of imidacloprid pesticide. J. Mol. Liq. 2018, 266, 453–459. [Google Scholar] [CrossRef]
- Tang, Y.; Zhang, X.; Ma, Y.; Wang, X.; Su, C.; Zhang, D.; Pu, X.; Geng, Y. One-dimensional core-shell Zn0.1Cd0.9S/Snln4S8 heterojunction for enhanced visible light photocatalytic degradation. Sep. Purif. Technol. 2020, 230, 115896. [Google Scholar] [CrossRef]
- Sun, Y.; Meng, P.; Liu, X. Self-assembly of tungstophosphoric acid/acidified carbon nitride hybrids with enhanced visible-light-driven photocatalytic activity for the degradation of imidacloprid and acetamiprid. Appl. Surf. Sci. 2018, 456, 259–269. [Google Scholar] [CrossRef]
- Soltani-Nezhad, F.; Saljooqi, A.; Shamspur, T.; Mostafavi, A. Photocatalytic degradation of imidacloprid using GO/Fe3O4/TiO2-NiO under visible radiation: Optimization by response level method. Polyhedron 2019, 165, 188–196. [Google Scholar] [CrossRef]
- Sarro, M.; Gule, N.P.; Laurenti, E.; Gamberini, R.; Paganini, M.C.; Mallon, P.E.; Calza, P. ZnO-based materials and enzymes hybrid systems as highly efficient catalysts for recalcitrant pollutants abatement. Chem. Eng. J. 2018, 334, 2530–2538. [Google Scholar] [CrossRef]
- Papoutsakis, S.; Pulgarin, C.; Oller, I.; Sánchez-Moreno, R.; Malato, S. Enhancement of the Fenton and photo-Fenton processes by components found in wastewater from the industrial processing of natural products: The possibilities of cork boiling wastewater reuse. Chem. Eng. J. 2016, 304, 890–896. [Google Scholar] [CrossRef]
- Papoutsakis, S.; Brites-Nóbrega, F.F.; Pulgarin, C.; Malato, S. Benefits and limitations of using Fe(III)-EDDS for the treatment of highly contaminated water at near-neutral pH. J. Photochem. Photobiol. Chem. 2015, 303, 1–7. [Google Scholar] [CrossRef]
- Liang, S.; Zhang, D.; Pu, X.; Yao, X.; Han, R.; Yin, J.; Ren, X. A novel Ag2O/g-C3N4 p-n heterojunction photocatalysts with enhanced visible and near-infrared light activity. Sep. Purif. Technol. 2019, 210, 786–797. [Google Scholar] [CrossRef]
- Vela, N.; Fenoll, J.; Navarro, G.; Garrido, I.; Navarro, S. Trial of solar heating methods (solarization and biosolarization) to reduce persistence of neonicotinoid and diamide insecticides in a semiarid Mediterranean soil. Sci. Total Environ. 2017, 590, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Bourgin, M.; Violleau, F.; Debrauwer, L.; Albet, J. Ozonation of imidacloprid in aqueous solutions: Reaction monitoring and identification of degradation products. J. Hazard. Mater. 2011, 190, 60–68. [Google Scholar] [CrossRef]
- Chen, C.; Shan, T.; Liu, Y.; Wang, C.; Shi, X.; Gao, X. Identification and functional analysis of a cytochrome P450 gene involved in imidacloprid resistance in Bradysia odoriphaga. Pestic. Biochem. Phys. 2019, 153, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, O.; Gómez, J.; Pazos, M.; Sanromán, M.Á. Electro-Fenton oxidation of imidacloprid by Fe alginate gel beads. Appl. Catal. B Environ. 2014, 144, 416–424. [Google Scholar] [CrossRef]
- Malato, S.; Caceres, J. Degradation of imidacloprid in water by Photo-Fenton and TiO2 photocatalysis at a solar pilot plant: A comparative study. Environ. Sci. Technol. 2001, 35, 4359–4366. [Google Scholar] [CrossRef]
- Raut-Jadhav, S.; Saharan, V.K.; Pinjari, D.; Sonawane, S.; Saini, D.; Pandit, A. Synergetic effect of combination of AOP’s (hydrodynamic cavitation and H2O2) on the degradation of neonicotinoid class of insecticide. J. Hazard. Mater. 2013, 261, 139–147. [Google Scholar] [CrossRef]
- Zhou, X.; Tian, Y.; Liu, X.; Huang, L.; Wen, Y. Reduction of imidacloprid by sponge iron and identification of its degradation products. Water Environ. Res. 2018, 90, 2049–2055. [Google Scholar] [CrossRef]
- Chen, S.; Dong, Y.H.; Chang, C.; Deng, Y.; Zhang, X.F.; Zhong, G.; Song, H.; Hu, M.; Zhang, L.H. Characterization of a novel cyfluthrin-degrading bacterial strain Brevibacterium aureum and its biochemical degradation pathway. Bioresour. Technol. 2013, 132, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Zhan, H.; Feng, Y.; Fan, X.; Chen, S. Recent advances in glyphosate biodegradation. Appl. Microbiol. Biotechnol. 2018, 102, 5033–5043. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, P.; Huang, Y.; Zhang, W.; Sharma, A.; Chen, S. Enhanced cypermethrin degradation kinetics and metabolic pathway in Bacillus thuringiensis strain SG4. Microorganisms 2020, 8, 233. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Huang, Y.; Zhan, H.; Bhatt, P.; Chen, S. An overview of strobilurin fungicide degradation: Current status and future perspective. Front. Microbiol. 2020, 11, 389. [Google Scholar] [CrossRef]
- Arora, P.K.; Srivastava, A.; Garg, S.K.; Singh, V.P. Recent advances in degradation of chloronitrophenols. Bioresour. Technol. 2018, 250, 902–909. [Google Scholar] [CrossRef]
- Erguven, G.O.; Yildirim, N. The evaluation of imidacloprid remediation in soil media by two bacterial strains. Curr. Microbiol. 2019, 76, 1461–1466. [Google Scholar] [CrossRef]
- Bhatt, P.; Bhatt, K.; Huang, Y.; Lin, Z.; Chen, S. Esterase is a powerful tool for the biodegradation of pyrethroid insecticides. Chemosphere 2020, 244, 125507. [Google Scholar] [CrossRef]
- Lin, Z.; Zhang, W.; Pang, S.; Huang, Y.; Mishra, S.; Bhatt, P.; Chen, S. Current approaches to and future perspectives on methomyl degradation in contaminated soil/water environments. Molecules 2020, 25, 738. [Google Scholar] [CrossRef]
- Zhang, W.; Lin, Z.; Pang, S.; Bhatt, P.; Chen, S. Insights into the biodegradation of lindane (γ-hexachlorocyclohexane) using a microbial system. Front. Microbiol. 2020, 11, 522. [Google Scholar] [CrossRef]
- Bhatt, P.; Zhang, W.; Lin, Z.; Pang, S.; Huang, Y.; Chen, S. Biodegradation of allethrin by a novel fungus Fusarium proliferatum strain CF2, isolated from contaminated soils. Microorganisms 2020, 8, 593. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, W.; Pang, S.; Lin, Z.; Zhang, Y.; Huang, Y.; Bhatt, P.; Chen, S. Kinetics and new mechanism of azoxystrobin biodegradation by an Ochrobactrum anthropi strain SH14. Microorganisms 2020, 8, 625. [Google Scholar] [CrossRef] [PubMed]
- Zhan, H.; Huang, Y.; Lin, Z.; Bhatt, P.; Chen, S. New insights into the microbial degradation and catalytic mechanism of synthetic pyrethroids. Environ. Res. 2020, 182, 109138. [Google Scholar] [CrossRef] [PubMed]
- Anhalt, J.C.; Moorman, T.B.; Koskinen, W.C. Biodegradation of imidacloprid by an isolated soil microorganism. J. Environ. Sci. Health B 2007, 42, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Geng, P.; Xiao, Y.; Hu, M. Bioremediation of beta-cypermethrin and 3-phenoxybenzaldehyde contaminated soils using Streptomyces aureus HP-S-01. Appl Microbiol Biotechnol. 2012, 94, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Pang, S.; Zhang, W.; Mishra, S.; Bhatt, P.; Chen, S. Degradation of acephate and its intermediate methamidophos: Mechanisms and biochemical pathways. Front Microbiol. 2020, 11, 2045. [Google Scholar] [CrossRef]
- Xiao, Y.; Chen, S.; Gao, Y.; Hu, W.; Hu, M.; Zhong, G. Isolation of a novel beta-cypermethrin degrading strain Bacillus subtilis BSF01 and its biodegradation pathway. Appl. Microbiol. Biotechnol. 2015, 99, 2849–2859. [Google Scholar] [CrossRef]
- Bhatt, P.; Rene, E.R.; Kumar, A.J.; Zhang, W.; Chen, S. Binding interaction of allethrin with esterase: Bioremediation potential and mechanism. Bioresour. Technol. 2020, 315, 123845. [Google Scholar] [CrossRef]
- Bhatt, P.; Verma, A.; Verma, S.; Anwar, M.S.; Prasher, P.; Mudila, H.; Chen, S. Understanding phytomicrobiome: A potential reservoir for better crop management. Sustainability 2020, 12, 5446. [Google Scholar] [CrossRef]
- Gupta, M.; Mathur, S.; Sharma, T.K.; Rana, M.; Gairola, A.; Navani, N.K.; Pathania, R. A study on metabolic prowess of Pseudomonas sp. RPT 52 to degrade imidacloprid, endosulfan and coragen. J. Hazard. Mater. 2016, 301, 250–258. [Google Scholar] [CrossRef]
- Wu, P.; Zhang, X.; Niu, T.; Wang, Y.; Liu, R.; Zhang, Y. The imidacloprid remediation, soil fertility enhancement and microbial community change in soil by Rhodopseudomonas capsulata using effluent as carbon source. Environ. Pollut. 2020, 257, 114254. [Google Scholar] [CrossRef]
- Pandey, G.; Dorrian, S.J.; Russell, R.J.; Oakeshott, J.G. Biotransformation of the neonicotinoid insecticides imidacloprid and thiamethoxam by Pseudomonas sp. 1G. Biochem. Biophys. Res. Commun. 2009, 380, 710–714. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhai, S.; Mao, S.Y.; Sun, S.L.; Wang, Y.; Liu, Z.H.; Dai, Y.J.; Yuan, S. Co-metabolic transformation of the neonicotinoid insecticide imidacloprid by the new soil isolate Pseudoxanthomonas indica CGMCC 6648. J. Environ. Sci. Health B. 2014, 49, 661–670. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Deng, Y.; Chang, C.; Lee, J.; Cheng, Y.; Cui, Z.; Zhou, J.; He, F.; Hu, M.; Zhang, L.H. Pathway and kinetics of cyhalothrin biodegradation by Bacillus thuringiensis strain ZS-19. Sci. Rep. 2015, 5, 8784. [Google Scholar] [CrossRef] [PubMed]
- Zhan, H.; Wang, H.; Liao, L.; Feng, Y.; Fan, X.; Zhang, L.; Chen, S. Kinetics and novel degradation pathway of permethrin in Acinetobacter baumannii ZH-14. Front. Microbiol. 2018, 9, 98. [Google Scholar] [CrossRef]
- Wang, M.; Yao, Y.; Yang, G.; Min, H.; Wang, X.; Lu, Z. Nicotine degradation by two novel bacterial isolatesof Acinetobacter sp. TW and Sphingomonas sp. TY and their responses in the presence of neonicotinoid insecticides. World J. Microbiol. Biotechnol. 2011, 27, 1633–1640. [Google Scholar] [CrossRef]
- Ferreira, L.; Rosales, E.; Danko, A.S.; Sanromán, M.A.; Pazos, M.M. Bacillus thuringiensis a promising bacterium for degrading emerging pollutants. Process. Saf. Environ. 2016, 101, 19–26. [Google Scholar] [CrossRef]
- Amaresh, Y.S.; Chennappa, G.; Avinash, S.; Naik, M.K.; Sreenivasa, M.Y. Chapter 15—Trichoderma—A new strategy in combating agriculture problems. In New and Future Developments in Microbial Biotechnology and Bioengineering; Singh, J.S., Ed.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 235–244. [Google Scholar]
- Hu, G.; Zhao, Y.; Liu, B.; Song, F.; You, M. Isolation of an indigenous imidacloprid-degrading bacterium and imidacloprid bioremediation under simulated in situ and ex situ conditions. J. Microbiol. Biotechnol. 2013, 23, 1617–1626. [Google Scholar] [CrossRef]
- Smriti, S.; Balwinder, S.; Gupta, V. K. Assessment of imidacloprid degradation by soil-isolated Bacillus alkalinitrilicus. Environ. Monit. Assess. 2014, 186, 7183–7193. [Google Scholar]
- Shettigar, M.; Pearce, S.; Pandey, R.; Khan, F.; Dorrian, S.J.; Balotra, S.; Russell, R.J.; Oakeshott, J.G.; Pandey, G. Cloning of a novel 6-chloronicotinic acid chlorohydrolase from the newly isolated 6-chloronicotinic acid mineralizing Bradyrhizobiaceae strain SG-6C. PLoS ONE 2012, 7, e51162. [Google Scholar] [CrossRef]
- Dai, Y.J.; Yuan, S.; Ge, F.; Chen, T.; Xu, S.C.; Ni, J.P. Microbial hydroxylation of imidacloprid for the synthesis of highly insecticidal olefin imidacloprid. Appl. Microbiol. Biotechnol. 2006, 71, 927–934. [Google Scholar] [CrossRef]
- Kandil, M.M.; Trigo, C.; Koskinen, W.C.; Sadowsky, M.J. Isolation and characterization of a novel imidacloprid-degrading Mycobacterium sp. strain MK6 from an Egyptian soil. J. Agric. Food Chem. 2015, 63, 4721–4727. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Dai, Z.; Guo, J.; Yang, W.; Ge, F.; Dai, Y. Oligotrophic bacterium Hymenobacter latericoloratus CGMCC 16346 degrades the neonicotinoid imidacloprid in surface water. AMB Express. 2020, 10, 7. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Lian, G. Study on culture conditions of an imidacloprid-degrading bacterium. Ochrobactum BB-1. Biotechnology 2011, 21, 75–79. [Google Scholar]
- Romila, A.; Balwinder, S. Metabolic degradation of imidacloprid in paddy field soil. Environ. Monit. Assess. 2014, 186, 5977–5984. [Google Scholar]
- Phugare, S.S.; Kalyani, D.C.; Gaikwad, Y.B.; Jadhav, J.P. Microbial degradation of imidacloprid and toxicological analysis of its biodegradation metabolites in silkworm (Bombyx mori). Chem. Eng. J. 2013, 230, 27–35. [Google Scholar] [CrossRef]
- Sabourmoghaddam, N.; Zakaria, M.P.; Omar, D. Evidence for the microbial degradation of imidacloprid in soils of Cameron Highlands. J. Saudi Soc. Agr. Sci. 2015, 14, 182–188. [Google Scholar] [CrossRef]
- Naqqash, M.N.; Gökçe, A.; Aksoy, E.; Bakhsh, A. Downregulation of imidacloprid resistant genes alters the biological parameters in Colorado potato beetle, Leptinotarsa decemlineata Say (Chrysomelidae: Coleoptera). Chemosphere 2020, 240, 124857. [Google Scholar] [CrossRef]
- Hirata, K.; Jouraku, A.; Kuwazaki, S.; Kanazawa, J.; Iwasa, T. The R81T mutation in the nicotinic acetylcholine receptor of Aphis gossypii is associated with neonicotinoid insecticide resistance with differential effects for cyano- and nitro-substituted neonicotinoids. Pestic. Biochem. Phys. 2017, 143, 57–65. [Google Scholar] [CrossRef]
- Kalsi, M.; Palli, S.R. Transcription factor cap n collar C regulates multiple cytochrome P450 genes conferring adaptation to potato plant allelochemicals and resistance to imidacloprid in Leptinotarsa decemlineata (Say). Insect Biochem. Molec. 2017, 83, 1–12. [Google Scholar] [CrossRef]
- Hamada, A.; Wahl, G.D.; Nesterov, A.; Nakao, T.; Kawashima, M.; Banba, S. Differential metabolism of imidacloprid and dinotefuran by Bemisia tabaci CYP6CM1 variants. Pestic. Biochem. Phys. 2019, 159, 27–33. [Google Scholar] [CrossRef]
- Hamada, A.; Stam, L.; Nakao, T.; Kawashima, M.; Banba, S. Differential metabolism of neonicotinoids by brown planthopper, Nilaparvata lugens, CYP6ER1 variants. Pestic. Biochem. Phys. 2020, 165, 104538. [Google Scholar] [CrossRef] [PubMed]
- Elzaki, M.E.A.; Zhang, W.; Feng, A.; Qiou, X.; Zhao, W.; Han, Z. Constitutive overexpression of cytochrome P450 associated with imidacloprid resistance in Laodelphax striatellus (Fallén). Pest. Manag. Sci. 2016, 72, 1051–1058. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Cao, Q.; Li, G.; Ma, D. Role of several cytochrome P450s in the resistance and cross-resistance against imidacloprid and acetamiprid of Bemisia tabaci (Hemiptera: Aleyrodidae) MEAM1 cryptic species in Xinjiang, China. Pestic. Biochem. Phys. 2020, 163, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Kishk, A.; Hijaz, F.; Anber, H.A.I.; AbdEl-Raof, T.K.; El-Sherbeni, A.-H.D.; Hamed, S.; Killiny, N. RNA interference of acetylcholinesterase in the Asian citrus psyllid, Diaphorina citri, increases its susceptibility to carbamate and organophosphate insecticides. Pestic. Biochem. Phys. 2017, 143, 81–89. [Google Scholar] [CrossRef]
- Lu, K.; Chen, X.; Liu, W.; Zhang, Z.; Wang, Y.; You, K.; Li, Y.; Zhang, R.; Zhou, Q. Characterization of heat shock protein 70 transcript from Nilaparvata lugens (Stål): Its response to temperature and insecticide stresses. Pestic. Biochem. Phys. 2017, 142, 102–110. [Google Scholar] [CrossRef]
- Wan, Y.; Yuan, G.; He, B.; Xu, B.; Xie, W.; Wang, S.; Zhang, Y.; Wu, Q.; Zhou, X. Foccα6, a truncated nAChR subunit, positively correlates with spinosad resistance in the western flower thrips, Frankliniella occidentalis (Pergande). Insect Biochem. Molec. 2018, 99, 1–10. [Google Scholar] [CrossRef]
- Zhang, B.; Su, X.; Xie, L.; Zhen, C.; Hu, G.; Jiang, K.; Huang, Z.Y.; Liu, R.Q.; Gao, Y.F.; Chen, X.L.; et al. Multiple detoxification genes confer imidacloprid resistance to Sitobion avenae Fabricius. Crop. Prot. 2020, 128, 105014. [Google Scholar] [CrossRef]
- Tian, F.; Wang, Z.; Li, C.; Liu, J.; Zeng, X. UDP-Glycosyltransferases are involved in imidacloprid resistance in the Asian citrus psyllid, Diaphorina citri (Hemiptera: Lividae). Pestic. Biochem. Phys. 2019, 154, 23–31. [Google Scholar] [CrossRef]
- He, C.; Liang, J.; Liu, S.; Wang, S.; Wu, Q.; Xie, W.; Zhang, Y. Changes in the expression of four ABC transporter genes in response to imidacloprid in Bemisia tabaci Q (Hemiptera: Aleyrodidae). Pestic. Biochem. Phys. 2019, 153, 136–143. [Google Scholar] [CrossRef]
- Liu, X.; Jiang, H.; Xiong, Y.; Peng, P.; Li, H.F.; Yuan, G.R.; Dou, W.; Wang, J.J. Genome-wide identification of ATP-binding cassette transporters and expression profiles in the Asian citrus psyllid, Diaphorina citri, exposed to imidacloprid. Comp. Biochem. Phys. D Genom. Proteom. 2019, 30, 305–311. [Google Scholar] [CrossRef]
- He, C.; Liang, J.; Liu, S.; Zeng, Y.; Wang, S.; Wu, Q.; Xie, W.; Zhang, Y. Molecular characterization of an NADPH cytochrome P450 reductase from Bemisia tabaci Q: Potential involvement in susceptibility to imidacloprid. Pestic. Biochem. Phys. 2020, 162, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Huang, L.; Wang, Y.; Zhang, Y.; Fang, S.; Liu, Z. No cross-resistance between imidacloprid and pymetrozine in the brown planthopper: Status and mechanisms. Pestic. Biochem. Phys. 2016, 130, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Casida, J.E. Neonicotinoid metabolism: Compounds, substituents, pathways, enzymes, organisms, and relevance. J. Agric. Food Chem. 2011, 59, 2923–2931. [Google Scholar] [CrossRef]
- Schulz-Jander, D.A.; Casida, J.E. Imidacloprid insecticide metabolism: Human cytochrome P450 isozymes differ in selectivity for imidazolidine oxidation versus nitroimine reduction. Toxicol. Lett. 2002, 132, 65–77. [Google Scholar] [CrossRef]
| S. NO. | Sources | Imidacloprid Concentration/Median Lethal Concentration (LC50)/Median Lethal Dose (LD50) | Results | References |
|---|---|---|---|---|
| 1. | Podisus nigrispinus | LC50 = 3.75 mg·L−1 | Histological changes are observed in the mesenteric epithelium | [22] |
| 2. | Humans | 0.05 mg·L−1 | Leads to severe respiratory failure and a drop in consciousness, induces lymphocyte apoptosis | [23,24,25,26,27] |
| 3. | Rats | 1.0 mg·kg−1 | Disorders the pancreatic tissue structure, decreases the expression of insulin, and disrupts blood glucose regulation | [18] |
| 4. | Insect-eating bats | LD50 = 115 mg·kg−1 | Interferes with the echolocation system, reduces the expression of auditory-related proteins in the cochlea | [19] |
| 5. | Earthworm (Eisenia fetida) | LD50 = 3.05 mg·kg−1 | Inhibits fecundity and cellulase activity, and damages the epidermal and midgut cells of earthworm | [21] |
| 6. | Lizards (Eremias argus) | 20 mg·kg−1 | Inhibits thyroid hormone secretion, causes abnormal alignment of the spermatogenic epithelium, epididymal hyperplasia, and oligospermia in male lizards | [28,29,30] |
| 7. | Brown shrimp | 0.5 g·L−1 | Sublethal effect, especially in dehulling delay and growth reduction | [20,31] |
| 8. | Zebrafish (Danio rerio) | 400 µg·L−1 | Sublethal effect, especially in dehulling delay and growth reduction | [20] |
| 9. | Medaka (Japanese rice fish) | 0.2 µg·L−1 | Causes deformity and reduces growth | [20] |
| 10. | Sydney rock oyster | 2 mg·L−1 | Affects the biochemical processes and metabolites, such as the parotid and digestive glands and other metabolic active tissue | [32] |
| 11. | Asian freshwater (Corbicula fluminea) | 2000 µg·L−1 | Causes oxidative stress, cell detoxification, and multixenobiotic resistance systems | [33] |
| 12. | Neotropical fish (Prochilodus lineatus) | 5.45–359 µg·L−1 | Alters the biotransformation and the antioxidant enzyme activity in different tissues | [34] |
| 13. | Aquatic macrophyte (Bidens laevis) | 1–1000 µg·L−1 | Alters the mitotic dynamics and causes aneugenic and clastogenic effects | [35] |
| 14. | Chironomus riparius | LC50 = 31.5 mg·L−1 | Leads to an imbalance between reducing and oxidizing glutathione | [36] |
| 15. | Asian tiger mosquito (Aedes albopictus) | LC50 = 47.5 µg·L−1 | Decreases the sensitivity to imidacloprid | [37] |
| 16. | Engytatus varians | 1400 mg·L−1 | Dies at a concentration of 100% of the maximum field registration concentration | [38] |
| 17. | Aphids | LC50 = 3.186 mg·L−1 | The LC50 of imidacloprid is 6.4 × 10−3 times lower | [39] |
| 18. | Hippodamia variegata | LC10 = 3.92 mg·L−1; LC30 = 8.69 mg·L−1 | Decreases adult longevity and survival rate | [40] |
| 19. | Apis mellifera | LD5048h = 0.0037 lg/bee | Poses a major threat to bee biodiversity, affects the individual immunity of bees | [41,42,43] |
| S. NO. | Physicochemical Methods | Reaction Conditions | Results | References |
|---|---|---|---|---|
| 1. | The photocatalyst ZnO/CoFe2O4 magnetic nanocomposite | Room temperature and pH of 10 | 0.05 g of the photocatalyst can completely degrade the concentration of imidacloprid at 5 mg·L−1 for 45 min | [74] |
| 2. | Based on (SnO2)-Cu2ZnSnS4-TiO2 coating heteropair methylene blue | Visible infra spectroscopy (VIS) | Improves the removal of imidacloprid (10 mg·L−1) compared to the reference TiO2 film | [73] |
| 3. | Ru/TiO2 | Molecularly imprinted titanium dioxide photocatalyst synthesized by the sol–gel method | 9.5 × 10−5 M of imidacloprid is completely degraded in 300 min | [66] |
| 4. | Bamboo vinegar | Under the irradiation of a high-pressure mercury lamp | The half-lives of imidacloprid is 17.6 min at a concentration of 5 mg·L−1 | [83] |
| 5. | A modified carbon nitride/tungstophosphoric acid composite | - | The degradation rate constant(0.70 h−1) of imidacloprid (10 mg·L−1) is 6.4 times than that of carbon nitride 450 | [78] |
| 6. | UV-activated persulfate (UV/PS) and UV-activated peroxymonosulfate (UV/PMS) | UV | UV/PS and UV/PMS systems achieve great removal efficiency of imidacloprid (2.5 mg·L−1) | [80] |
| 7. | Type III photocatalytic composite heterojunction H3PW12O40/TiO2-In2S3 | Under visible light irradiation (λ = 400 nm) | Higher photocatalytic degradation activity (82.7%) | [81] |
| 8. | Double continuous process | The electrolytic oxidation of ozone treatment | Completely removes imidacloprid (5 mg·L−1) in 120 min | [85] |
| 9. | Heterogeneous structure of Ag2O/g-C3N4 | Visible light for 30 min and near-infrared light for 120 min | The degradation rate of imidacloprid (10 mg·L−1) is 66% in 120 min | [94] |
| 10. | Fe(III)-ethylenediamine-N,N-disuccinic acid (EDDS) | Solar radiation | Removes more than 90% of imidacloprid (60 mg·L−1) | [93] |
| 11. | Cork boiling wastewater | pH 5 | Removes more than 95% of imidacloprid (70 mg·L−1) | [92] |
| 12. | Combined advanced oxidation process based on hydrodynamic cavitation | Hydrogen peroxide | Completely removes imidacloprid (60 mg·L−1) in 120 min | [67] |
| 13. | Combined advanced oxidation process based on hydrodynamic cavitation | The Fenton process | Completely removes imidacloprid (60 mg·L−1) in 60 min | [67] |
| 14. | Zinc-based materials and an enzyme hybrid system | Using attenuated total reflection Fourier transform infrared spectroscopy | Degrades 85% of imidacloprid (10 mg·L−1) within 24 h | [91] |
| 15. | GO/Fe3O4/TiO2-NiO | At pH = 9, in visible light | 0.08 g can catalyze 97.34% of imidacloprid (10 mg·L−1) in 30 min | [90] |
| 16. | Tungstophosphoric acid and acidified carbon nitride | Under visible light exposure (λ > 400 nm) | Tungstophosphoric acid and acidified carbon nitride imidacloprid is degraded 16 times higher than that of acidified carbon nitride (ACN) imidacloprid | [89] |
| 17. | Zn0.1Cd0.9s/SnIn4S8 | In visible light | 55% imidacloprid (5 mg·L−1) is degraded within 240 min | [88] |
| 18. | Solar heating technology | Solarization and biosolarization | The disappearance rate of imidacloprid is increased | [95] |
| 19. | Irradiation | UV | The degradation rate of imidacloprid (0.0255 mg·mL−1) is 98.43% after ultraviolet irradiation for 10 min | [77] |
| 20. | Permanganate oxidation | - | It can remove 0.001–0.05 mM of imidacloprid and mainly hydrolyzes the C–H bond | [71] |
| 21. | Sodium percarbonate | Artificial neural network and response surface methodology—Box-Behnken design | 99.54% of imidacloprid (1 mM) is removed at optimum conditions | [69] |
| 22. | Ti/PbO2-Tb | 8 mA·cm−2 current density, pH 9, temperature 30 °C, and 7.0 g·L−1 NaCl electrolyte | 76.07% of imidacloprid (150 mg·L−1) is removed in 5 h; has energy-saving performance and good repeatable use | [70] |
| 23. | Polypyrrole, polyaniline, and sodium alginate composites with peanut husk | pH, pesticide concentration, compound dose, contact time, and temperature | Can effectively remove imidacloprid (25 mg·L−1) | [64] |
| 24. | Fe4O3-GO-β-CD | Metal organic frameworks | Better improves the adsorption capacity for imidacloprid (100 mg·L−1) | [75] |
| S. No. | Microorganisms | Isolation Sources | Handling Methods | Results | References |
|---|---|---|---|---|---|
| 1. | Hymenobacter latericoloratus CGMCC 16346 | Water sediment | High-performance liquid chromatography (HPLC) and liquid chromatography–mass spectrometry (LC–MS) | Degrades 64.4% of imidacloprid (100 mg·L−1) within six days | [134] |
| 2. | Ochrobactum sp. BB-1 | Tea plantation with long-term application of pesticides | Ultrasonic cell -break method and HPLC | The optimal nitrogen source is 16 g·L−1 of yeast extract; the optimal growth conditions include pH 7 and 30 °C | [135] |
| 3. | Rhodopseudomonas capsulata | Wastewater | HPLC | Degrades 97% of imidacloprid in five days | [121] |
| 4. | Acinetobacter sp. TW | Solid tobacco waste | Morphology, physiological and biochemical tests, Biolog analysis, and 16S rDNA sequencing | Degrades acetamiprid and imidacloprid under broad pH and temperature conditions | [126] |
| 5. | Sphingomonas sp. TY | Solid tobacco waste | Morphology, physiological and biochemical tests, Biolog analysis, and 16S rDNA sequencing | Degrades acetamiprid and imidacloprid under broad pH and temperature conditions | [126] |
| 6. | Pseudomonas sp. 1G | Neonicotinoid-exposed golf course soil | HPLC and LC-MS | 28 °C, microaerophilic | [122] |
| 7. | Bacillus aerophilus | Sugarcane field soils | HPLC | Degrades imidacloprid in sandy loam soil | [136] |
| 8. | Bacillus alkalinitrilicus | Sugarcane fields | 16S rRNA sequence homology and amplification of 16S rRNA gene region | Degrades more than 90% of imidacloprid in 56 days | [130] |
| 9. | Bradyrhizobiaceae SG-6C | Soil | HPLC and LC–MS | Hydrolyzes imidacloprid to dechlorinated 6-chloronicotinic acid to 6-hydroxynicotinic acid | [131] |
| 10. | Ochrobactrum BCL-1 | Tea rhizosphere soil | HPLC | Degrades 78% of imidacloprid within seven days under 30 °C | [129] |
| 11. | Klebsiella pneumonia BCH-1 | Pesticide-contaminated agricultural field | HPLC | pH 7, 30 °C, static condition | [137] |
| 12. | Leifsonia sp. PC-21 | Soil | HPLC and LC–MS | Degrades 37–58% of imidacloprid in full strength tryptic soy broth | [114] |
| 13. | Mycobacterium sp. MK6 | Agricultural soil | HPLC and an enrichment technique | Liquid minimal medium | [133] |
| 14. | Pseudoxanthomonas indica CGMCC 6648 | Rhizospheric soils | 16S rRNA gene analysis, LC–MS, and nuclear magnetic resonance analysis | Different organic acids have substantial effects on olefin imidacloprid production | [123] |
| 15. | Rhizobium sp. | Vegetable farming areas | HPLC | Degrades 25.36–45.48% of imidacloprid (25 mg·L−1) in limited media | [138]) |
| 16. | Aspergillus terreus YESM3 | Agricultural wastewater | HPLC | Degrades 85% of imidacloprid (25 mg·L−1) in Czapek Dox broth medium at pH 4 and 28 °C for 6 days under static conditions | [16] |
| 17. | Stenotrophomonas maltophilia CGMCC 1.1788 | China General Microbiological Culture Collection Center | HPLC | Optimal conditions for the reaction are 30 °C and pH 7.2 | [132] |
| 18. | Pseudomonas sp. RPT 52 | Pesticide-contaminated agricultural field | HPLC | Degrades 46.5% of imidacloprid (0.5 mM) within 40 h; follows first-order kinetics | [120] |
| S. No. | Genes | Resources | Results | References |
|---|---|---|---|---|
| 1. | CYP6ER1 | Nilaparvata lugens | Cross-resistant; low level of resistance to dinotefuran | [143] |
| 2. | CYP6FV12 | Bradysia odoriphaga | Most expressed in the fourth instar larvae; when the gene is knocked out, mortality increases by 28.62 | [97] |
| 3. | CYP353D1v2 | Bacillus striatum | Short-term imidacloprid resistance | [144] |
| 4. | CYP4C71v2 | Bacillus striatum | Long-term imidacloprid resistance | [144] |
| 5. | CYP4C72 | Bacillus striatum | Long-term imidacloprid resistance | [144] |
| 6. | CYP6AY3v2 | Bacillus striatum | Long-term imidacloprid resistance | [144] |
| 7. | CYP6CM1 | Bemisia tabaci | Causes no or a low level of cross-resistance of dinotefuran | [142] |
| 8. | CYP6BJa/b | Colorado potato beetle | Mediated by exogenous transcription factors CncC and Maf | [141] |
| 9. | CYP6BJ1v1 | Colorado potato beetle | Mediated by exogenous transcription factors CncC and Maf | [141] |
| 10. | CYP9Z25 | Colorado potato beetle | Mediated by exogenous transcription factors CncC and Maf | [141] |
| 11. | CYP9Z29 | Colorado potato beetle | Mediated by exogenous transcription factors CncC and Maf | [141] |
| 12. | AChE | Diaphorina citri | RNA interference from acetylcholinesterase | [141] |
| 13. | ChE | Diaphorina citri | RNA interference from acetylcholinesterase | [141] |
| 14. | Nlhsp70 | Nilaparvata lugens | Full-length, 2805 bp; open reading frame (ORF), 1896 bp | [147] |
| 15. | UGT375A1 | Diaphorina citri | Significantly improves the group resistance | [150] |
| 16. | UGT383A1 | Diaphorina citri | Significantly improves the group resistance | [150] |
| 17. | UGT383B1 | Diaphorina citri | Significantly improves the group resistance | [150] |
| 18. | UGT384A1 | Diaphorina citri | Significantly improves the group resistance | [150] |
| 19. | Foccα6 | The western flower thrips | Mutations in nicotinic acetylcholine receptor (nAChRs) | [148] |
| 20. | CPM | Rhodopseudomonas capsulata | Controls the synthesis of P450 | [121] |
| 21. | CYP6A14-1 | Sitobion avenae Fabricius | Increases resistance to imidacloprid | [149] |
| 22. | CYP307A1 | Sitobion avenae Fabricius | Increases resistance to imidacloprid | [149] |
| 23. | GST1-1-1 | Sitobion avenae Fabricius | Increases resistance to imidacloprid | [149] |
| 24. | COE2 | Sitobion avenae Fabricius | Increases resistance to imidacloprid | [149] |
| 25. | CYP4CS3 | Bemisia tabaci | Shows cross resistance to imidacloprid and acetamiprid | [145] |
| 26. | CYP6CX5 | Bemisia tabaci | Shows cross resistance to imidacloprid and acetamiprid | [145] |
| 27. | CYP6DW2 | Bemisia tabaci | Shows cross resistance to imidacloprid and acetamiprid | [145] |
| 28. | CYP6CM1 | Bemisia tabaci | Shows cross resistance to imidacloprid and acetamiprid | [145] |
| 29. | cch2 | Bradyrhizobiaceae | A candidate gene encoding the initial dechlorination step and is a member of the metal-dependent hydrolase superfamily | [131] |
| S. No. | Proteins | Resources | Results | References |
|---|---|---|---|---|
| 1. | ABCG3 | Bemisia tabaci | Higher expression level in females than in males | [151] |
| 2. | BtCPR | Bemisia tabaci | Higher expression level in males and head tissues | [153] |
| 3. | R81T | Myzus persicae and aphid cotton | High resistance to imidacloprid; different effects on cyano-nitrosubstituted neonicotinoids and sulfoniloxalic acid | [140] |
| 4. | Aldehyde oxidase | New Zealand rabbits | Plays a more important lethal role than cytochrome P450 | [31] |
| 5. | Aldehyde oxidase | Chinese lizard | Key enzymes in nitro reduction | [50] |
| 6. | CYP2C9 | Chinese lizard | Key enzymes in nitro reduction | [50] |
| 7. | CYP3A4 | Chinese lizard | Hydroxylation and desaturation | [50] |
| 8. | CYP6AY1 | Nilaparvata lugens | Effectively metabolizes imidacloprid | [154] |
| 9. | CYP6ER1 | Nilaparvata lugens | Effectively metabolizes imidacloprid | [154] |
| 10. | DcitABC | Diaphorina citri | Forty-four DcitABC, expressed in multiple D. citri sites | [152] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pang, S.; Lin, Z.; Zhang, Y.; Zhang, W.; Alansary, N.; Mishra, S.; Bhatt, P.; Chen, S. Insights into the Toxicity and Degradation Mechanisms of Imidacloprid Via Physicochemical and Microbial Approaches. Toxics 2020, 8, 65. https://doi.org/10.3390/toxics8030065
Pang S, Lin Z, Zhang Y, Zhang W, Alansary N, Mishra S, Bhatt P, Chen S. Insights into the Toxicity and Degradation Mechanisms of Imidacloprid Via Physicochemical and Microbial Approaches. Toxics. 2020; 8(3):65. https://doi.org/10.3390/toxics8030065
Chicago/Turabian StylePang, Shimei, Ziqiu Lin, Yuming Zhang, Wenping Zhang, Nasser Alansary, Sandhya Mishra, Pankaj Bhatt, and Shaohua Chen. 2020. "Insights into the Toxicity and Degradation Mechanisms of Imidacloprid Via Physicochemical and Microbial Approaches" Toxics 8, no. 3: 65. https://doi.org/10.3390/toxics8030065
APA StylePang, S., Lin, Z., Zhang, Y., Zhang, W., Alansary, N., Mishra, S., Bhatt, P., & Chen, S. (2020). Insights into the Toxicity and Degradation Mechanisms of Imidacloprid Via Physicochemical and Microbial Approaches. Toxics, 8(3), 65. https://doi.org/10.3390/toxics8030065