Knockdown of Butyrylcholinesterase but Not Inhibition by Chlorpyrifos Alters Early Differentiation Mechanisms in Human Neural Stem Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. iPSC and NSC Cultures
2.2. NSC Differentiation
2.3. Chlorpyrifos Exposure
2.4. Lentivirus Production and shRNA Knockdown
2.5. Quantitative Real-Time Polymerase Chain Reaction
2.6. Butyrylcholinesterase and Acetylcholinesterase Enzyme Assays
2.7. Immunocytochemistry and Microscopy
2.8. Statistical Analysis
3. Results
3.1. NSC Model of Early Neuronal Differentiation
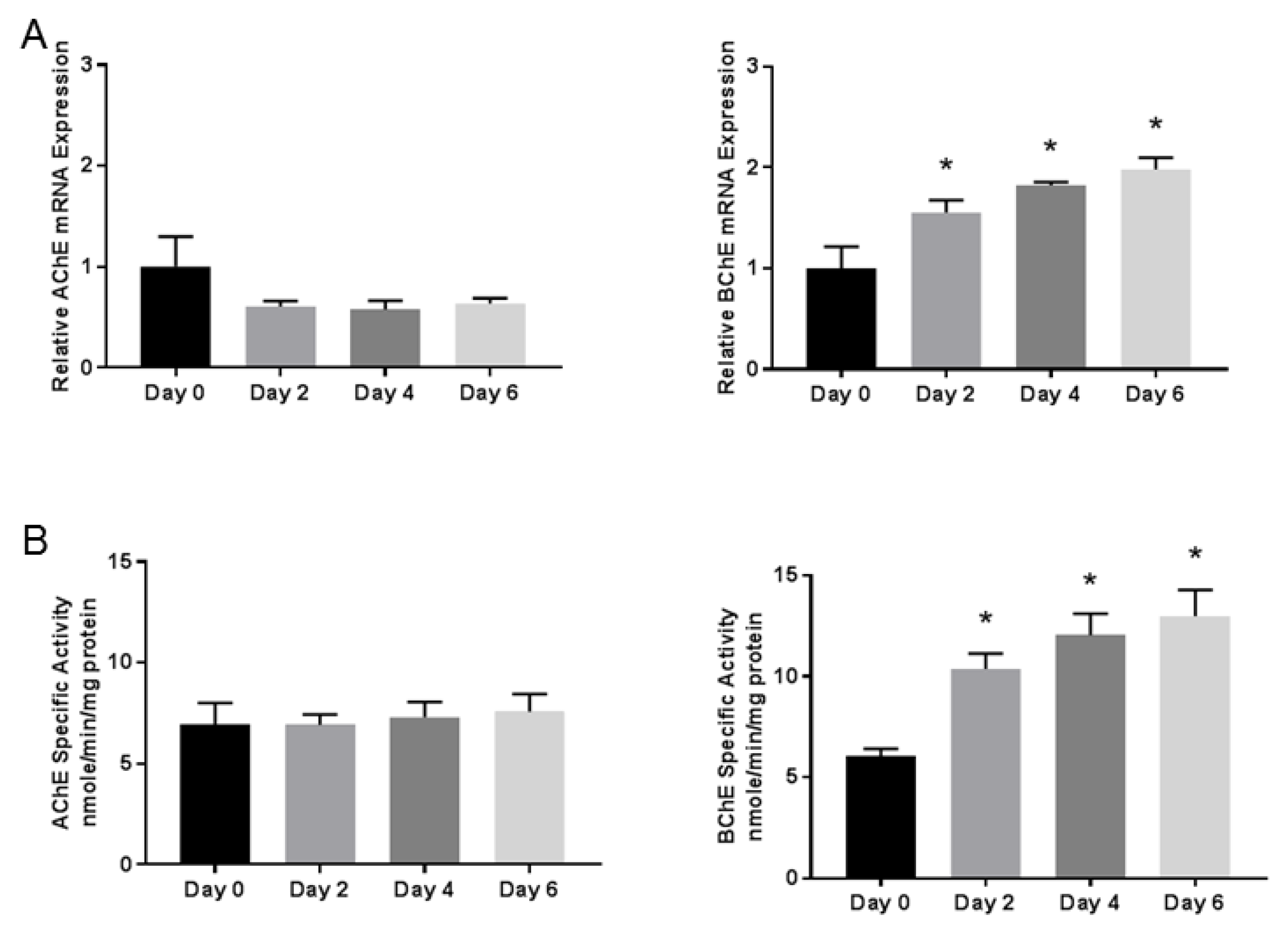
3.2. Butyrylcholinesterase mRNA Expression and Activity Progressively Increases During Differentiation
3.3. Cholinesterase Inhibition by Chlorpyrifos Does Not Affect Neuronal Marker Expression
3.4. Knockdown of BChE mRNA Alters HES5 mRNA Expression
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Eaton, D.L.; Daroff, R.B.; Autrup, H.; Bridges, J.; Buffler, P.; Costa, L.G.; Coyle, J.; McKhann, G.; Mobley, W.C.; Nadel, L.; et al. Review of the toxicology of chlorpyrifos with an emphasis on human exposure and neurodevelopment. Crit. Rev. Toxicol. 2008, 38 (Suppl. S2), 1–125. [Google Scholar] [CrossRef] [PubMed]
- Rauh, V.; Arunajadai, S.; Horton, M.; Perera, F.; Hoepner, L.; Barr, D.B.; Whyatt, R. Seven-year neurodevelopmental scores and prenatal exposure to chlorpyrifos, a common agricultural pesticide. Environ. Health Perspect. 2011, 119, 1196–1201. [Google Scholar] [CrossRef] [PubMed]
- Berkowitz, G.S.; Wetmur, J.G.; Birman-Deych, E.; Obel, J.; Lapinski, R.H.; Godbold, J.H.; Holzman, I.R.; Wolff, M.S. In utero pesticide exposure, maternal paraoxonase activity, and head circumference. Environ. Health Perspect. 2004, 112, 388–391. [Google Scholar] [CrossRef] [PubMed]
- Gunier, R.B.; Bradman, A.; Harley, K.G.; Kogut, K.; Eskenazi, B. Prenatal residential proximity to agricultural pesticide use and IQ in 7-year-old children. Environ. Health Perspect. 2017, 125. [Google Scholar] [CrossRef] [PubMed]
- Richardson, J.; Chambers, J. Effects of gestational exposure to chlorpyrifos on postnatal central and peripheral cholinergic neurochemistry. J. Toxicol. Environ. Health A 2003, 66, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Richardson, J.R.; Chambers, J.E. Effects of repeated oral postnatal exposure to chlorpyrifos on cholinergic neurochemistry in developing rats. Toxicol. Sci. 2005, 84, 352–359. [Google Scholar] [CrossRef] [PubMed]
- Venerosi, A.; Calamandrei, G.; Ricceri, L. A social recognition test for female mice reveals behavioral effects of developmental chlorpyrifos exposure. Neurotoxicol. Teratol. 2006, 28, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Venerosi, A.; Ricceri, L.; Tait, S.; Calamandrei, G. Sex dimorphic behaviors as markers of neuroendocrine disruption by environmental chemicals: The case of chlorpyrifos. Neurotoxicology 2012, 33, 1420–1426. [Google Scholar] [CrossRef] [PubMed]
- De Felice, A.; Venerosi, A.; Ricceri, L.; Sabbioni, M.; Scattoni, M.L.; Chiarotti, F.; Calamandrei, G. Sex-dimorphic effects of gestational exposure to the organophosphate insecticide chlorpyrifos on social investigation in mice. Neurotoxicol. Teratol. 2014, 46, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Venerosi, A.; Tait, S.; Stecca, L.; Chiarotti, F.; De Felice, A.; Cometa, M.F.; Volpe, M.T.; Calamandrei, G.; Ricceri, L. Effects of maternal chlorpyrifos diet on social investigation and brain neuroendocrine markers in the offspring—A mouse study. Environ. Health 2015, 14, 32. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.P.; Jin, J.L.; Li, C.W.; Wang, F.S. Effects of low concentration of chlorpyrifos prenatal exposure on generation mouse brain hippocampus and somatosensory cortex. Chin. J. Ind. Hyg. Occup. Dis. 2009, 27, 557–559. [Google Scholar]
- Roy, T.S.; Sharma, V.; Seidler, F.J.; Slotkin, T.A. Quantitative morphological assessment reveals neuronal and glial deficits in hippocampus after a brief subtoxic exposure to chlorpyrifos in neonatal rats. Dev. Brain Res. 2005, 155, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Rauh, V.A.; Perera, F.P.; Horton, M.K.; Whyatt, R.M.; Bansal, R.; Hao, X.; Liu, J.; Barr, D.B.; Slotkin, T.A.; Peterson, B.S. Brain anomalies in children exposed prenatally to a common organophosphate pesticide. Proc. Natl. Acad. Sci. USA 2012, 109, 7871–7876. [Google Scholar] [CrossRef] [PubMed]
- Costa, L.G.; Giordano, G.; Guizzetti, M.; Vitalone, A. Neurotoxicity of pesticides: A brief review. Front. Biosci. 2008, 13, 1240–1249. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Howard, A.; Bruun, D.; Ajua-Alemanj, M.; Pickart, C.; Lein, P.J. Chlorpyrifos and chlorpyrifos-oxon inhibit axonal growth by interfering with the morphogenic activity of acetylcholinesterase. Toxicol. Appl. Pharmacol. 2008, 228, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Slotkin, T.A.; Seidler, F.J.; Fumagalli, F. Targeting of neurotrophic factors, their receptors, and signaling pathways in the developmental neurotoxicity of organophosphates in vivo and in vitro. Brain Res. Bull. 2008, 76, 424–438. [Google Scholar] [CrossRef] [PubMed]
- Paraoanu, L.E.; Layer, P.G. Acetylcholinesterase in cell adhesion, neurite growth and network formation. FEBS J. 2008, 275, 618–624. [Google Scholar] [CrossRef] [PubMed]
- Johnson, G.; Moore, S.W. Human acetylcholinesterase binds to mouse laminin-1 and human collagen IV by an electrostatic mechanism at the peripheral anionic site. Neurosci. Lett. 2003, 337, 37–40. [Google Scholar] [CrossRef]
- Vogel-Hopker, A.; Sperling, L.E.; Layer, P.G. Co-opting functions of cholinesterases in neural, limb and stem cell development. Protein Pept. Lett. 2012, 19, 155–164. [Google Scholar] [PubMed]
- Drews, U. Cholinesterase in embryonic development. Prog. Histochem. Cytochem. 1975, 7, 1–52. [Google Scholar] [CrossRef]
- Layer, P.G.; Sporns, O. Spatiotemporal relationship of embryonic cholinesterases with cell proliferation in chicken brain and eye. Proc. Natl. Acad. Sci. USA 1987, 84, 284–288. [Google Scholar] [CrossRef] [PubMed]
- Layer, P.G.; Willbold, E. Novel functions of cholinesterases in development, physiology and disease. Prog. Histochem. Cytochem. 1995, 29, 1–94. [Google Scholar] [CrossRef]
- Robitzki, A.; Mack, A.; Chatonnet, A.; Layer, P.G. Transfection of reaggregating embryonic chicken retinal cells with an antisense 5′-DNA butyrylcholinesterase expression vector inhibits proliferation and alters morphogenesis. J. Neurochem. 1997, 69, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Robitzki, A.; Doll, F.; Richter-Landsberg, C.; Layer, P.G. Regulation of the rat oligodendroglia cell line OLN-93 by antisense transfection of butyrylcholinesterase. Glia 2000, 31, 195–205. [Google Scholar] [CrossRef]
- Mack, A.; Robitzki, A. The key role of butyrylcholinesterase during neurogenesis and neural disorders: An antisense-5′butyrylcholinesterase-DNA study. Prog. Neurobiol. 2000, 60, 607–628. [Google Scholar] [CrossRef]
- Zimmerman, G.; Soreq, H. Termination and beyond: Acetylcholinesterase as a modulator of synaptic transmission. Cell Tissue Res. 2006, 326, 655–669. [Google Scholar] [CrossRef] [PubMed]
- Layer, P.G.; Klaczinski, J.; Salfelder, A.; Sperling, L.E.; Thangaraj, G.; Tuschl, C.; Vogel-Hopker, A. Cholinesterases in development: Ache as a firewall to inhibit cell proliferation and support differentiation. Chem. Biol. Interact. 2013, 203, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Gage, F.H.; Temple, S. Neural stem cells: Generating and regenerating the brain. Neuron 2013, 80, 588–601. [Google Scholar] [CrossRef] [PubMed]
- Breier, J.M.; Gassmann, K.; Kayser, R.; Stegeman, H.; De Groot, D.; Fritsche, E.; Shafer, T.J. Neural progenitor cells as models for high-throughput screens of developmental neurotoxicity: State of the science. Neurotoxicol. Teratol. 2010, 32, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, B.Z.; Lehmann, M.; Gutbier, S.; Nembo, E.; Noel, S.; Smirnova, L.; Forsby, A.; Hescheler, J.; Avci, H.X.; Hartung, T.; et al. In vitro acute and developmental neurotoxicity screening: An overview of cellular platforms and high-throughput technical possibilities. Arch. Toxicol. 2017, 91, 1–33. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, T.E.; Bergendahl, V.; Levenstein, M.E.; Yu, J.; Probasco, M.D.; Thomson, J.A. Feeder-independent culture of human embryonic stem cells. Nat. Methods 2006, 3, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.C.; Sheldon, M.H.; Hart, R.P. Biobanking in the Era of the Stem Cell: A Technical and Operational Guide; Morgan & Claypool Life Sciences: San Rafael, CA, USA, 2012; p. 78. [Google Scholar]
- Goff, L.A.; Davila, J.; Swerdel, M.R.; Moore, J.C.; Cohen, R.I.; Wu, H.; Sun, Y.E.; Hart, R.P. Ago2 immunoprecipitation identifies predicted microRNAs in human embryonic stem cells and neural precursors. PLoS ONE 2009, 4, e7192. [Google Scholar] [CrossRef] [PubMed]
- Gerrard, L.; Rodgers, L.; Cui, W. Differentiation of human embryonic stem cells to neural lineages in adherent culture by blocking bone morphogenetic protein signaling. Stem Cells 2005, 23, 1234–1241. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta c(t)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Sauvageau, M.; Goff, L.A.; Lodato, S.; Bonev, B.; Groff, A.F.; Gerhardinger, C.; Sanchez-Gomez, D.B.; Hacisuleyman, E.; Li, E.; Spence, M.; et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife 2013, 2, e01749. [Google Scholar] [CrossRef] [PubMed]
- Kageyama, R.; Ohtsuka, T. The Notch-Hes pathway in mammalian neural development. Cell Res. 1999, 9, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Peng, I.; Binder, L.I.; Black, M.M. Cultured neurons contain a variety of microtubule-associated proteins. Brain Res. 1985, 361, 200–211. [Google Scholar] [CrossRef]
- Bai, J.; Ramos, R.L.; Ackman, J.B.; Thomas, A.M.; Lee, R.V.; LoTurco, J.J. RNAi reveals doublecortin is required for radial migration in rat neocortex. Nat. Neurosci. 2003, 6, 1277–1283. [Google Scholar] [CrossRef] [PubMed]
- Buzanska, L.; Sypecka, J.; Nerini-Molteni, S.; Compagnoni, A.; Hogberg, H.T.; del Torchio, R.; Domanska-Janik, K.; Zimmer, J.; Coecke, S. A human stem cell-based model for identifying adverse effects of organic and inorganic chemicals on the developing nervous system. Stem Cells 2009, 27, 2591–2601. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.Y.; Wegner, S.H.; Van Ness, K.P.; Park, J.J.; Pacheco, S.E.; Workman, T.; Hong, S.; Griffith, W.; Faustman, E.M. Differential epigenetic effects of chlorpyrifos and arsenic in proliferating and differentiating human neural progenitor cells. Reprod. Toxicol. 2016, 65, 212–223. [Google Scholar] [CrossRef] [PubMed]
- Zarei, M.H.; Soodi, M.; Qasemian-Lemraski, M.; Jafarzadeh, E.; Taha, M.F. Study of the chlorpyrifos neurotoxicity using neural differentiation of adipose tissue-derived stem cells. Environ. Toxicol. 2016, 31, 1510–1519. [Google Scholar] [CrossRef] [PubMed]
- Slotkin, T.A.; Skavicus, S.; Card, J.; Levin, E.D.; Seidler, F.J. Diverse neurotoxicants target the differentiation of embryonic neural stem cells into neuronal and glial phenotypes. Toxicology 2016, 372, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Christen, V.; Rusconi, M.; Crettaz, P.; Fent, K. Developmental neurotoxicity of different pesticides in PC-12 cells in vitro. Toxicol. Appl. Pharmacol. 2017, 325, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Sindi, R.A.; Harris, W.; Arnott, G.; Flaskos, J.; Lloyd Mills, C.; Hargreaves, A.J. Chlorpyrifos- and chlorpyrifos oxon-induced neurite retraction in pre-differentiated N2a cells is associated with transient hyperphosphorylation of neurofilament heavy chain and ERK 1/2. Toxicol. Appl. Pharmacol. 2016, 308, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Kageyama, R.; Ohtsuka, T.; Hatakeyama, J.; Ohsawa, R. Roles of bHLH genes in neural stem cell differentiation. Exp. Cell Res. 2005, 306, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Ohtsuka, T.; Ishibashi, M.; Gradwohl, G.; Nakanishi, S.; Guillemot, F.; Kageyama, R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 1999, 18, 2196–2207. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, M.; Isaka, F.; Ishibashi, M.; Tomita, K.; Tsuda, H.; Nakanishi, S.; Kageyama, R. Structure, chromosomal locus, and promoter of mouse Hes2 Gene, a homologue of Drosophila hairy and Enhancer of split. Genomics 1998, 49, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, J.; Bessho, Y.; Katoh, K.; Ookawara, S.; Fujioka, M.; Guillemot, F.; Kageyama, R. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development 2004, 131, 5539–5550. [Google Scholar] [CrossRef] [PubMed]
- Lavado, A.; Oliver, G. Jagged1 is necessary for postnatal and adult neurogenesis in the dentate gyrus. Dev. Biol. 2014, 388, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Wilhelmsson, U.; Faiz, M.; de Pablo, Y.; Sjoqvist, M.; Andersson, D.; Widestrand, A.; Potokar, M.; Stenovec, M.; Smith, P.L.; Shinjyo, N.; et al. Astrocytes negatively regulate neurogenesis through the Jagged1-mediated Notch pathway. Stem Cells 2012, 30, 2320–2329. [Google Scholar] [CrossRef] [PubMed]
- Scholl, F.G.; Scheiffele, P. Making connections: Cholinesterase-domain proteins in the CNS. Trends Neurosci. 2003, 26, 618–624. [Google Scholar] [CrossRef] [PubMed]
- Brazzolotto, X.; Wandhammer, M.; Ronco, C.; Trovaslet, M.; Jean, L.; Lockridge, O.; Renard, P.Y.; Nachon, F. Human butyrylcholinesterase produced in insect cells: Huprine-based affinity purification and crystal structure. FEBS J. 2012, 279, 2905–2916. [Google Scholar] [CrossRef] [PubMed]
- Bodur, E.; Layer, P.G. Counter-regulation of cholinesterases: Differential activation of PKC and ERK signaling in retinal cells through BChE knockdown. Biochimie 2011, 93, 469–476. [Google Scholar] [CrossRef] [PubMed]



| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| ACHE | GGGGCTCAGCAGTACGTTAG | CGGTGGCGCTGAGCAATTT |
| BCHE | CTTTGTTGCAGAGAATCGGAAATC | CGGTGGCGCTGAGCAATTT |
| DCX | GACTCAGCAAACGGAACCTCC | GAATCACCAAGCGAGTCCGA |
| MAP2 | TGCGCTGATTCTTCAGCTTG | TGTGTCGTGTTCTCAAAGGGT |
| HES5 | CGGCACCAGCCCAACTCCAA | GCGACGAAGGCTTTGCTGTGC |
| HES1 | GAAAGATAGCTCGCGGCATT | TACTTCCCCAGCACACTTGG |
| HES3 | CTGATGGAGAAAAAGCGCCG | TTCCGGATCTGGTGCGAGTA |
| DELTA4 | TGTGCAAGAAGCGCAATGAC | AAGACAGATAGGCTGTTGGCA |
| JAG1 | GTCTCAACGGGGGAACTTGT | GCGTGCTCAGCAATTTCACA |
| TBP | AAGACCATTGCACTTCGTGCCC | TGGACTGTTCTTCACTCTTGGCTCC |
| CYP3A7 | TTGAAACACGTCTTTGGGGC | TGAGAGAACGAATGGATCTAATGGA |
| POR | TGCCAGCGTTTCATGATCAAC | GAGACCCACGATGAGCGAAA |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tiethof, A.K.; Richardson, J.R.; Hart, R.P. Knockdown of Butyrylcholinesterase but Not Inhibition by Chlorpyrifos Alters Early Differentiation Mechanisms in Human Neural Stem Cells. Toxics 2018, 6, 52. https://doi.org/10.3390/toxics6030052
Tiethof AK, Richardson JR, Hart RP. Knockdown of Butyrylcholinesterase but Not Inhibition by Chlorpyrifos Alters Early Differentiation Mechanisms in Human Neural Stem Cells. Toxics. 2018; 6(3):52. https://doi.org/10.3390/toxics6030052
Chicago/Turabian StyleTiethof, Angela K., Jason R. Richardson, and Ronald P. Hart. 2018. "Knockdown of Butyrylcholinesterase but Not Inhibition by Chlorpyrifos Alters Early Differentiation Mechanisms in Human Neural Stem Cells" Toxics 6, no. 3: 52. https://doi.org/10.3390/toxics6030052
APA StyleTiethof, A. K., Richardson, J. R., & Hart, R. P. (2018). Knockdown of Butyrylcholinesterase but Not Inhibition by Chlorpyrifos Alters Early Differentiation Mechanisms in Human Neural Stem Cells. Toxics, 6(3), 52. https://doi.org/10.3390/toxics6030052






