A Review of Quantitative Structure–Activity Relationship (QSAR) Models to Predict Thyroid Hormone System Disruption by Chemical Substances
Highlights
- This review maps the landscape of QSAR models developed between 2010 and 2024 for predicting the disruption of the thyroid hormone system, induced by chemicals, within an AOP framework.
- This review shows progress in modelling key MIEs (e.g., TR, TTR) but reveals that many other MIEs are poorly addressed or entirely overlooked.
- This review identifies a preference for classification-based approaches, a frequent reliance on simple algorithms, insufficient AD definitions, and limited mechanistic interpretations and chemical space coverage, challenging model confidence and/or its broader application.
- This review provides a state-of-the-art resource to guide future QSAR development for thyroid hormone system disruption by consolidating existing knowledge and identifying critical research gaps.
- The findings suggest a clear need for future research to address overlooked MIEs, to expand the range of chemical classes studied, and to develop new QSARs with explicitly defined ADs and improved mechanistic interpretability.
Abstract
1. Introduction
2. Materials and Methods
Criteria of Inclusion and Exclusion and Literature Collection
3. Results and Discussion
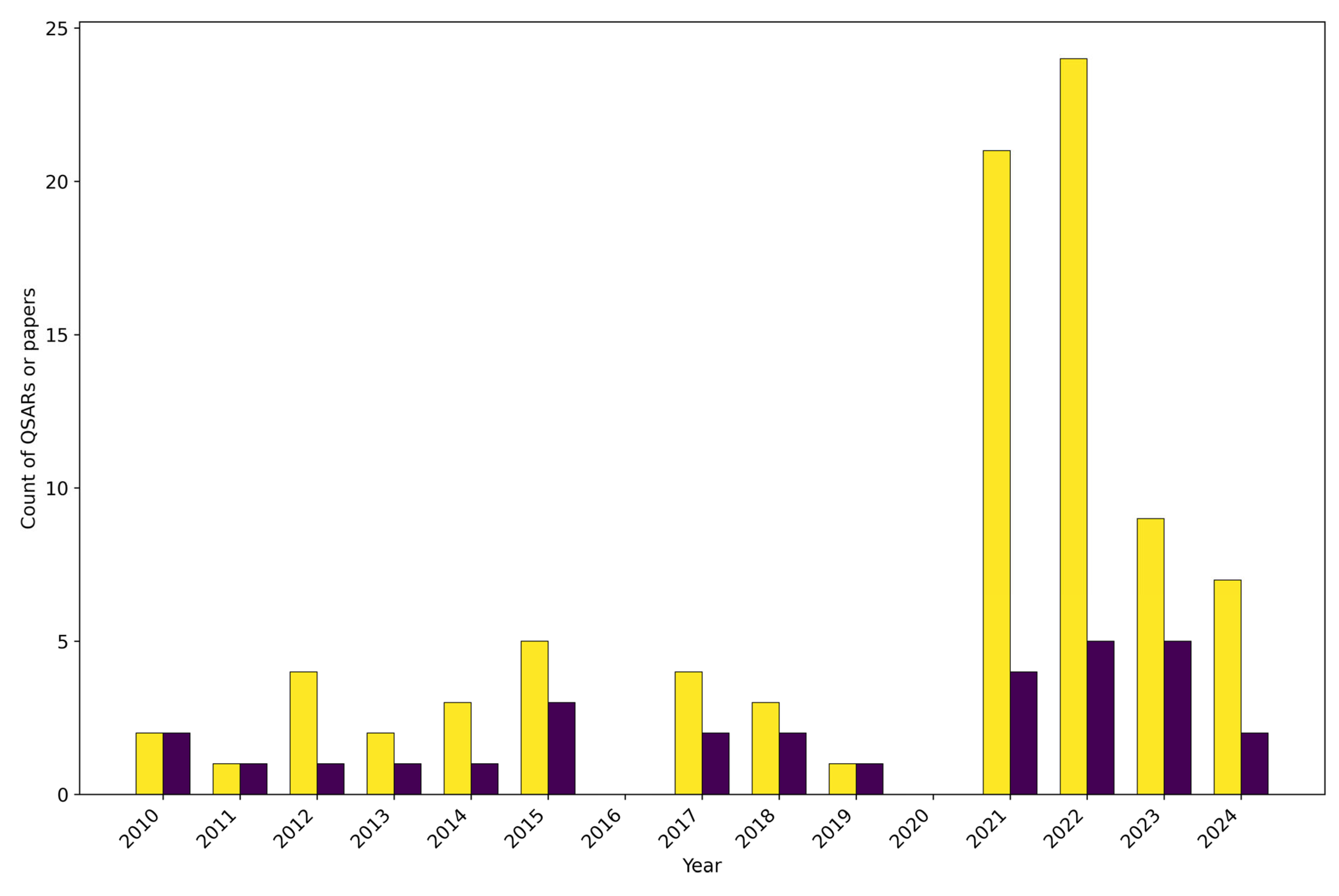
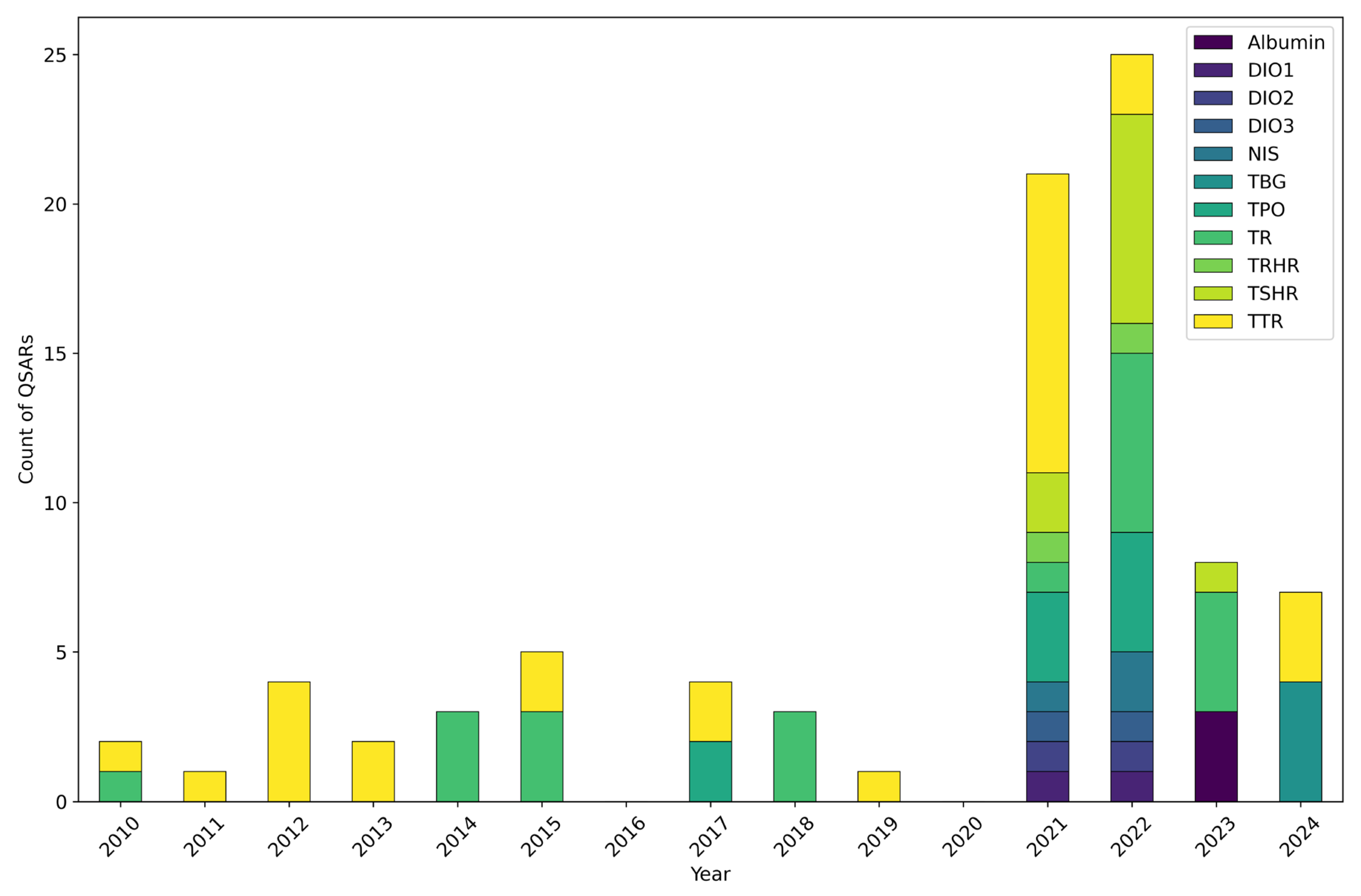
3.1. Temporal Trend
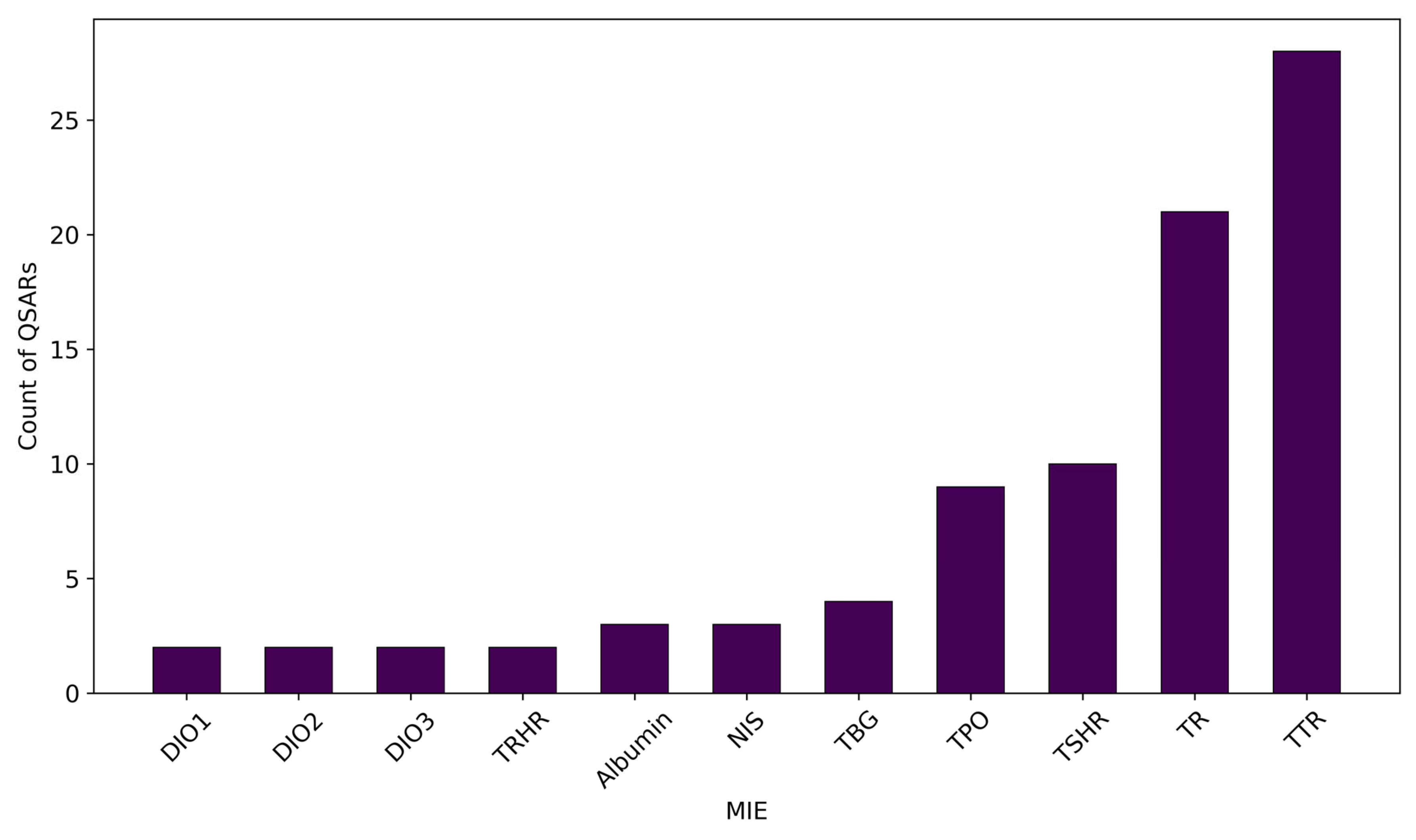
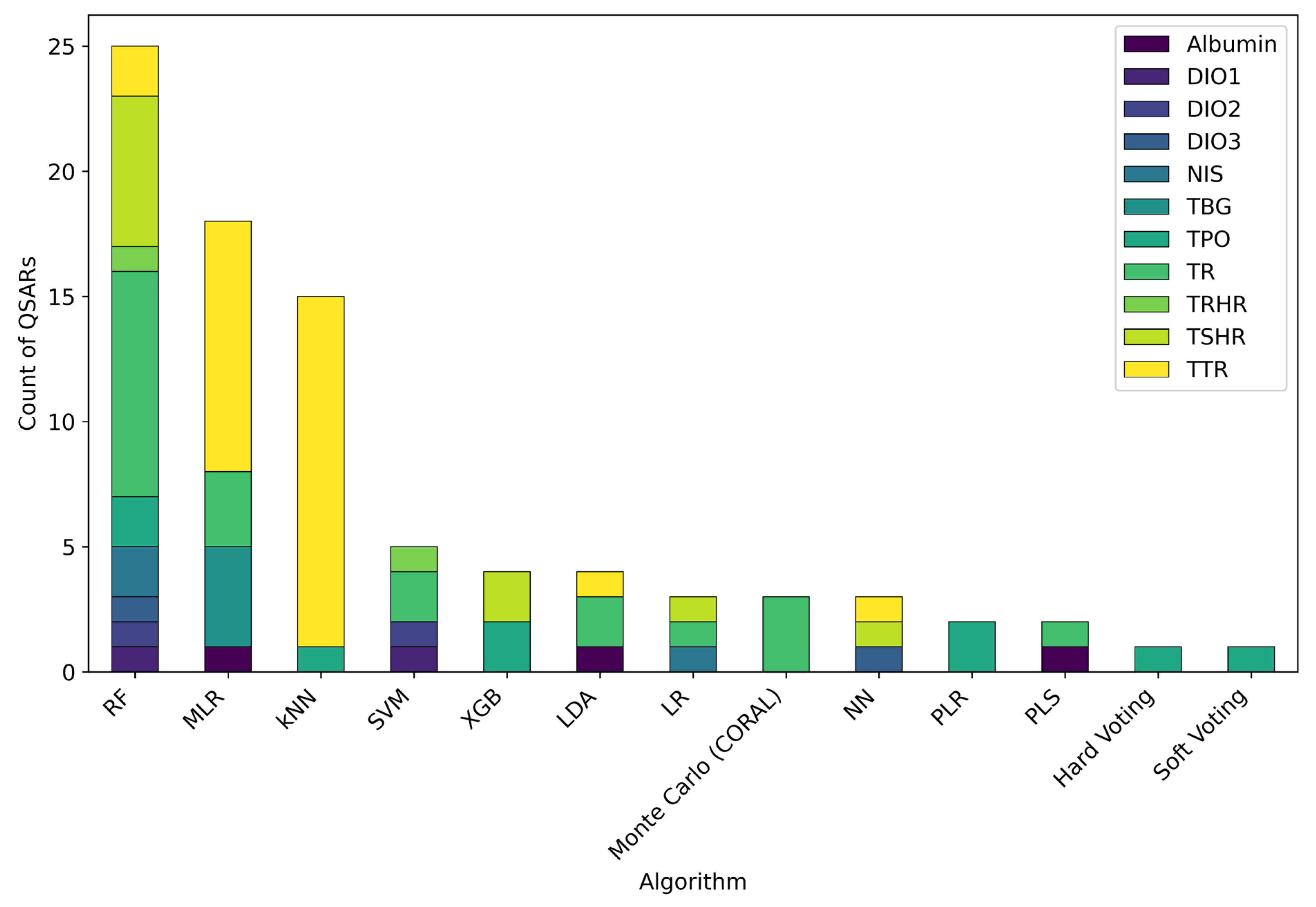
3.2. Modelled MIEs
3.3. Data Sources
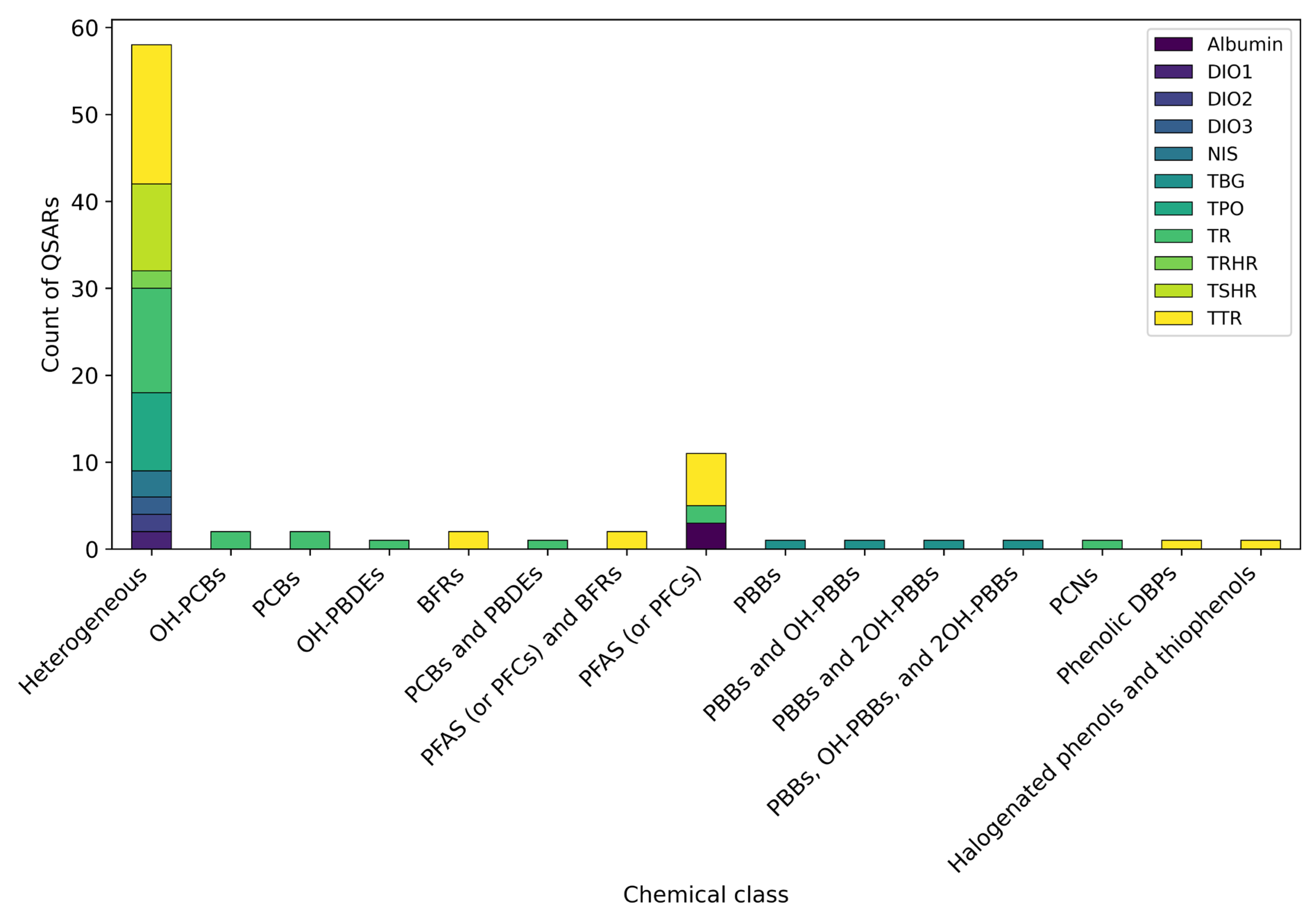
3.4. Chemical Classes
3.5. Modelling Approaches
3.6. Validation Strategies
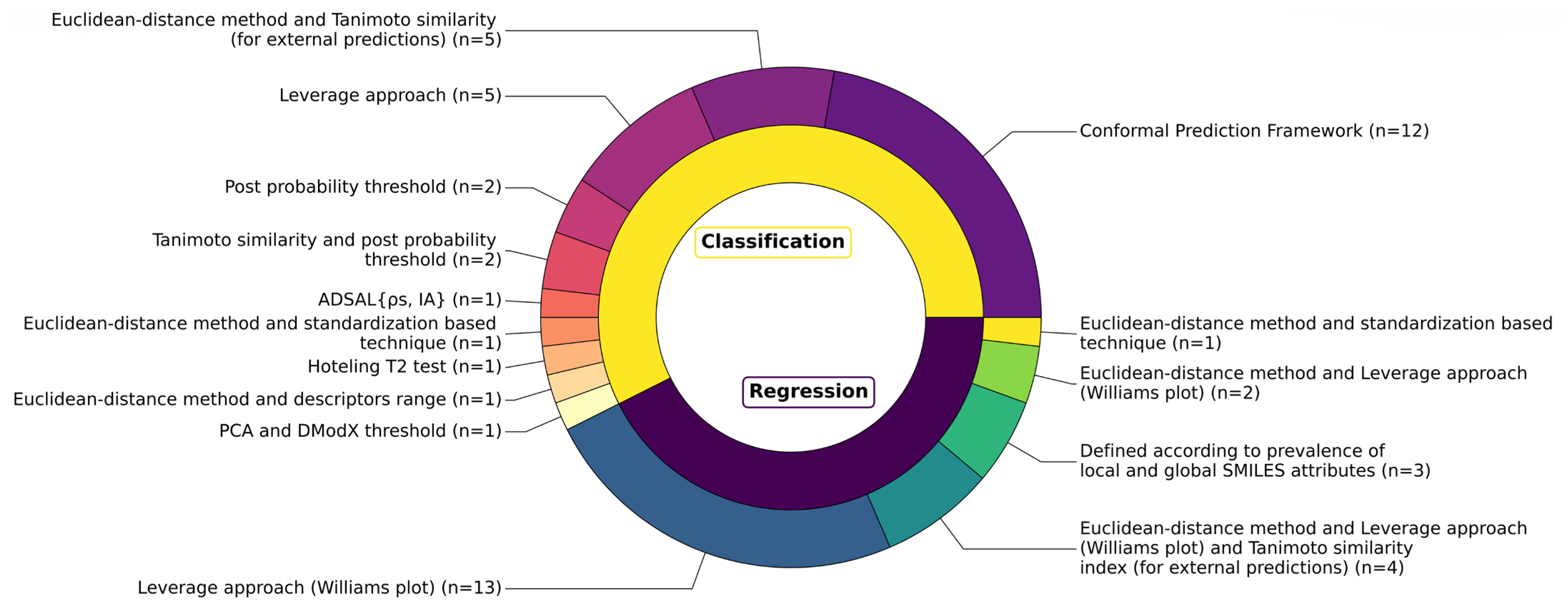
3.7. Applicability Domains
3.8. Molecular Descriptors: Mechanistic Interpretations and Feature Importance
3.9. Recent Advances: 2025
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AD | Applicability domain |
| AdaB | Adaptive Boosting |
| AhR | Aryl hydrocarbon receptor |
| AOP | Adverse outcome pathway |
| ASNNs | Associative neural networks |
| BFR | Brominated flame retardant |
| BMC | Biopartitioning micellar chromatography |
| CAR | Constitutive androstane receptor |
| CART | Classification and regression trees |
| CP | Conformal prediction |
| DBP | Phenolic disinfection byproduct |
| DIO | Iodothyronine deiodinase |
| DIO1 | Type 1 deiodinase |
| DIO2 | Type 2 deiodinase |
| DIO3 | Type 3 deiodinase |
| DTC | Decision Tree Classifier |
| DUOX | Dual oxidase |
| EDC | Endocrine-disrupting chemical |
| EU | European Union |
| EURL ECVAM | European Union Reference Laboratory for Alternatives to Animal Testing |
| FAIR | Findable, Accessible, Interoperable, Reusable |
| FP | Fingerprint |
| HPA | Hypothalamic–pituitary–adrenal |
| HPG | Hypothalamic–pituitary–gonadal |
| HPT | Hypothalamic–pituitary–thyroid |
| HTS | High-throughput screening |
| IYD | Iodotyrosine deiodinase |
| kNN | k-nearest neighbours |
| LDA | Linear discriminant analysis |
| LMO | Leave more out |
| LOO | Leave one out |
| LR | Logistic regression |
| MCT | Monocarboxylate transporter |
| MCT10 | Monocarboxylate transporter 10 |
| MCT8 | Monocarboxylate transporter 8 |
| MDR1 | Multidrug resistance protein 1 |
| MIE | Molecular initiating event |
| MLR | Multiple linear regression |
| MRM | Multiple regression model |
| MRP2 | Multidrug resistance-associated protein 2 |
| MW | Molecular weight |
| NAMs | New approach methodologies |
| NIS | Sodium iodide symporter |
| NN | Neural network |
| OATP | Organic anion transporter polypeptide |
| OATP1A4 | Organic anion transporter polypeptide 1A4 |
| OATP1C1 | Organic anion transporter polypeptide 1C1 |
| OECD | Organisation for Economic Co-operation and Development |
| PBB | Polybrominated biphenyl |
| PBDE | Polybrominated diphenyl ether |
| PCA | Principal component analysis |
| PCB | Polychlorinated biphenyl |
| PCN | Polychlorinated naphthalene |
| PFAS | Per- and polyfluoroalkyl substances |
| PFC | Poly- and perfluorinated compound |
| PLR | Partial logistic regression |
| PLS | Partial least squares |
| PLS-DA | Partial least squares discriminant analysis |
| PPAR | Peroxisome proliferator-activated receptor |
| PS | Prediction entropy |
| PXR | Pregnane X receptor |
| QSAR | Quantitative structure–activity relationship |
| RF | Random forest |
| SHAP | Shapley additive explanation |
| SMOTEENN | Synthetic minority over-sampling technique-edited nearest neighbours |
| SVM | Support vector machine |
| T3 | Triiodothyronine |
| T4 | Thyroxine |
| TBG | Thyroid-binding globulin |
| TH | Thyroid hormone |
| THSDC | Thyroid hormone system-disrupting chemical |
| TPO | Thyroperoxidase |
| TR | Thyroid hormone receptor |
| TRHR | Thyrotropin-releasing hormone receptor |
| TSHR | Thyroid-stimulating hormone receptor |
| TTR | Transthyretin |
| XGB | Extreme gradient boosting |
References
- Gore, A.C.; La Merrill, M.A.; Patisaul, H.B.; Sargis, R. Endocrine Disrupting Chemicals: Threats to Human Health. The Endocrine Society and IPEN. 2024. Available online: https://ipen.org/sites/default/files/documents/edc_report-2024-final-compressed.pdf (accessed on 22 June 2025).
- World Health Organization/International Programme on Chemical Safety (WHO/IPCS). Global Assessment on the State of the Science of Endocrine Disruptors; World Health Organization: Geneva, Switzerland, 2002. [Google Scholar]
- Ahn, C.; Jeung, E.-B. Endocrine-Disrupting Chemicals and Disease Endpoints. Int. J. Mol. Sci. 2023, 24, 5342. [Google Scholar] [CrossRef]
- World Health Organization/International Programme on Chemical Safety (WHO/IPCS). State of the Science of Endocrine Disrupting Chemicals 2012; World Health Organization: Geneva, Switzerland, 2012. [Google Scholar]
- de Oliveira Santos, A.D.; do Nascimento, M.T.L.; de Freitas, A.d.S.; de Carvalho, D.G.; Bila, D.M.; Hauser-Davis, R.A.; Monteiro da Fonseca, E.; Baptista Neto, J.A. The Evolution of Endocrine Disruptor Chemical Assessments Worldwide in the Last Three Decades. Mar. Pollut. Bull. 2023, 197, 115727. [Google Scholar] [CrossRef]
- FAO. Exposure to Endocrine Disrupting Chemicals—Changes from 2002 to 2024; Food Safety and Quality Series, No. 30; FAO: Rome, Italy, 2024. [Google Scholar] [CrossRef]
- Diamanti-Kandarakis, E.; Bourguignon, J.-P.; Giudice, L.C.; Hauser, R.; Prins, G.S.; Soto, A.M.; Zoeller, R.T.; Gore, A.C. Endocrine-Disrupting Chemicals: An Endocrine Society Scientific Statement. Endocr. Rev. 2009, 30, 293–342. [Google Scholar] [CrossRef]
- Feldt-Rasmussen, U.; Effraimidis, G.; Klose, M. The Hypothalamus-Pituitary-Thyroid (HPT)-Axis and Its Role in Physiology and Pathophysiology of Other Hypothalamus-Pituitary Functions. Mol. Cell Endocrinol. 2021, 525, 111173. [Google Scholar] [CrossRef]
- Fekete, C.; Lechan, R.M. Central Regulation of Hypothalamic-Pituitary-Thyroid Axis Under Physiological and Pathophysiological Conditions. Endocr. Rev. 2014, 35, 159–194. [Google Scholar] [CrossRef] [PubMed]
- Sabatino, L.; Vassalle, C. Thyroid Hormones and Metabolism Regulation: Which Role on Brown Adipose Tissue and Browning Process? Biomolecules 2025, 15, 361. [Google Scholar] [CrossRef] [PubMed]
- Mullur, R.; Liu, Y.-Y.; Brent, G.A. Thyroid Hormone Regulation of Metabolism. Physiol. Rev. 2014, 94, 355–382. [Google Scholar] [CrossRef] [PubMed]
- De Luca, R.; Davis, P.J.; Lin, H.-Y.; Gionfra, F.; Percario, Z.A.; Affabris, E.; Pedersen, J.Z.; Marchese, C.; Trivedi, P.; Anastasiadou, E.; et al. Thyroid Hormones Interaction With Immune Response, Inflammation and Non-Thyroidal Illness Syndrome. Front. Cell Dev. Biol. 2021, 8, 614030. [Google Scholar] [CrossRef]
- Sawicka-Gutaj, N.; Zawalna, N.; Gut, P.; Ruchała, M. Relationship between Thyroid Hormones and Central Nervous System Metabolism in Physiological and Pathological Conditions. Pharmacol. Rep. 2022, 74, 847–858. [Google Scholar] [CrossRef]
- Bassett, J.H.D.; Williams, G.R. Role of Thyroid Hormones in Skeletal Development and Bone Maintenance. Endocr. Rev. 2016, 37, 135–187. [Google Scholar] [CrossRef]
- Silva, J.F.; Ocarino, N.M.; Serakides, R. Thyroid Hormones and Female Reproduction. Biol. Reprod. 2018, 99, 907–921. [Google Scholar] [CrossRef] [PubMed]
- Yamakawa, H.; Kato, T.S.; Noh, J.Y.; Yuasa, S.; Kawamura, A.; Fukuda, K.; Aizawa, Y. Thyroid Hormone Plays an Important Role in Cardiac Function: From Bench to Bedside. Front. Physiol. 2021, 12, 606931. [Google Scholar] [CrossRef] [PubMed]
- Brent, G.A. Mechanisms of Thyroid Hormone Action. J. Clin. Investig. 2012, 122, 3035–3043. [Google Scholar] [CrossRef]
- Alcaide Martin, A.; Mayerl, S. Local Thyroid Hormone Action in Brain Development. Int. J. Mol. Sci. 2023, 24, 12352. [Google Scholar] [CrossRef]
- Giannocco, G.; Kizys, M.M.L.; Maciel, R.M.; de Souza, J.S. Thyroid Hormone, Gene Expression, and Central Nervous System: Where We Are. Semin. Cell Dev. Biol. 2021, 114, 47–56. [Google Scholar] [CrossRef]
- Adu-Gyamfi, E.A.; Wang, Y.-X.; Ding, Y.-B. The Interplay between Thyroid Hormones and the Placenta: A Comprehensive Review. Biol. Reprod. 2020, 102, 8–17. [Google Scholar] [CrossRef]
- Moog, N.K.; Entringer, S.; Heim, C.; Wadhwa, P.D.; Kathmann, N.; Buss, C. Influence of Maternal Thyroid Hormones during Gestation on Fetal Brain Development. Neuroscience 2017, 342, 68–100. [Google Scholar] [CrossRef]
- Köhrle, J.; Frädrich, C. Thyroid Hormone System Disrupting Chemicals. Best Pract. Res. Clin. Endocrinol. Metab. 2021, 35, 101562. [Google Scholar] [CrossRef]
- Kortenkamp, A.; Axelstad, M.; Baig, A.H.; Bergman, Å.; Bornehag, C.-G.; Cenijn, P.; Christiansen, S.; Demeneix, B.; Derakhshan, A.; Fini, J.-B.; et al. Removing Critical Gaps in Chemical Test Methods by Developing New Assays for the Identification of Thyroid Hormone System-Disrupting Chemicals—The ATHENA Project. Int. J. Mol. Sci. 2020, 21, 3123. [Google Scholar] [CrossRef]
- Oliveira, K.J.; Chiamolera, M.I.; Giannocco, G.; Pazos-Moura, C.C.; Ortiga-Carvalho, T.M. Thyroid Function Disruptors: From Nature to Chemicals. J. Mol. Endocrinol. 2019, 62, R1–R19. [Google Scholar] [CrossRef]
- Calsolaro, V.; Pasqualetti, G.; Niccolai, F.; Caraccio, N.; Monzani, F. Thyroid Disrupting Chemicals. Int. J. Mol. Sci. 2017, 18, 2583. [Google Scholar] [CrossRef]
- Boas, M.; Feldt-Rasmussen, U.; Main, K.M. Thyroid Effects of Endocrine Disrupting Chemicals. Mol. Cell Endocrinol. 2012, 355, 240–248. [Google Scholar] [CrossRef]
- Salazar, P.; Villaseca, P.; Cisternas, P.; Inestrosa, N.C. Neurodevelopmental Impact of the Offspring by Thyroid Hormone System-Disrupting Environmental Chemicals during Pregnancy. Environ. Res. 2021, 200, 111345. [Google Scholar] [CrossRef] [PubMed]
- Alsen, M.; Sinclair, C.; Cooke, P.; Ziadkhanpour, K.; Genden, E.; van Gerwen, M. Endocrine Disrupting Chemicals and Thyroid Cancer: An Overview. Toxics 2021, 9, 14. [Google Scholar] [CrossRef] [PubMed]
- Olanrewaju, O.A.; Asghar, R.; Makwana, S.; Yahya, M.; Kumar, N.; Khawar, M.H.; Ahmed, A.; Islam, T.; Kumari, K.; Shadmani, S.; et al. Thyroid and Its Ripple Effect: Impact on Cardiac Structure, Function, and Outcomes. Cureus 2024, 16, e51574. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.D.L.; Obeng-Gyasi, B.; Hall, J.E.; Shekhar, S. The Thyroid Hormone Axis and Female Reproduction. Int. J. Mol. Sci. 2023, 24, 9815. [Google Scholar] [CrossRef]
- Murk, A.J.; Rijntjes, E.; Blaauboer, B.J.; Clewell, R.; Crofton, K.M.; Dingemans, M.M.L.; David Furlow, J.; Kavlock, R.; Köhrle, J.; Opitz, R.; et al. Mechanism-Based Testing Strategy Using in Vitro Approaches for Identification of Thyroid Hormone Disrupting Chemicals. Toxicol. Vitr. 2013, 27, 1320–1346. [Google Scholar] [CrossRef]
- Crofton, K.M. Thyroid Disrupting Chemicals: Mechanisms and Mixtures. Int. J. Androl. 2008, 31, 209–223. [Google Scholar] [CrossRef]
- Bernasconi, C.; Sampani, S.; Beronius, A.; Coecke, S.; Langezaal, I.; Pistollato, F.; Paini, A.; Muñoz, A.; Asturiol, D.; Kienzler, A.; et al. Chemical Selection for the Thyroid Validation Study Coordinated by EURL ECVAM and Involving EU-NETVAL Laboratories. ALTEX—Altern. Anim. Exp. 2025. [Google Scholar] [CrossRef]
- European Commission. The European Green Deal 2019; European Commission: Brussels, Belgium, 2019. [Google Scholar]
- European Commission. Chemicals Strategy for Sustainability Towards a Toxic-Free Environment 2020; European Commission: Brussels, Belgium, 2020. [Google Scholar]
- Holmer, M.L.; Holmberg, R.D.; Despicht, C.; Bouftas, N.; Axelstad, M.; Beronius, A.; Zilliacus, J.; Van Duursen, M.; Svingen, T. Assessment of Endocrine Disruptors in the European Union: Current Regulatory Framework, Use of New Approach Methodologies (NAMs) and Recommendations for Improvements. Regul. Toxicol. Pharmacol. 2025, 162, 105883. [Google Scholar] [CrossRef]
- Ramhøj, L.; Axelstad, M.; Baert, Y.; Cañas-Portilla, A.I.; Chalmel, F.; Dahmen, L.; De La Vieja, A.; Evrard, B.; Haigis, A.-C.; Hamers, T.; et al. New Approach Methods to Improve Human Health Risk Assessment of Thyroid Hormone System Disruption–a PARC Project. Front. Toxicol. 2023, 5, 1189303. [Google Scholar] [CrossRef] [PubMed]
- ECHA. Key Areas of Regulatory Challenge. 2025. Available online: https://echa.europa.eu/documents/10162/17228/key_areas_regulatory_challenge_2025_en.pdf/da33bf25-2b75-1fe9-c308-53043f9b9a28?t=1749466525527 (accessed on 22 June 2025).
- OECD. Introduction. In Revised Guidance Document 150 on Standardised Test Guidelines for Evaluating Chemicals for Endocrine Disruption; OECD publishing: Paris, France, 2018; pp. 19–39. [Google Scholar]
- European Commission. Commission Delegated Regulation (EU) 2017/2100 of 4 September 2017 Setting out Scientific Criteria for the Determination of Endocrine-Disrupting Properties Pursuant to Regulation (EU) No 528/2012 of the European Parliament and Council (Text with EEA Relevance); European Commission: Brussels, Belgium, 2017. [Google Scholar]
- European Commission. Commission Regulation (EU) 2018/605 of 19 April 2018 Amending Annex II to Regulation (EC) No 1107/2009 by Setting out Scientific Criteria for the Determination of Endocrine Disrupting Properties (Text with EEA Relevance); European Commission: Brussels, Belgium, 2018. [Google Scholar]
- European Commission. Commission Delegated Regulation (EU) 2023/707 of 19 December 2022 Amending Regulation (EC) No 1272/2008 as Regards Hazard Classes and Criteria for the Classification, Labelling and Packaging of Substances and Mixtures (Text with EEA Relevance); European Commission: Brussels, Belgium, 2022. [Google Scholar]
- Ankley, G.T.; Bennett, R.S.; Erickson, R.J.; Hoff, D.J.; Hornung, M.W.; Johnson, R.D.; Mount, D.R.; Nichols, J.W.; Russom, C.L.; Schmieder, P.K.; et al. Adverse Outcome Pathways: A Conceptual Framework to Support Ecotoxicology Research and Risk Assessment. Environ. Toxicol. Chem. 2010, 29, 730–741. [Google Scholar] [CrossRef] [PubMed]
- Svingen, T.; Schwartz, C.L.; Rosenmai, A.K.; Ramhøj, L.; Johansson, H.K.L.; Hass, U.; Draskau, M.K.; Davidsen, N.; Christiansen, S.; Ballegaard, A.-S.R.; et al. Using Alternative Test Methods to Predict Endocrine Disruption and Reproductive Adverse Outcomes: Do We Have Enough Knowledge? Environ. Pollut. 2022, 304, 119242. [Google Scholar] [CrossRef] [PubMed]
- Cronin, M.T.D.; Richarz, A.-N. Relationship Between Adverse Outcome Pathways and Chemistry-Based In Silico Models to Predict Toxicity. Appl. Vitr. Toxicol. 2017, 3, 286–297. [Google Scholar] [CrossRef]
- Muratov, E.N.; Bajorath, J.; Sheridan, R.P.; Tetko, I.V.; Filimonov, D.; Poroikov, V.; Oprea, T.I.; Baskin, I.I.; Varnek, A.; Roitberg, A.; et al. QSAR without Borders. Chem. Soc. Rev. 2020, 49, 3525–3564. [Google Scholar] [CrossRef]
- Noyes, P.D.; Friedman, K.P.; Browne, P.; Haselman, J.T.; Gilbert, M.E.; Hornung, M.W.; Barone, S.; Crofton, K.M.; Laws, S.C.; Stoker, T.E.; et al. Evaluating Chemicals for Thyroid Disruption: Opportunities and Challenges with in Vitro Testing and Adverse Outcome Pathway Approaches. Environ. Health Perspect. 2019, 127, 095001. [Google Scholar] [CrossRef]
- Bernasconi, C.; Langezaal, I.; Bartnicka, J.; Asturiol, D.; Bowe, G.; Coecke, S.; Kienzler, A.; Liska, R.; Milcamps, A.; Munoz-Pineiro, M.A.; et al. Validation of a Battery of Mechanistic Methods Relevant for the Detection of Chemicals That Can Disrupt the Thyroid Hormone System; Publications Office of the European Union: Luxembourg, 2023. [Google Scholar]
- Sellami, A.; Reau, M.; Montes, M.; Lagarde, N. Review of in Silico Studies Dedicated to the Nuclear Receptor Family: Therapeutic Prospects and Toxicological Concerns. Front. Endocrinol. 2022, 13, 986016. [Google Scholar] [CrossRef]
- Vergauwen, L.; Bajard, L.; Tait, S.; Langezaal, I.; Sosnowska, A.; Roncaglioni, A.; Hessel, E.; van den Brand, A.D.; Haigis, A.-C.; Novák, J.; et al. A 2024 Inventory of Test Methods Relevant to Thyroid Hormone System Disruption for Human Health and Environmental Regulatory Hazard Assessment. Open Res. Eur. 2024, 4, 242. [Google Scholar] [CrossRef]
- Dracheva, E.; Norinder, U.; Rydén, P.; Engelhardt, J.; Weiss, J.M.; Andersson, P.L. In Silico Identification of Potential Thyroid Hormone System Disruptors among Chemicals in Human Serum and Chemicals with a High Exposure Index. Environ. Sci. Technol. 2022, 56, 8363–8372. [Google Scholar] [CrossRef]
- Yang, L.; Sun, P.; Tao, L.; Zhao, X. An in Silico Study on Human Carcinogenicity Mechanism of Polybrominated Biphenyls Exposure. Chem.-Biol. Interact. 2024, 397, 111075. [Google Scholar] [CrossRef]
- Evangelista, M.; Chirico, N.; Papa, E. In Silico Models for the Screening of Human Transthyretin Disruptors. J. Hazard. Mater. 2024, 480, 136188. [Google Scholar] [CrossRef]
- Cao, J.; Lin, Y.; Guo, L.-H.; Zhang, A.-Q.; Wei, Y.; Yang, Y. Structure-Based Investigation on the Binding Interaction of Hydroxylated Polybrominated Diphenyl Ethers with Thyroxine Transport Proteins. Toxicology 2010, 277, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Montaño, M.; Cocco, E.; Guignard, C.; Marsh, G.; Hoffmann, L.; Bergman, Å.; Gutleb, A.C.; Murk, A.J. New Approaches to Assess the Transthyretin Binding Capacity of Bioactivated Thyroid Hormone Disruptors. Toxicol. Sci. 2012, 130, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Grimm, F.A.; Lehmler, H.-J.; He, X.; Robertson, L.W.; Duffel, M.W. Sulfated Metabolites of Polychlorinated Biphenyls Are High-Affinity Ligands for the Thyroid Hormone Transport Protein Transthyretin. Environ. Health Perspect. 2013, 121, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Ou, W.; Xi, Y.; Chen, J.; Liu, H. Emerging Polar Phenolic Disinfection Byproducts Are High-Affinity Human Transthyretin Disruptors: An in Vitro and in Silico Study. Environ. Sci. Technol. 2019, 53, 7019–7028. [Google Scholar] [CrossRef]
- Xi, Y.; Yang, X.; Zhang, H.; Liu, H.; Watson, P.; Yang, F. Binding Interactions of Halo-Benzoic Acids, Halo-Benzenesulfonic Acids and Halo-Phenylboronic Acids with Human Transthyretin. Chemosphere 2020, 242, 125135. [Google Scholar] [CrossRef]
- Yang, X.; Ou, W.; Zhao, S.; Wang, L.; Chen, J.; Kusko, R.; Hong, H.; Liu, H. Human Transthyretin Binding Affinity of Halogenated Thiophenols and Halogenated Phenols: An in Vitro and in Silico Study. Chemosphere 2021, 280, 130627. [Google Scholar] [CrossRef]
- Rosenmai, A.K.; Winge, S.B.; Möller, M.; Lundqvist, J.; Wedebye, E.B.; Nikolov, N.G.; Lilith Johansson, H.K.; Vinggaard, A.M. Organophosphate Ester Flame Retardants Have Antiandrogenic Potential and Affect Other Endocrine Related Endpoints in Vitro and in Silico. Chemosphere 2021, 263, 127703. [Google Scholar] [CrossRef]
- Ren, X.M.; Guo, L.-H. Assessment of the Binding of Hydroxylated Polybrominated Diphenyl Ethers to Thyroid Hormone Transport Proteins Using a Site-Specific Fluorescence Probe. Environ. Sci. Technol. 2012, 46, 4633–4640. [Google Scholar] [CrossRef]
- Ren, X.-M.; Qin, W.-P.; Cao, L.-Y.; Zhang, J.; Yang, Y.; Wan, B.; Guo, L.-H. Binding Interactions of Perfluoroalkyl Substances with Thyroid Hormone Transport Proteins and Potential Toxicological Implications. Toxicology 2016, 366–367, 32–42. [Google Scholar] [CrossRef]
- Ouyang, X.; Froment, J.; Leonards, P.E.G.; Christensen, G.; Tollefsen, K.-E.; de Boer, J.; Thomas, K.V.; Lamoree, M.H. Miniaturization of a Transthyretin Binding Assay Using a Fluorescent Probe for High Throughput Screening of Thyroid Hormone Disruption in Environmental Samples. Chemosphere 2017, 171, 722–728. [Google Scholar] [CrossRef]
- Qin, W.-P.; Li, C.-H.; Guo, L.-H.; Ren, X.-M.; Zhang, J.-Q. Binding and Activity of Polybrominated Diphenyl Ether Sulfates to Thyroid Hormone Transport Proteins and Nuclear Receptors. Environ. Sci. Process. Impacts 2019, 21, 950–956. [Google Scholar] [CrossRef]
- Ren, X.-M.; Yao, L.; Xue, Q.; Shi, J.; Zhang, Q.; Wang, P.; Fu, J.; Zhang, A.; Qu, G.; Jiang, G. Binding and Activity of Tetrabromobisphenol A Mono-Ether Structural Analogs to Thyroid Hormone Transport Proteins and Receptors. Environ. Health Perspect. 2020, 128, 107008. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Wang, X.; Zhang, H.; Zeng, L.; Zhang, X.; Wang, B.; Zhou, Y.; Jing, T. Structure-Directed Screening and Analysis of Thyroid-Disrupting Chemicals Targeting Transthyretin Based on Molecular Recognition and Chromatographic Separation. Environ. Sci. Technol. 2020, 54, 5437–5445. [Google Scholar] [CrossRef]
- Hamers, T.; Kortenkamp, A.; Scholze, M.; Molenaar, D.; Cenijn, P.H.; Weiss, J.M. Transthyretin-Binding Activity of Complex Mixtures Representing the Composition of Thyroid-Hormone Disrupting Contaminants in House Dust and Human Serum. Environ. Health Perspect. 2020, 128, 017015. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, K.J. Interaction of Chlorinated Phenols with Thyroxine Binding Sites of Human Transthyretin, Albumin and Thyroid Binding Globulin. Chem. Biol. Interact. 1990, 76, 63–75. [Google Scholar] [CrossRef] [PubMed]
- den Besten, C.; Vet, J.J.R.M.; Besselink, H.T.; Kiel, G.S.; van Berkel, B.J.M.; Beems, R.; van Bladeren, P.J. The Liver, Kidney, and Thyroid Toxicity of Chlorinated Benzenes. Toxicol. Appl. Pharmacol. 1991, 111, 69–81. [Google Scholar] [CrossRef]
- Lans, M.C.; Klasson-Wehler, E.; Willemsen, M.; Meussen, E.; Safe, S.; Brouwer, A. Structure-Dependent, Competitive Interaction of Hydroxy-Polychlorobiphenyls, -Dibenzo-p-Dioxins and -Dibenzofurans with Human Transthyretin. Chem. Biol. Interact. 1993, 88, 7–21. [Google Scholar] [CrossRef]
- Cheek, A.O.; Kow, K.; Chen, J.; McLachlan, J.A. Potential Mechanisms of Thyroid Disruption in Humans: Interaction of Organochlorine Compounds with Thyroid Receptor, Transthyretin, and Thyroid-Binding Globulin. Environ. Health Perspect. 1999, 107, 273–278. [Google Scholar] [CrossRef]
- Meerts, I.A.T.M.; van Zanden, J.J.; Luijks, E.A.C.; van Leeuwen-Bol, I.; Marsh, G.; Jakobsson, E.; Bergman, Å.; Brouwer, A. Potent Competitive Interactions of Some Brominated Flame Retardants and Related Compounds with Human Transthyretin in Vitro. Toxicol. Sci. 2000, 56, 95–104. [Google Scholar] [CrossRef]
- Sandau, C.D.; Meerts, I.A.T.M.; Letcher, R.J.; McAlees, A.J.; Chittim, B.; Brouwer, A.; Norstrom, R.J. Identification of 4-Hydroxyheptachlorostyrene in Polar Bear Plasma and Its Binding Affinity to Transthyretin: A Metabolite of Octachlorostyrene? Environ. Sci. Technol. 2000, 34, 3871–3877. [Google Scholar] [CrossRef]
- Chauhan, K.R.; Kodavanti, P.R.S.; McKinney, J.D. Assessing the Role of Ortho-Substitution on Polychlorinated Biphenyl Binding to Transthyretin, a Thyroxine Transport Protein. Toxicol. Appl. Pharmacol. 2000, 162, 10–21. [Google Scholar] [CrossRef]
- Legler, J.; Cenijn, P.H.; Malmberg, T.; Bergman, A.; Brouwer, A. Determination of the Endocrine Disrupting Potency of Hydroxylated PCB’s and Flame Retardants with in Vitro Bioassays. Organohalogen Compd. 2002, 53–56. Available online: https://research.vu.nl/en/publications/determination-of-the-endocrine-disrupting-potency-of-hydroxylated (accessed on 16 April 2023).
- Meerts, I.A.T.M.; Assink, Y.; Cenijn, P.H.; van den Berg, J.H.J.; Weijers, B.M.; Bergman, Å.; Koeman, J.H.; Brouwer, A. Placental Transfer of a Hydroxylated Polychlorinated Biphenyl and Effects on Fetal and Maternal Thyroid Hormone Homeostasis in the Rat. Toxicol. Sci. 2002, 68, 361–371. [Google Scholar] [CrossRef]
- Maia, F.; Almeida, M.d.R.; Gales, L.; Kijjoa, A.; Pinto, M.M.M.; Saraiva, M.J.; Damas, A.M. The Binding of Xanthone Derivatives to Transthyretin. Biochem. Pharmacol. 2005, 70, 1861–1869. [Google Scholar] [CrossRef] [PubMed]
- Hamers, T.; Kamstra, J.H.; Sonneveld, E.; Murk, A.J.; Kester, M.H.A.; Andersson, P.L.; Legler, J.; Brouwer, A. In Vitro Profiling of the Endocrine-Disrupting Potency of Brominated Flame Retardants. Toxicol. Sci. 2006, 92, 157–173. [Google Scholar] [CrossRef] [PubMed]
- Harju, M.; Hamers, T.; Kamstra, J.H.; Sonneveld, E.; Boon, J.P.; Tysklind, M.; Andersson, P.L. Quantitative Structure–activity Relationship Modeling on in Vitro Endocrine Effects and Metabolic Stability Involving 26 Selected Brominated Flame Retardants. Environ. Toxicol. Chem. 2007, 26, 816–826. [Google Scholar] [CrossRef] [PubMed]
- Hamers, T.; Kamstra, J.H.; Sonneveld, E.; Murk, A.J.; Visser, T.J.; Van Velzen, M.J.M.; Brouwer, A.; Bergman, Å. Biotransformation of Brominated Flame Retardants into Potentially Endocrine-Disrupting Metabolites, with Special Attention to 2,2′,4,4′-Tetrabromodiphenyl Ether (BDE-47). Mol. Nutr. Food Res. 2008, 52, 284–298. [Google Scholar] [CrossRef]
- Gales, L.; Almeida, M.R.; Arsequell, G.; Valencia, G.; Saraiva, M.J.; Damas, A.M. Iodination of Salicylic Acid Improves Its Binding to Transthyretin. Biochim. Biophis. Acta 2008, 1784, 512–517. [Google Scholar] [CrossRef]
- Weiss, J.M.; Andersson, P.L.; Lamoree, M.H.; Leonards, P.E.G.; van Leeuwen, S.P.J.; Hamers, T. Competitive Binding of Poly- and Perfluorinated Compounds to the Thyroid Hormone Transport Protein Transthyretin. Toxicol. Sci. 2009, 109, 206–216. [Google Scholar] [CrossRef]
- Hamers, T.; Kamstra, J.H.; Cenijn, P.H.; Pencikova, K.; Palkova, L.; Simeckova, P.; Vondracek, J.; Andersson, P.L.; Stenberg, M.; Machala, M. In Vitro Toxicity Profiling of Ultrapure Non–Dioxin-like Polychlorinated Biphenyl Congeners and Their Relative Toxic Contribution to PCB Mixtures in Humans. Toxicol. Sci. 2011, 121, 88–100. [Google Scholar] [CrossRef]
- Simon, E.; Bytingsvik, J.; Jonker, W.; Leonards, P.E.G.; de Boer, J.; Jenssen, B.M.; Lie, E.; Aars, J.; Hamers, T.; Lamoree, M.H. Blood Plasma Sample Preparation Method for the Assessment of Thyroid Hormone-Disrupting Potency in Effect-Directed Analysis. Environ. Sci. Technol. 2011, 45, 7936–7944. [Google Scholar] [CrossRef] [PubMed]
- Simon, E.; van Velzen, M.; Brandsma, S.H.; Lie, E.; Løken, K.; de Boer, J.; Bytingsvik, J.; Jenssen, B.M.; Aars, J.; Hamers, T.; et al. Effect-Directed Analysis To Explore the Polar Bear Exposome: Identification of Thyroid Hormone Disrupting Compounds in Plasma. Environ. Sci. Technol. 2013, 47, 8902–8912. [Google Scholar] [CrossRef] [PubMed]
- Viluksela, M.; Heikkinen, P.; van der Ven, L.T.M.; Rendel, F.; Roos, R.; Esteban, J.; Korkalainen, M.; Lensu, S.; Miettinen, H.M.; Savolainen, K.; et al. Toxicological Profile of Ultrapure 2,2′,3,4,4′,5,5′-Heptachlorbiphenyl (PCB 180) in Adult Rats. PLoS ONE 2014, 9, e104639. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kamstra, J.H.; Ghorbanzadeh, M.; Weiss, J.M.; Hamers, T.; Andersson, P.L. In Silico Approach To Identify Potential Thyroid Hormone Disruptors among Currently Known Dust Contaminants and Their Metabolites. Environ. Sci. Technol. 2015, 49, 10099–10107. [Google Scholar] [CrossRef]
- Weiss, J.M.; Andersson, P.L.; Zhang, J.; Simon, E.; Leonards, P.E.G.; Hamers, T.; Lamoree, M.H. Tracing Thyroid Hormone-Disrupting Compounds: Database Compilation and Structure–activity Evaluation for an Effect-Directed Analysis of Sediment. Anal. Bioanal. Chem. 2015, 407, 5625–5634. [Google Scholar] [CrossRef]
- Hill, K.L.; Mortensen, Å.-K.; Teclechiel, D.; Willmore, W.G.; Sylte, I.; Jenssen, B.M.; Letcher, R.J. In Vitro and in Silico Competitive Binding of Brominated Polyphenyl Ether Contaminants with Human and Gull Thyroid Hormone Transport Proteins. Environ. Sci. Technol. 2018, 52, 1533–1541. [Google Scholar] [CrossRef]
- Kowalska, D.; Sosnowska, A.; Bulawska, N.; Stępnik, M.; Besselink, H.; Behnisch, P.; Puzyn, T. How the Structure of Per- and Polyfluoroalkyl Substances (PFAS) Influences Their Binding Potency to the Peroxisome Proliferator-Activated and Thyroid Hormone Receptors—An In Silico Screening Study. Molecules 2023, 28, 479. [Google Scholar] [CrossRef]
- Akinola, L.K.; Uzairu, A.; Shallangwa, G.A.; Abechi, S.E. Development of Binary Classification Models for Grouping Hydroxylated Polychlorinated Biphenyls into Active and Inactive Thyroid Hormone Receptor Agonists. SAR QSAR Environ. Res. 2023, 34, 267–284. [Google Scholar] [CrossRef]
- Arulmozhiraja, S.; Shiraishi, F.; Okumura, T.; Iida, M.; Takigami, H.; Edmonds, J.S.; Morita, M. Structural Requirements for the Interaction of 91 Hydroxylated Polychlorinated Biphenyls with Estrogen and Thyroid Hormone Receptors. Toxicol. Sci. 2005, 84, 49–62. [Google Scholar] [CrossRef]
- Gallagher, A.; Kar, S.; Sepúlveda, M.S. Computational Modeling of Human Serum Albumin Binding of Per- and Polyfluoroalkyl Substances Employing QSAR, Read-Across, and Docking. Molecules 2023, 28, 5375. [Google Scholar] [CrossRef] [PubMed]
- Jackson, T.W.; Scheibly, C.M.; Polera, M.E.; Belcher, S.M. Rapid Characterization of Human Serum Albumin Binding for Per- and Polyfluoroalkyl Substances Using Differential Scanning Fluorimetry. Environ. Sci. Technol. 2021, 55, 12291–12301. [Google Scholar] [CrossRef]
- Liu, W.; Wang, Z.; Chen, J.; Tang, W.; Wang, H. Machine Learning Model for Screening Thyroid Stimulating Hormone Receptor Agonists Based on Updated Datasets and Improved Applicability Domain Metrics. Chem. Res. Toxicol. 2023, 36, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Neumann, S.; Huang, W.; Titus, S.; Krause, G.; Kleinau, G.; Alberobello, A.T.; Zheng, W.; Southall, N.T.; Inglese, J.; Austin, C.P.; et al. Small-Molecule Agonists for the Thyrotropin Receptor Stimulate Thyroid Function in Human Thyrocytes and Mice. Proc. Natl. Acad. Sci. USA 2009, 106, 12471–12476. [Google Scholar] [CrossRef]
- Jäschke, H.; Neumann, S.; Moore, S.; Thomas, C.J.; Colson, A.-O.; Costanzi, S.; Kleinau, G.; Jiang, J.-K.; Paschke, R.; Raaka, B.M.; et al. A Low Molecular Weight Agonist Signals by Binding to the Transmembrane Domain of Thyroid-Stimulating Hormone Receptor (TSHR) and Luteinizing Hormone/Chorionic Gonadotropin Receptor (LHCGR). J. Biol. Chem. 2006, 281, 9841–9844. [Google Scholar] [CrossRef]
- Titus, S.; Neumann, S.; Zheng, W.; Southall, N.; Michael, S.; Klumpp, C.; Yasgar, A.; Shinn, P.; Thomas, C.J.; Inglese, J.; et al. Quantitative High-Throughput Screening Using a Live-Cell cAMP Assay Identifies Small-Molecule Agonists of the TSH Receptor. SLAS Discov. 2008, 13, 120–127. [Google Scholar] [CrossRef]
- Huang, R.; Xia, M.; Sakamuru, S.; Zhao, J.; Shahane, S.A.; Attene-Ramos, M.; Zhao, T.; Austin, C.P.; Simeonov, A. Modelling the Tox21 10 K Chemical Profiles for in Vivo Toxicity Prediction and Mechanism Characterization. Nat. Commun. 2016, 7, 10425. [Google Scholar] [CrossRef]
- Huang, R.; Xia, M.; Sakamuru, S.; Zhao, J.; Lynch, C.; Zhao, T.; Zhu, H.; Austin, C.P.; Simeonov, A. Expanding Biological Space Coverage Enhances the Prediction of Drug Adverse Effects in Human Using in Vitro Activity Profiles. Sci. Rep. 2018, 8, 3783. [Google Scholar] [CrossRef]
- Olker, J.H.; Korte, J.J.; Denny, J.S.; Hartig, P.C.; Cardon, M.C.; Knutsen, C.N.; Kent, P.M.; Christensen, J.P.; Degitz, S.J.; Hornung, M.W. Screening the ToxCast Phase 1, Phase 2, and E1k Chemical Libraries for Inhibitors of Iodothyronine Deiodinases. Toxicol. Sci. 2019, 168, 430–442. [Google Scholar] [CrossRef]
- Wang, J.; Hallinger, D.R.; Murr, A.S.; Buckalew, A.R.; Lougee, R.R.; Richard, A.M.; Laws, S.C.; Stoker, T.E. High-Throughput Screening and Chemotype-Enrichment Analysis of ToxCast Phase II Chemicals Evaluated for Human Sodium-Iodide Symporter (NIS) Inhibition. Environ. Int. 2019, 126, 377–386. [Google Scholar] [CrossRef]
- Paul Friedman, K.; Watt, E.D.; Hornung, M.W.; Hedge, J.M.; Judson, R.S.; Crofton, K.M.; Houck, K.A.; Simmons, S.O. Tiered High-Throughput Screening Approach to Identify Thyroperoxidase Inhibitors Within the ToxCast Phase I and II Chemical Libraries. Toxicol. Sci. 2016, 151, 160–180. [Google Scholar] [CrossRef] [PubMed]
- Gadaleta, D.; Spînu, N.; Roncaglioni, A.; Cronin, M.T.D.; Benfenati, E. Prediction of the Neurotoxic Potential of Chemicals Based on Modelling of Molecular Initiating Events Upstream of the Adverse Outcome Pathways of (Developmental) Neurotoxicity. Int. J. Mol. Sci. 2022, 23, 3053. [Google Scholar] [CrossRef] [PubMed]
- Gaulton, A.; Hersey, A.; Nowotka, M.; Bento, A.P.; Chambers, J.; Mendez, D.; Mutowo, P.; Atkinson, F.; Bellis, L.J.; Cibrián-Uhalte, E.; et al. The ChEMBL Database in 2017. Nucleic Acids Res. 2017, 45, D945–D954. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Wang, C.; Gui, B.; Yuan, X.; Li, C.; Zhao, Y.; Martyniuk, C.J.; Su, L. Application of Machine Learning to Predict the Inhibitory Activity of Organic Chemicals on Thyroid Stimulating Hormone Receptor. Environ. Res. 2022, 212, 113175. [Google Scholar] [CrossRef]
- Seo, M.; Lim, C.; Kwon, H. In Silico Prediction Models for Thyroid Peroxidase Inhibitors and Their Application to Synthetic Flavors. Food Sci. Biotechnol. 2022, 31, 483–495. [Google Scholar] [CrossRef]
- Carvalho, D.P.; Ferreira, A.C.F.; Coelho, S.M.; Moraes, J.M.; Camacho, M.A.S.; Rosenthal, D. Thyroid Peroxidase Activity Is Inhibited by Amino Acids. Braz. J. Med. Biol. Res. 2000, 33, 355–361. [Google Scholar] [CrossRef]
- Divi, R.L.; Doerge, D.R. Inhibition of Thyroid Peroxidase by Dietary Flavonoids. Chem. Res. Toxicol. 1996, 9, 16–23. [Google Scholar] [CrossRef]
- Habza-Kowalska, E.; Kaczor, A.A.; Żuk, J.; Matosiuk, D.; Gawlik-Dziki, U. Thyroid Peroxidase Activity Is Inhibited by Phenolic Compounds—Impact of Interaction. Molecules 2019, 24, 2766. [Google Scholar] [CrossRef]
- Lee, J. Conversion of the Organic Breakdown Products of Glucosinolate to Thiocyanate Anions and Their Effects on Thyroid Hormone Production. Ph.D. Thesis, Seoul National University, Seoul, Republic of Korea, 2015. [Google Scholar]
- Li, X.; Gu, W.; Zhang, B.; Xin, X.; Kang, Q.; Yang, M.; Chen, B.; Li, Y. Insights into Toxicity of Polychlorinated Naphthalenes to Multiple Human Endocrine Receptors: Mechanism and Health Risk Analysis. Environ. Int. 2022, 165, 107291. [Google Scholar] [CrossRef]
- Sapounidou, M.; Norinder, U.; Andersson, P.L. Predicting Endocrine Disruption Using Conformal Prediction—A Prioritization Strategy to Identify Hazardous Chemicals with Confidence. Chem. Res. Toxicol. 2023, 36, 53–65. [Google Scholar] [CrossRef]
- Yang, X.; Ou, W.; Zhao, S.; Xi, Y.; Wang, L.; Liu, H. Rapid Screening of Human Transthyretin Disruptors through a Tiered in Silico Approach. ACS Sustain. Chem. Eng. 2021, 9, 5661–5672. [Google Scholar] [CrossRef]
- Van den Berg, K.J.; van Raaij, J.A.G.M.; Bragt, P.C.; Notten, W.R.F. Interactions of Halogenated Industrial Chemicals with Transthyretin and Effects on Thyroid Hormone Levels In Vivo. Arch. Toxicol. 1991, 65, 15–19. [Google Scholar] [CrossRef]
- Marchesini, G.R.; Meulenberg, E.; Haasnoot, W.; Mizuguchi, M.; Irth, H. Biosensor Recognition of Thyroid-Disrupting Chemicals Using Transport Proteins. Anal. Chem. 2006, 78, 1107–1114. [Google Scholar] [CrossRef]
- Marchesini, G.R.; Meimaridou, A.; Haasnoot, W.; Meulenberg, E.; Albertus, F.; Mizuguchi, M.; Takeuchi, M.; Irth, H.; Murk, A.J. Biosensor Discovery of Thyroxine Transport Disrupting Chemicals. Toxicol. Appl. Pharmacol. 2008, 232, 150–160. [Google Scholar] [CrossRef]
- Purkey, H.E.; Palaninathan, S.K.; Kent, K.C.; Smith, C.; Safe, S.H.; Sacchettini, J.C.; Kelly, J.W. Hydroxylated Polychlorinated Biphenyls Selectively Bind Transthyretin in Blood and Inhibit Amyloidogenesis: Rationalizing Rodent PCB Toxicity. Chem. Biol. 2004, 11, 1719–1728. [Google Scholar] [CrossRef]
- Garcia de Lomana, M.; Weber, A.G.; Birk, B.; Landsiedel, R.; Achenbach, J.; Schleifer, K.-J.; Mathea, M.; Kirchmair, J. In Silico Models to Predict the Perturbation of Molecular Initiating Events Related to Thyroid Hormone Homeostasis. Chem. Res. Toxicol. 2021, 34, 396–411. [Google Scholar] [CrossRef]
- Gadaleta, D.; d’Alessandro, L.; Marzo, M.; Benfenati, E.; Roncaglioni, A. Quantitative Structure–activity Relationship Modeling of the Amplex Ultrared Assay to Predict Thyroperoxidase Inhibitory Activity. Front. Pharmacol. 2021, 12, 713037. [Google Scholar] [CrossRef]
- Rosenberg, S.A.; Watt, E.D.; Judson, R.S.; Simmons, S.O.; Paul Friedman, K.; Dybdahl, M.; Nikolov, N.G.; Wedebye, E.B. QSAR Models for Thyroperoxidase Inhibition and Screening of U.S. and EU Chemical Inventories. Comput. Toxicol. 2017, 4, 11–21. [Google Scholar] [CrossRef]
- Bai, X.; Yan, L.; Ji, C.; Zhang, Q.; Dong, X.; Chen, A.; Zhao, M. A Combination of Ternary Classification Models and Reporter Gene Assays for the Comprehensive Thyroid Hormone Disruption Profiles of 209 Polychlorinated Biphenyls. Chemosphere 2018, 210, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Zhang, Q.; Huang, F.; Nie, W.-W.; Hu, C.-Q.; Ying, H.-Z.; Dong, X.-W.; Zhao, M.-R. Ternary Classification Models for Predicting Hormonal Activities of Chemicals via Nuclear Receptors. Chem. Phys. Lett. 2018, 706, 360–366. [Google Scholar] [CrossRef]
- Nakamura, N.; Matsubara, K.; Sanoh, S.; Ohta, S.; Uramaru, N.; Kitamura, S.; Yamaguchi, M.; Sugihara, K.; Fujimoto, N. Cell Type-Dependent Agonist/Antagonist Activities of Polybrominated Diphenyl Ethers. Toxicol. Lett. 2013, 223, 192–197. [Google Scholar] [CrossRef]
- Ren, X.-M.; Guo, L.-H. Molecular Toxicology of Polybrominated Diphenyl Ethers: Nuclear Hormone Receptor Mediated Pathways. Environ. Sci. Process. Impacts 2013, 15, 702–708. [Google Scholar] [CrossRef]
- Amano, I.; Miyazaki, W.; Iwasaki, T.; Shimokawa, N.; Koibuchi, N. The Effect of Hydroxylated Polychlorinated Biphenyl (OH-PCB) on Thyroid Hormone Receptor (TR)-Mediated Transcription through Native-Thyroid Hormone Response Element (TRE). Ind. Health 2010, 48, 115–118. [Google Scholar] [CrossRef]
- Du, G.; Shen, O.; Sun, H.; Fei, J.; Lu, C.; Song, L.; Xia, Y.; Wang, S.; Wang, X. Assessing Hormone Receptor Activities of Pyrethroid Insecticides and Their Metabolites in Reporter Gene Assays. Toxicol. Sci. 2010, 116, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ma, M.; Wang, Z. A Two-hybrid Yeast Assay to Quantify the Effects of Xenobiotics on Thyroid Hormone-mediated Gene Expression. Environ. Toxicol. Chem. 2008, 27, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Shen, O.-X.; Wang, X.-R.; Zhou, L.; Zhen, S.; Chen, X. Anti-Thyroid Hormone Activity of Bisphenol A, Tetrabromobisphenol A and Tetrachlorobisphenol A in an Improved Reporter Gene Assay. Toxicol. Vitr. 2009, 23, 950–954. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Liu, H.; Sun, H.; Shen, O.; Wang, X.; Lam, M.H.W.; Giesy, J.P.; Zhang, X.; Yu, H. Endocrine Effects of Methoxylated Brominated Diphenyl Ethers in Three in Vitro Models. Mar. Pollut. Bull. 2011, 62, 2356–2361. [Google Scholar] [CrossRef]
- Liu, H.; Hu, W.; Sun, H.; Shen, O.; Wang, X.; Lam, M.H.W.; Giesy, J.P.; Zhang, X.; Yu, H. In Vitro Profiling of Endocrine Disrupting Potency of 2,2′,4,4′-Tetrabromodiphenyl Ether (BDE47) and Related Hydroxylated Analogs (HO-PBDEs). Mar. Pollut. Bull. 2011, 63, 287–296. [Google Scholar] [CrossRef]
- Kojima, H.; Takeuchi, S.; Uramaru, N.; Sugihara, K.; Yoshida, T.; Kitamura, S. Nuclear Hormone Receptor Activity of Polybrominated Diphenyl Ethers and Their Hydroxylated and Methoxylated Metabolites in Transactivation Assays Using Chinese Hamster Ovary Cells. Environ. Health Perspect. 2009, 117, 1210–1218. [Google Scholar] [CrossRef]
- Kar, S.; Sepúlveda, M.S.; Roy, K.; Leszczynski, J. Endocrine-Disrupting Activity of per- and Polyfluoroalkyl Substances: Exploring Combined Approaches of Ligand and Structure Based Modeling. Chemosphere 2017, 184, 514–523. [Google Scholar] [CrossRef]
- Dix, D.J.; Houck, K.A.; Martin, M.T.; Richard, A.M.; Setzer, R.W.; Kavlock, R.J. The ToxCast Program for Prioritizing Toxicity Testing of Environmental Chemicals. Toxicol. Sci. 2007, 95, 5–12. [Google Scholar] [CrossRef]
- Richard, A.M.; Judson, R.S.; Houck, K.A.; Grulke, C.M.; Volarath, P.; Thillainadarajah, I.; Yang, C.; Rathman, J.; Martin, M.T.; Wambaugh, J.F.; et al. ToxCast Chemical Landscape: Paving the Road to 21st Century Toxicology. Chem. Res. Toxicol. 2016, 29, 1225–1251. [Google Scholar] [CrossRef]
- EDSP21 Work Plan. The Incorporation of In Silico Models and In Vitro High Throughput Assays in the Endocrine Disruptor Screening Program (EDSP) for Prioritization and Screening. 2011. Available online: https://www.epa.gov/sites/default/files/2015-07/documents/edsp21_work_plan_summary_overview_final.pdf (accessed on 13 March 2017).
- Rybacka, A.; Rudén, C.; Tetko, I.V.; Andersson, P.L. Identifying Potential Endocrine Disruptors among Industrial Chemicals and Their Metabolites—Development and Evaluation of in Silico Tools. Chemosphere 2015, 139, 372–378. [Google Scholar] [CrossRef]
- Toropova, A.P.; Toropov, A.A.; Benfenati, E. CORAL: Prediction of Binding Affinity and Efficacy of Thyroid Hormone Receptor Ligands. Eur. J. Med. Chem. 2015, 101, 452–461. [Google Scholar] [CrossRef]
- Politi, R.; Rusyn, I.; Tropsha, A. Prediction of Binding Affinity and Efficacy of Thyroid Hormone Receptor Ligands Using QSAR and Structure-Based Modeling Methods. Toxicol. Appl. Pharmacol. 2014, 280, 177–189. [Google Scholar] [CrossRef]
- Gaulton, A.; Bellis, L.J.; Bento, A.P.; Chambers, J.; Davies, M.; Hersey, A.; Light, Y.; McGlinchey, S.; Michalovich, D.; Al-Lazikani, B.; et al. ChEMBL: A Large-Scale Bioactivity Database for Drug Discovery. Nucleic Acids Res. 2012, 40, D1100–D1107. [Google Scholar] [CrossRef]
- Arnold, L.A.; Kosinski, A.; Estébanez-Perpiñá, E.; Guy, R.K. Inhibitors of the Interaction of a Thyroid Hormone Receptor and Coactivators: Preliminary Structure−Activity Relationships. J. Med. Chem. 2007, 50, 5269–5280. [Google Scholar] [CrossRef]
- Hwang, J.Y.; Arnold, L.A.; Zhu, F.; Kosinski, A.; Mangano, T.J.; Setola, V.; Roth, B.L.; Guy, R.K. Improvement of Pharmacological Properties of Irreversible Thyroid Receptor Coactivator Binding Inhibitors. J. Med. Chem. 2009, 52, 3892–3901. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hwang, J.Y.; Attia, R.R.; Zhu, F.; Yang, L.; Lemoff, A.; Jeffries, C.; Connelly, M.C.; Guy, R.K. Synthesis and Evaluation of Sulfonylnitrophenylthiazoles (SNPTs) as Thyroid Hormone Receptor–Coactivator Interaction Inhibitors. J. Med. Chem. 2012, 55, 2301–2310. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Papa, E.; Kovarich, S.; Gramatica, P. QSAR Prediction of the Competitive Interaction of Emerging Halogenated Pollutants with Human Transthyretin. SAR QSAR Environ. Res. 2013, 24, 333–349. [Google Scholar] [CrossRef] [PubMed]
- Kovarich, S.; Papa, E.; Li, J.; Gramatica, P. QSAR Classification Models for the Screening of the Endocrine-Disrupting Activity of Perfluorinated Compounds. SAR QSAR Environ. Res. 2012, 23, 207–220. [Google Scholar] [CrossRef]
- Kovarich, S.; Papa, E.; Gramatica, P. QSAR Classification Models for the Prediction of Endocrine Disrupting Activity of Brominated Flame Retardants. J. Hazard. Mater. 2011, 190, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Papa, E.; Kovarich, S.; Gramatica, P. QSAR Modeling and Prediction of the Endocrine-Disrupting Potencies of Brominated Flame Retardants. Chem. Res. Toxicol. 2010, 23, 946–954. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Xie, Q.; Li, X.; Li, N.; Chi, P.; Chen, J.; Wang, Z.; Hao, C. Hormone Activity of Hydroxylated Polybrominated Diphenyl Ethers on Human Thyroid Receptor-β: In Vitro and In Silico Investigations. Environ. Health Perspect. 2010, 118, 602–606. [Google Scholar] [CrossRef]
- OECD. New Scoping Document on In Vitro and Ex Vivo Assays for the Identification of Modulators of Thyroid Hormone Signalling; OECD Publishing: Paris, France, 2014. [Google Scholar]
- OECD. Thyroid in Vitro Methods: Assessment Reports by the Thyroid Disruption Methods Expert Group: Reports Assessing the Validation Status of Assays from the EU-NETVAL Activities. In OECD Series on Testing and Assessment, No. 403; OECD Publishing: Paris, France, 2024. [Google Scholar]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.-W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for Scientific Data Management and Stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, M.E.; O’Shaughnessy, K.L.; Axelstad, M. Regulation of Thyroid-Disrupting Chemicals to Protect the Developing Brain. Endocrinology 2020, 161, bqaa106. [Google Scholar] [CrossRef]
- Pirhadi, S.; Shiri, F.; Ghasemi, J.B. Multivariate Statistical Analysis Methods in QSAR. RSC Adv. 2015, 5, 104635–104665. [Google Scholar] [CrossRef]
- Schür, C.; Gasser, L.; Perez-Cruz, F.; Schirmer, K.; Baity-Jesi, M. A Benchmark Dataset for Machine Learning in Ecotoxicology. Sci. Data 2023, 10, 718. [Google Scholar] [CrossRef]
- Schür, C.; Schirmer, K.; Baity-Jesi, M. On the Comparability between Studies in Predictive Ecotoxicology. Comput. Toxicol. 2025, 35, 100367. [Google Scholar] [CrossRef]
- Wassenaar, P.N.H.; Minnema, J.; Vriend, J.; Peijnenburg, W.J.G.M.; Pennings, J.L.A.; Kienhuis, A. The Role of Trust in the Use of Artificial Intelligence for Chemical Risk Assessment. Regul. Toxicol. Pharmacol. 2024, 148, 105589. [Google Scholar] [CrossRef]
- OECD. Guidance Document on the Validation of (Quantitative) Structure–activity Relationship [(Q)SAR] Models; OECD Publishing: Paris, France, 2014. [Google Scholar]
- OECD. (Q)SAR Assessment Framework: Guidance for the Regulatory Assessment of (Quantitative) Structure Activity Relationship Models and Predictions; OECD Series on Testing and Assessment, No. 386; OECD Publishing: Paris, France, 2023. [Google Scholar]
- Tropsha, A. Best Practices for QSAR Model Development, Validation, and Exploitation. Mol. Inform. 2010, 29, 476–488. [Google Scholar] [CrossRef] [PubMed]
- Golbraikh, A.; Tropsha, A. Beware of q2! J. Mol. Graph. Model. 2002, 20, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Golbraikh, A.; Tropsha, A. Predictive QSAR Modeling Based on Diversity Sampling of Experimental Datasets for the Training and Test Set Selection. Mol. Divers. 2000, 5, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction, 2nd ed.; Springer: New York, NY, USA, 2009. [Google Scholar]
- Todeschini, R.; Consonni, V.; Maiocchi, A. The K Correlation Index: Theory Development and Its Application in Chemometrics. Chemometr. Intell. Lab. 1999, 46, 13–29. [Google Scholar] [CrossRef]
- Tropsha, A.; Gramatica, P.; Gombar, V.K. The Importance of Being Earnest: Validation Is the Absolute Essential for Successful Application and Interpretation of QSPR Models. QSAR Comb. Sci. 2003, 22, 69–77. [Google Scholar] [CrossRef]
- Wehrens, R.; Putter, H.; Buydens, L.M.C. The Bootstrap: A Tutorial. Chemom. Intell. Lab. Syst. 2000, 54, 35–52. [Google Scholar] [CrossRef]
- Ambure, P.; Gajewicz-Skretna, A.; Cordeiro, M.N.D.S.; Roy, K. New Workflow for QSAR Model Development from Small Data Sets: Small Dataset Curator and Small Dataset Modeler. Integration of Data Curation, Exhaustive Double Cross-Validation, and a Set of Optimal Model Selection Techniques. J. Chem. Inf. Model. 2019, 59, 4070–4076. [Google Scholar] [CrossRef]
- Raste, S.; Singh, R.; Vaughan, J.; Nair, V.N. Quantifying Inherent Randomness in Machine Learning Algorithms. arXiv 2022, arXiv:2206.12353. [Google Scholar]
- Li, J.; Zhao, T.; Yang, Q.; Du, S.; Xu, L. A Review of Quantitative Structure–activity Relationship: The Development and Current Status of Data Sets, Molecular Descriptors and Mathematical Models. Chemom. Intell. Lab. Syst. 2025, 256, 105278. [Google Scholar] [CrossRef]
- Khan, A.A. Balanced Split: A New Train-Test Data Splitting Strategy for Imbalanced Datasets. arXiv 2022, arXiv:2212.11116. [Google Scholar] [CrossRef]
- An, C.; Park, Y.W.; Ahn, S.S.; Han, K.; Kim, H.; Lee, S.-K. Radiomics Machine Learning Study with a Small Sample Size: Single Random Training-Test Set Split May Lead to Unreliable Results. PLoS ONE 2021, 16, e0256152. [Google Scholar] [CrossRef]
- Golbraikh, A.; Shen, M.; Xiao, Z.; Xiao, Y.-D.; Lee, K.-H.; Tropsha, A. Rational Selection of Training and Test Sets for the Development of Validated QSAR Models. J. Comput. Aided Mol. Des. 2003, 17, 241–253. [Google Scholar] [CrossRef]
- Kennard, R.W.; Stone, L.A. Computer Aided Design of Experiments. Technometrics 1969, 11, 137–148. [Google Scholar] [CrossRef]
- Roy, K.; Kar, S.; Ambure, P. On a Simple Approach for Determining Applicability Domain of QSAR Models. Chemom. Intell. Lab. Syst. 2015, 145, 22–29. [Google Scholar] [CrossRef]
- Netzeva, T.I.; Worth, A.P.; Aldenberg, T.; Benigni, R.; Cronin, M.T.D.; Gramatica, P.; Jaworska, J.S.; Kahn, S.; Klopman, G.; Marchant, C.A.; et al. Current Status of Methods for Defining the Applicability Domain of (Quantitative) Structure–activity Relationships: The Report and Recommendations of ECVAM Workshop 52. Altern. Lab. Anim. 2005, 33, 155–173. [Google Scholar] [CrossRef] [PubMed]
- Sahigara, F.; Mansouri, K.; Ballabio, D.; Mauri, A.; Consonni, V.; Todeschini, R. Comparison of Different Approaches to Define the Applicability Domain of QSAR Models. Molecules 2012, 17, 4791–4810. [Google Scholar] [CrossRef]
- Klingspohn, W.; Mathea, M.; ter Laak, A.; Heinrich, N.; Baumann, K. Efficiency of Different Measures for Defining the Applicability Domain of Classification Models. J. Cheminform. 2017, 9, 44. [Google Scholar] [CrossRef]
- Toropova, A.P.; Toropov, A.A.; Rallo, R.; Leszczynska, D.; Leszczynski, J. Optimal Descriptor as a Translator of Eclectic Data into Prediction of Cytotoxicity for Metal Oxide Nanoparticles under Different Conditions. Ecotoxicol. Environ. Saf. 2015, 112, 39–45. [Google Scholar] [CrossRef]
- Todeschini, R.; Consonni, V. Molecular Descriptors for Chemoinformatics; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2009; ISBN 978-3-527-62876-6. [Google Scholar]
- Vasilev, B.; Atanasova, M. A (Comprehensive) Review of the Application of Quantitative Structure–Activity Relationship (QSAR) in the Prediction of New Compounds with Anti-Breast Cancer Activity. Appl. Sci. 2025, 15, 1206. [Google Scholar] [CrossRef]
- Yap, C.W. PaDEL-Descriptor: An Open Source Software to Calculate Molecular Descriptors and Fingerprints. J. Comput. Chem. 2011, 32, 1466–1474. [Google Scholar] [CrossRef]
- Todeschini, R.; Consonni, V. Handbook of Molecular Descriptors; WILEY-VCH: Weinheim, Germany, 2000. [Google Scholar]
- Mauri, A. alvaDesc: A Tool to Calculate and Analyze Molecular Descriptors and Fingerprints. In Ecotoxicological QSARs; Roy, K., Ed.; Springer: New York, NY, USA, 2020; pp. 801–820. ISBN 978-1-07-160150-1. [Google Scholar]
- Mauri, A.; Bertola, M. Alvascience: A New Software Suite for the QSAR Workflow Applied to the Blood–Brain Barrier Permeability. Int. J. Mol. Sci. 2022, 23, 12882. [Google Scholar] [CrossRef]
- Matveieva, M.; Polishchuk, P. Benchmarks for Interpretation of QSAR Models. J. Cheminform. 2021, 13, 41. [Google Scholar] [CrossRef]
- Gião, T.; Saavedra, J.; Cotrina, E.; Quintana, J.; Llop, J.; Arsequell, G.; Cardoso, I. Undiscovered Roles for Transthyretin: From a Transporter Protein to a New Therapeutic Target for Alzheimer’s Disease. Int. J. Mol. Sci. 2020, 21, 2075. [Google Scholar] [CrossRef]
- Janicka, M.; Sztanke, M.; Sztanke, K. Biomimetic Chromatography/QSAR Investigations in Modeling Properties Influencing the Biological Efficacy of Phenoxyacetic Acid-Derived Congeners. Molecules 2025, 30, 688. [Google Scholar] [CrossRef]
- Charest, N.; Sinclair, G.; Eytcheson, S.A.; Chang, D.T.; Martin, T.M.; Lowe, C.N.; Paul Friedman, K.; Williams, A.J. Combined In Vitro and In Silico Workflow to Deliver Robust, Transparent, and Contextually Rigorous Models of Bioactivity. J. Chem. Inf. Model. 2025, 65, 4426–4441. [Google Scholar] [CrossRef]
- Eytcheson, S.A.; Zosel, A.D.; Olker, J.H.; Hornung, M.W.; Degitz, S.J. Screening the ToxCast Chemical Libraries for Binding to Transthyretin. Chem. Res. Toxicol. 2024, 37, 1670–1681. [Google Scholar] [CrossRef] [PubMed]
- Evangelista, M.; Chirico, N.; Papa, E. New QSAR Models to Predict Human Transthyretin Disruption by Per- and Polyfluoroalkyl Substances (PFAS): Development and Application. Toxics 2025, 13, 590. [Google Scholar] [CrossRef] [PubMed]
- Degitz, S.J.; Olker, J.H.; Denny, J.S.; Degoey, P.P.; Hartig, P.C.; Cardon, M.C.; Eytcheson, S.A.; Haselman, J.T.; Mayasich, S.A.; Hornung, M.W. In Vitro Screening of per- and Polyfluorinated Substances (PFAS) for Interference with Seven Thyroid Hormone System Targets across Nine Assays. Toxicol. Vitr. 2024, 95, 105762. [Google Scholar] [CrossRef] [PubMed]
- Sosnowska, A.; Mudlaff, M.; Mombelli, E.; Behnisch, P.; Zdybel, S.; Besselink, H.; Kuckelkorn, J.; Bulawska, N.; Kepka, K.; Kowalska, D.; et al. Identification of New PFAS for Severe Interference with Thyroid Hormone Transport: A Combined in Vitro/Silico Approach. J. Hazard. Mater. 2025, 491, 137949. [Google Scholar] [CrossRef]
- Mansouri, K.; Grulke, C.M.; Judson, R.S.; Williams, A.J. OPERA Models for Predicting Physicochemical Properties and Environmental Fate Endpoints. J. Cheminform. 2018, 10, 10. [Google Scholar] [CrossRef]
- Cirino, T.; Pinto, L.; Iwan, M.; Dougha, A.; Lučić, B.; Kraljević, A.; Navoyan, Z.; Tevosyan, A.; Yeghiazaryan, H.; Khondkaryan, L.; et al. Consensus Modeling Strategies for Predicting Transthyretin Binding Affinity from Tox24 Challenge Data. Chem. Res. Toxicol. 2025, 38, 1061–1071. [Google Scholar] [CrossRef]
- Tetko, I.V. Tox24 Challenge. Chem. Res. Toxicol. 2024, 37, 825–826. [Google Scholar] [CrossRef]

| Model ID | Ref. | Year | MIE | Algorithm | C or R | Chemical Class | Data Source Type | Data Source Literature Reference(s) |
|---|---|---|---|---|---|---|---|---|
| ID_1 | [52] | 2024 | TBG | MLR | R | PBBs | Primary | [52] |
| ID_2 | [52] | 2024 | TBG | MLR | R | PBBs and OH-PBBs | Primary | [52] |
| ID_3 | [52] | 2024 | TBG | MLR | R | PBBs and 2OH-PBBs | Primary | [52] |
| ID_4 | [52] | 2024 | TBG | MLR | R | PBBs, OH-PBBs, and 2OH-PBBs | Primary | [52] |
| ID_5 | [53] | 2024 | TTR | MLR | R | Heterogeneous | Secondary | [54,55,56,57,58,59,60] |
| ID_6 | [53] | 2024 | TTR | MLR | R | Heterogeneous | Secondary | [61,62,63,64,65,66,67] |
| ID_7 | [53] | 2024 | TTR | MLR | R | Heterogeneous | Secondary | [68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89] |
| ID_8 | [90] | 2023 | TR α | MLR | R | PFAS | Primary | [90] |
| ID_9 | [90] | 2023 | TR β | MLR | R | PFAS | Primary | [90] |
| ID_10 | [91] | 2023 | TR n.s. | LDA | C | OH-PCBs | Secondary | [92] |
| ID_11 | [91] | 2023 | TR n.s. | LR | C | OH-PCBs | Secondary | [92] |
| ID_12 | [93] | 2023 | Albumin | PLS | R | PFAS | Secondary | [94] |
| ID_13 | [93] | 2023 | Albumin | LDA | C | PFAS | Secondary | [94] |
| ID_14 | [93] | 2023 | Albumin | MLR | R | PFAS | Secondary | [94] |
| ID_15 | [95] | 2023 | TSHR | RF | C | Heterogeneous | Tox21 database and secondary | [96,97,98] |
| ID_16 | [51] | 2022 | TTR | RF | C | Heterogeneous | Secondary | [87] |
| ID_17 | [51] | 2022 | TR β | RF | C | Heterogeneous | Tox21 database * | [99,100] |
| ID_18 | [51] | 2022 | TR β | RF | C | Heterogeneous | Tox21 database * | [99,100] |
| ID_19 | [51] | 2022 | TSHR | RF | C | Heterogeneous | Tox21 database * | [99,100] |
| ID_20 | [51] | 2022 | TSHR | RF | C | Heterogeneous | Tox21 database * | [99,100] |
| ID_21 | [51] | 2022 | TRHR | RF | C | Heterogeneous | Tox21 database * | [99,100] |
| ID_22 | [51] | 2022 | DIO1 | RF | C | Heterogeneous | ToxCast database ** | [101] |
| ID_23 | [51] | 2022 | DIO2 | RF | C | Heterogeneous | ToxCast database ** | [101] |
| ID_24 | [51] | 2022 | DIO3 | RF | C | Heterogeneous | ToxCast database ** | [101] |
| ID_25 | [51] | 2022 | NIS | RF | C | Heterogeneous | ToxCast database ** | [102] |
| ID_26 | [51] | 2022 | TPO | RF | C | Heterogeneous | ToxCast database ** | [103] |
| ID_27 | [104] | 2022 | TTR | RF | C | Heterogeneous | ChEMBL database *** | [105] |
| ID_28 | [104] | 2022 | TR α | RF | C | Heterogeneous | ChEMBL database *** | [105] |
| ID_29 | [104] | 2022 | TR β | RF | C | Heterogeneous | ChEMBL database *** | [105] |
| ID_30 | [104] | 2022 | NIS | RF | C | Heterogeneous | ChEMBL database *** | [105] |
| ID_31 | [106] | 2022 | TSHR | RF | C | Heterogeneous | Tox21 database | https://tripod.nih.gov/tox21/assays/ |
| ID_32 | [106] | 2022 | TSHR | RF | C | Heterogeneous | Tox21 database | https://tripod.nih.gov/tox21/assays/ |
| ID_33 | [106] | 2022 | TSHR | XGB | C | Heterogeneous | Tox21 database | https://tripod.nih.gov/tox21/assays/ |
| ID_34 | [106] | 2022 | TSHR | LR | C | Heterogeneous | Tox21 database | https://tripod.nih.gov/tox21/assays/ |
| ID_35 | [106] | 2022 | TSHR | XGB | R | Heterogeneous | Tox21 database | https://tripod.nih.gov/tox21/assays/ |
| ID_36 | [107] | 2022 | TPO | XGB | C | Heterogeneous | ToxCast database and secondary ** | [103,108,109,110,111] |
| ID_37 | [107] | 2022 | TPO | Hard Voting | C | Heterogeneous | ToxCast database and secondary ** | [103,108,109,110,111] |
| ID_38 | [107] | 2022 | TPO | Soft Voting | C | Heterogeneous | ToxCast database and secondary ** | [103,108,109,110,111] |
| ID_39 | [112] | 2022 | TR β | MLR | R | PCNs | Primary | [112] |
| ID_40 | [113] | 2023 | TR β | RF | C | Heterogeneous | Tox21 database | National Center for Biotechnology Information. PubChem Database. Source = 824, AID = 743067, https://pubchem.ncbi.nlm.nih.gov/bioassay/743067 (accessed 13 May 2021) |
| ID_41 | [59] | 2021 | TTR | MLR | R | Halogenated phenols and thiophenols | Primary and Secondary | [57,59] |
| ID_42 | [114] | 2021 | TTR | kNN | C | Heterogeneous | Secondary | [54,55,56,57,58,59,61,62,64,68,70,71,72,73,75,78,79,80,81,82,83,84,85,86,87,88,89,115,116,117,118] |
| ID_43 | [114] | 2021 | TTR | kNN | C | Heterogeneous | Secondary | [54,55,56,57,58,59,61,62,64,68,70,71,72,73,75,78,79,80,81,82,83,84,85,86,87,88,89,115,116,117,118] |
| ID_44 | [114] | 2021 | TTR | kNN | C | Heterogeneous | Secondary | [54,55,56,57,58,59,61,62,64,68,70,71,72,73,75,78,79,80,81,82,83,84,85,86,87,88,89,115,116,117,118] |
| ID_45 | [114] | 2021 | TTR | kNN | C | Heterogeneous | Secondary | [54,55,56,57,58,59,61,62,64,68,70,71,72,73,75,78,79,80,81,82,83,84,85,86,87,88,89,115,116,117,118] |
| ID_46 | [114] | 2021 | TTR | kNN | C | Heterogeneous | Secondary | [54,55,56,57,58,59,61,62,64,68,70,71,72,73,75,78,79,80,81,82,83,84,85,86,87,88,89,115,116,117,118] |
| ID_47 | [114] | 2021 | TTR | MLR | R | Heterogeneous | Secondary | [61,70,71,72,73,75,78,79,80,81,82,83,84,85,86,87,88] |
| ID_48 | [114] | 2021 | TTR | MLR | R | Heterogeneous | Secondary | [57,58,59] |
| ID_49 | [114] | 2021 | TTR | kNN | R | Heterogeneous | Secondary | [61,70,71,72,73,75,78,79,80,81,82,83,84,85,86,87,88] |
| ID_50 | [114] | 2021 | TTR | kNN | R | Heterogeneous | Secondary | [57,58,59] |
| ID_51 | [119] | 2021 | TR n.s. | RF | C | Heterogeneous | ToxCast database | Cited as ToxCast and Tox21 Summary Files for invitroDBv3.2, U.S. EPA, Washington, DC. |
| ID_52 | [119] | 2021 | TSHR | RF | C | Heterogeneous | ToxCast database | Cited as ToxCast and Tox21 Summary Files for invitroDBv3.2, U.S. EPA, Washington, DC. |
| ID_53 | [119] | 2021 | TSHR | NN | C | Heterogeneous | ToxCast database | Cited as ToxCast and Tox21 Summary Files for invitroDBv3.2, U.S. EPA, Washington, DC. |
| ID_54 | [119] | 2021 | TPO | XGB | C | Heterogeneous | ToxCast database ** | Cited as ToxCast and Tox21 Summary Files for invitroDBv3.2, U.S. EPA, Washington, DC. and [103] |
| ID_55 | [119] | 2021 | TRHR | SVM | C | Heterogeneous | ToxCast database | Cited as ToxCast and Tox21 Summary Files for invitroDBv3.2, U.S. EPA, Washington, DC. |
| ID_56 | [119] | 2021 | DIO1 | SVM | C | Heterogeneous | ToxCast database ** | Cited as ToxCast and Tox21 Summary Files for invitroDBv3.2, U.S. EPA, Washington, DC. and [101] |
| ID_57 | [119] | 2021 | DIO2 | SVM | C | Heterogeneous | ToxCast database ** | [101] |
| ID_58 | [119] | 2021 | DIO3 | NN | C | Heterogeneous | ToxCast database ** | [101] |
| ID_59 | [119] | 2021 | NIS | LR | C | Heterogeneous | ToxCast database ** | Cited as ToxCast and Tox21 Summary Files for invitroDBv3.2, U.S. EPA, Washington, DC. and [102] |
| ID_60 | [120] | 2021 | TPO | kNN | C | Heterogeneous | ToxCast database ** | [103,121] |
| ID_61 | [120] | 2021 | TPO | RF | C | Heterogeneous | ToxCast database ** | [103,121] |
| ID_62 | [57] | 2019 | TTR | MLR | R | Phenolic DBPs | Primary | [57] |
| ID_63 | [122] | 2018 | TR β | SVM | C | PCBs | Primary | [122] |
| ID_64 | [122] | 2018 | TR β | LDA | C | PCBs | Primary | [122] |
| ID_65 | [123] | 2018 | TR n.s. | SVM | C | PCBs and PBDEs | Secondary | [124,125,126,127,128,129,130,131,132] |
| ID_66 | [133] | 2017 | TTR | LDA | C | PFCs | Secondary | [82] |
| ID_67 | [133] | 2017 | TTR | MLR | R | PFCs | Secondary | [82] |
| ID_68 | [121] | 2017 | TPO | PLR | C | Heterogeneous | ToxCast database ** | [103,134,135,136] |
| ID_69 | [121] | 2017 | TPO | PLR | C | Heterogeneous | ToxCast database ** | [103,134,135,136] |
| ID_70 | [87] | 2015 | TTR | kNN | C | Heterogeneous | Secondary | [88] |
| ID_71 | [137] | 2015 | TTR | ASNN | C | Heterogeneous | Secondary | [88] |
| ID_72 | [138] | 2015 | TR β | Monte Carlo | R | Heterogeneous | Secondary | [139] |
| ID_73 | [138] | 2015 | TR β | Monte Carlo | R | Heterogeneous | Secondary | [139] |
| ID_74 | [138] | 2015 | TR β | Monte Carlo | R | Heterogeneous | Secondary | [139] |
| ID_75 | [139] | 2014 | TR β | RF | R | Heterogeneous | ChEMBL database *** | [140] |
| ID_76 | [139] | 2014 | TR β | RF | R | Heterogeneous | Secondary | [141,142,143] |
| ID_77 | [139] | 2014 | TR β | RF | C | Heterogeneous | ChEMBL database *** | [140] |
| ID_78 | [144] | 2013 | TTR | kNN | C | PFCs and BFRs | Secondary | [78,80,82] |
| ID_79 | [144] | 2013 | TTR | MLR | R | PFCs and BFRs | Secondary | [78,80,82] |
| ID_80 | [145] | 2012 | TTR | kNN | C | PFCs | Secondary | [82] |
| ID_81 | [145] | 2012 | TTR | kNN | C | PFCs | Secondary | [82] |
| ID_82 | [145] | 2012 | TTR | kNN | C | PFCs | Secondary | [82] |
| ID_83 | [145] | 2012 | TTR | kNN | C | PFCs | Secondary | [82] |
| ID_84 | [146] | 2011 | TTR | kNN | C | BFRs | Secondary | [78,80] |
| ID_85 | [147] | 2010 | TTR | MLR | R | BFRs | Secondary | [78,80] |
| ID_86 | [148] | 2010 | TR β | PLS | R | OH-PBDEs | Primary | [148] |
| MIE | Ref. | Model ID | Chemical Class | Descriptors | Software |
|---|---|---|---|---|---|
| TTR | [53] | ID_5 | Heterogeneous | AATSC1c; PubchemFP381; ATSC2s; nX | PaDEL [180] |
| ID_6 | Heterogeneous | naasC; SpMin4_Bhs; VE3_Dzs | PaDEL [180] | ||
| ID_7 | Heterogeneous | PubchemFP590; SpMax1_Bhe; PubchemFP18; GATS5c; AATSC1e; AATS4v | PaDEL [180] | ||
| [51] | ID_16 | Heterogeneous | Calculation of 119 RDKit chemical descriptors | RDKit: Open-source cheminformatics. http://www.rdkit.org | |
| [104] | ID_27 | Heterogeneous | Calculation of extended fingerprints with a KNIME implementation of the CDK toolkit | CDK toolkit: https://cdk.github.io/ | |
| [59] | ID_41 | Halogenated phenols and thiophenols | logDOW(pH = 7.40); ωadj; dipoleadj | Marvin Sketch 15.6.29.0, 2015: ChemAxon, http://www.chemaxon.com); Gaussian 16; GsGrid 1.7 (http://gsgrid.codeplex.com) | |
| [114] | ID_42 | Heterogeneous | Vsadj; Πadj; μadj | Marvin Sketch 15.6.29.0, 2015: ChemAxon, http://www.chemaxon.com; GaussView 6.0; Gaussian 16; GsGrid 1.7, http://gsgrid.codeplex.com | |
| ID_43 | Heterogeneous | Vsadj; O-059; μadj | Marvin Sketch 15.6.29.0, 2015: ChemAxon, http://www.chemaxon.com; GaussView 6.0; Gaussian 16; GsGrid 1.7, http://gsgrid.codeplex.com | ||
| ID_44 | Heterogeneous | Vsadj; H-050; nCbH | Marvin Sketch 15.6.29.0, 2015: ChemAxon, http://www.chemaxon.com; GaussView 6.0; Gaussian 16; GsGrid 1.7, http://gsgrid.codeplex.com | ||
| ID_45 | Heterogeneous | nArOH; Vsadj; ωadj | Marvin Sketch 15.6.29.0, 2015: ChemAxon, http://www.chemaxon.com; GaussView 6.0; Gaussian 16; GsGrid 1.7, http://gsgrid.codeplex.com | ||
| ID_46 | Heterogeneous | Vsadj; C-024; nHDon | Marvin Sketch 15.6.29.0, 2015: ChemAxon, http://www.chemaxon.com; GaussView 6.0; Gaussian 16; GsGrid 1.7, http://gsgrid.codeplex.com | ||
| ID_47 | Heterogeneous | C-040; nCq; H-050; O-058; Πadj; O-056 | Marvin Sketch 15.6.29.0, 2015: ChemAxon, http://www.chemaxon.com; GaussView 6.0; Gaussian 16; GsGrid 1.7, http://gsgrid.codeplex.com | ||
| ID_48 | Heterogeneous | log DOW(pH = 7.40); nArOH; O-057; nArNO2 | Marvin Sketch 15.6.29.0, 2015: ChemAxon, http://www.chemaxon.com; GaussView 6.0; Gaussian 16; GsGrid 1.7, http://gsgrid.codeplex.com | ||
| ID_49 | Heterogeneous | EHOMO-adj; nArOH; H052; ωadj | Marvin Sketch 15.6.29.0, 2015: ChemAxon, http://www.chemaxon.com; GaussView 6.0; Gaussian 16; GsGrid 1.7, http://gsgrid.codeplex.com | ||
| ID_50 | Heterogeneous | log DOW(pH = 7.40); nArOH | Marvin Sketch 15.6.29.0, 2015: ChemAxon, http://www.chemaxon.com; GaussView 6.0; Gaussian 16; GsGrid 1.7, http://gsgrid.codeplex.com | ||
| [57] | ID_62 | Phenolic DBPs | log D; dipoleadj | Marvin Sketch 15.6.29.0, 2015: ChemAxon, http://www.chemaxon.com; Gaussian 16 | |
| [133] | ID_66 | PFCs | Me; nCsp2; H-050 | DRAGON Version 6.0, 2011, http://www.talete.mi.it/ | |
| ID_67 | PFCs | IC3; ∑β’S | DRAGON Version 6.0, 2011, http://www.talete.mi.it/ | ||
| [87] | ID_70 | Heterogeneous | Based on the following 14 molecular descriptors: TPSA; a_don; a_nOH; nX; PEOE_VSA_FNEG; PEOE_RPC-; density; PEOE_RPC+; diameter; PEOE_PC+; vsa_hyd; KierFlex; logP(o/w); opr_brigid | Molecular Operating Environment (MOE), 2013.08; Chemical Computing Group Inc.: Montreal, QC, Canada, 2015 | |
| [137] | ID_71 | Heterogeneous | nArOH; nHDon; nCb-; nCRX3; nCH2RX; ALogPS_logP; nArOR; nCrq; nCq; nCp; nCs; nCbH | DRAGON version 6 [181]. | |
| [144] | ID_78 | PFCs and BFRs | nArOH; F03(Br..Br); HATS6m | DRAGON Version 5.5 for Windows, Talete srl, Milan, Italy, 2007 | |
| ID_79 | PFCs and BFRs | R5u; F07[C-O]; nArOH | DRAGON Version 5.5 for Windows, Talete srl, Milan, Italy, 2007 | ||
| [145] | ID_80 | PFCs | AMW; HATS6m | DRAGON Version 5.5 for Windows, Talete srl, Milan, Italy, 2007 | |
| ID_81 | PFCs | nH; HATS6m | DRAGON Version 5.5 for Windows, Talete srl, Milan, Italy, 2007 | ||
| ID_82 | PFCs | nH; F06[C-O] | DRAGON Version 5.5 for Windows, Talete srl, Milan, Italy, 2007 | ||
| ID_83 | PFCs | T(F..F); HATS6m | DRAGON Version 5.5 for Windows, Talete srl, Milan, Italy, 2007 | ||
| [146] | ID_84 | BFRs | DISPe; nArOH | DRAGON Version 5.5 for Windows, Talete srl, Milan, Italy, 2008 | |
| [147] | ID_85 | BFRs | qpmax; MATS6v | DRAGON Version 5.5 for Windows, Talete srl, Milan, Italy | |
| TR α | [90] | ID_8 | PFAS | X%; ICR | AlvaDesc [182] |
| [104] | ID_28 | Heterogeneous | Calculation of extended fingerprints with a KNIME implementation of the CDK toolkit | CDK toolkit: https://cdk.github.io/ | |
| TR β | [90] | ID_9 | PFAS | X%; TPC | AlvaDesc [182] |
| [51] | ID_17 | Heterogeneous | Calculation of 119 RDKit chemical descriptors | RDKit: Open-source cheminformatics. http://www.rdkit.org | |
| ID_18 | Heterogeneous | Calculation of 119 RDKit chemical descriptors | RDKit: Open-source cheminformatics. http://www.rdkit.org | ||
| [104] | ID_29 | Heterogeneous | Calculation of extended fingerprints with a KNIME implementation of the CDK toolkit | CDK toolkit: https://cdk.github.io/ | |
| [112] | ID_39 | PCNs | ELUMO; ΔE; μ; Qxx; Qyy; Qyz; q+; logKow; NCl; No | Gaussian 09 software. | |
| [113] | ID_40 | Heterogeneous | Use of RDKit descriptors | RDKit: Open-source cheminformatics. http://www.rdkit.org | |
| [122] | ID_63 | PCBs | logKow; ω; BER; nCl; EEig13d; JGI4 | EPI Suite, version 4.1 (US EPA, 2012); DRAGON | |
| ID_64 | PCBs | logKow; ω; BER; nCl; EEig13d; JGI4 | EPI Suite, version 4.1 (US EPA, 2012); DRAGON | ||
| [138] | ID_72 | Heterogeneous | Molecular optimal descriptor DCW(3, 10) | CORAL software: http://www.insilico.eu/coral | |
| ID_73 | Heterogeneous | Molecular optimal descriptor DCW(1, 3) | CORAL software: http://www.insilico.eu/coral | ||
| ID_74 | Heterogeneous | Molecular optimal descriptor DCW(3, 4) | CORAL software: http://www.insilico.eu/coral | ||
| [139] | ID_75 | Heterogeneous | Thirty-five most statistically significant descriptors were identified: F04[N-Cl]; EEig03d; F06[C-Cl]; EEig08r; GATS7e; nArOH; EEig07r; EEig05d; EEig06d; TPSA(Tot); GGI1; BEHp4; SPI; C-026; ESpm01d; nCb-; Hy; GATS8v; T(O..O); BLTA96; IVDE; MATS1e; Ms; GATS6e; MATS6m; MATS5m; MATS2e; MATS1p; MATS8v; MATS6e; MATS8p; X4Av; X2Av; X0Av; Jhetp | Dragon software (version 5.4; Talete s.r.l., Milan, Italy) | |
| ID_76 | Heterogeneous | Twenty-seven most statistically significant descriptors were identified: F08[C-Cl]; T(N..Cl); C-006; EEig06d; SEigm; ATS3m; ATS4m; BEHm6; T(O..Cl); ATS5m; ATS7m; BEHm7; Uindex; EEig04d; BELe3; EEig08d; HVcpx; PHI; BELm3; GGI8; BIC5; BEHml; JGI6; JGI7; BELml; GATS3p; VEA2 | Dragon software (version 5.4; Talete s.r.l., Milan, Italy) | ||
| ID_77 | Heterogeneous | Thirty most statistically significant descriptors were identified: B05[O-O]; EEig03d; nArOH; GGI7; EEig05d; PW2; F04[C-N]; C-026; ESpm01d; AAC; GATS8p; Hy; PCR; GATS8v; F05[O-O]; O-057; MATS5v; IVDE; MATS1e; Ms; MATS5p; ARR; MATS5m; PHI; MATS8v; GATS1e; MATS8p; RBF; Jhetp; X1A | Dragon software (version 5.4; Talete s.r.l., Milan, Italy) | ||
| [148] | ID_86 | OH-PBDEs | nBr; logKow; IA; ELUMO; ω; μ2 | EPI Suite, version 4.0 (U.S. Environmental Protection Agency 2009); Gaussian 03 programs; DRAGON [181] | |
| TR n.s. | [91] | ID_10 | OH-PCBs | RDF35u; RDF55u; RDF85u; RDF65v | PaDEL [180] |
| ID_11 | OH-PCBs | RDF35u; RDF55u; RDF85u; RDF65v | PaDEL [180] | ||
| [119] | ID_51 | Heterogeneous | Calculation of count-based Morgan fingerprints with a radius of 2 bonds and a length of 2048 bits, and of all 119 one-dimensional and two-dimensional RDKit chemical descriptors | RDKit: Open-source cheminformatics. http://www.rdkit.org | |
| [123] | ID_65 | PCBs; PBDEs | DELS; MAXDN; Mor31v; Ms; RDF040e; BER | DRAGON 5.5 for Windows, Talete srl, Milan, Italy, 2008 | |
| TSHR | [106] | ID_31 | Heterogeneous | Thirty-nine descriptors were used, here sorted by their weight in descending order (top seven descriptors were used to build Model ID_32.): Sw < 0.1 mg/mL probability; LogSw; LogD(pH = 7.4); LogL; S; R2; E; LogS(pH = 7.4); logP; Solubility class; AAB/LogP; McGowan Volume; MW; Pi2; LogS(pH = 7.4)-; L; V; Sw < 1 mg/mL probability; No Of H Donors; Acid_pKa; LogSwLo; Sw > 10 mg/mL probability; Abraham’s Alfa; NoOfRotBonds; A; Bo; 0Form; B; Form+; No Of H Acceptors; LogSwHi; Rel_pKa_ac; Base_pKa; Abraham’s BetaH; Ertl TPSA; Form-; Rule of 5; Rel_pKa_bs; Form± | KOWWIN program (EPI Suite version 4.1.1, https://www.epa.gov/tsca-screening-tools/epi-suitetm-estimation-program-interface) to calculate logKow. Software for the calculation of the other molecular descriptors was not specified |
| ID_32 | Sw < 0.1 mg/mL probability; LogSw; LogD(pH = 7.4); LogL; S; R2; E | KOWWIN program (EPI Suite version 4.1.1, https://www.epa.gov/tsca-screening-tools/epi-suitetm-estimation-program-interface) to calculate logKow. Software for the calculation of the other molecular descriptors was not specified | |||
| ID_33 | The use of thirty-nine descriptors was reported in the study | KOWWIN program (EPI Suite version 4.1.1, https://www.epa.gov/tsca-screening-tools/epi-suitetm-estimation-program-interface) to calculate logKow. Software for the calculation of the other molecular descriptors was not specified | |||
| ID_34 | LogS, LogP, E | KOWWIN program (EPI Suite version 4.1.1, https://www.epa.gov/tsca-screening-tools/epi-suitetm-estimation-program-interface) to calculate logKow. Software for the calculation of the other molecular descriptors was not specified | |||
| ID_35 | Forty-one descriptors were used, here sorted by their weight in descending order: Base_pKa; V; Abraham’s Alfa; 0Form; AAB/LogP; CDocker Energy; NoOfRotBonds; S; LogSwLo; LogSwHi; CDocker Interaction Energy; Rel_pKa_bs; R2; E; LogD(pH = 7.4); LogS(pH = 7.4)-; Sw < 0.1 mg/mL probability; A; Sw > 10 mg/mL probability; Ertl TPSA; MW; logP; LogSw; Pi2; Abraham’s BetaH; Solubility class; B; LogL; Sw < 1 mg/mL probability; L; Acid_pKa; Rel_pKa_ac; No Of H Acceptors; Bo; No Of H Donors; McGowan Volume; LogS(pH = 7.4); Form+; Form-; Form±; Rule of 5 | KOWWIN program (EPI Suite version 4.1.1, https://www.epa.gov/tsca-screening-tools/epi-suitetm-estimation-program-interface) to calculate logKow. Software for the calculation of the other molecular descriptors was not specified | |||
| [51] | ID_19 | Heterogeneous | Calculation of 119 RDKit chemical descriptors | RDKit: Open-source cheminformatics. http://www.rdkit.org | |
| ID_20 | Heterogeneous | Calculation of 119 RDKit chemical descriptors | RDKit: Open-source cheminformatics. http://www.rdkit.org | ||
| [119] | ID_52 | Heterogeneous | Calculation of count-based Morgan fingerprints with a radius of 2 bonds and a length of 2048 bits, and of all 119 one-dimensional and two-dimensional RDKit chemical descriptors | RDKit: Open-source cheminformatics. http://www.rdkit.org | |
| ID_53 | Heterogeneous | Calculation of count-based Morgan fingerprints with a radius of 2 bonds and a length of 2048 bits, and of all 119 one-dimensional and two-dimensional RDKit chemical descriptors | RDKit: Open-source cheminformatics. http://www.rdkit.org | ||
| [95] | ID_15 | Heterogeneous | Top twenty FPs with positive SHAP (Shapley additive explanation) values: PubchemFP12, PubchemFP259, PubchemFP257, PubchemFP256, PubchemFP628, PubchemFP185, PubchemFP258, PubchemFP2, PubchemFP143, PubchemFP146, PubchemFP656, PubchemFP633, PubchemFP150, PubchemFP464, PubchemFP442, PubchemFP607, PubchemFP613, PubchemFP549, PubchemFP153, PubchemFP418 | PaDEL [180] | |
| TPO | [51] | ID_26 | Heterogeneous | Calculation of 119 RDKit chemical descriptors | RDKit: Open-source cheminformatics. http://www.rdkit.org |
| [107] | ID_36 | Heterogeneous | Use of Atom Pair Count (APC) fingerprints | PaDEL [180] | |
| ID_37 | Heterogeneous | Use of Atom Pair Count (APC) fingerprints | PaDEL [180] | ||
| ID_38 | Heterogeneous | Use of Atom Pair Count (APC) fingerprints | PaDEL [180] | ||
| [119] | ID_54 | Heterogeneous | Calculation of count-based Morgan fingerprints with a radius of 2 bonds and a length of 2048 bits, and of all 119 one-dimensional and two-dimensional RDKit chemical descriptors | RDKit: Open-source cheminformatics. http://www.rdkit.org | |
| [120] | ID_60 | Heterogeneous | The top twenty ranked descriptors identified in the kNN model: GATS1e; NArOH; CATS2D_02_DL; MATS1e; MATS1s; C-026; CATS2D_03_DL; B10 [C-C]; MATS1m; ‘SpMax2_Bh(s); MATS1p; nCb-; NX; Uc; ‘P_VSA_i_1’; SpMAD_B(v); NCbH; GATS1s; MLOGP; Eta_C_A’ | DRAGON v7.0.8., 2017: https://chm.kode-solutions.net/products_dragon.php | |
| ID_61 | Heterogeneous | Based on 160 molecular descriptors | DRAGON v7.0.8., 2017: https://chm.kode-solutions.net/products_dragon.php | ||
| [121] | ID_68 | Heterogeneous | Based on scaffolds and structural features | Leadscope Predictive Data Miner (LPDM), Leadscope, Inc., (2016): http://www.leadscope.com/ | |
| ID_69 | Heterogeneous | The top ten most common structural features linked to active compounds: benzene, 1,3-dihydroxy-; Scaffold 288; benzene, 1-alkyl-,4-amino(NH2)-; benzene, 1,2-dihydroxy-; Scaffold 297; alcohol, alkenyl-; Scaffold 576; benzene, 1-alkoxy-,4-hydroxy-; Scaffold 306; Scaffold 574. The top ten most commons structural features linked to inactive compounds: Scaffold 110; Scaffold 342; Scaffold 210; Scaffold 253; Scaffold 303; Scaffold 108; benzene, 1-alkyl-,4-halo-; halide, benzyl-; Scaffold 454; Scaffold 194 | Leadscope Predictive Data Miner (LPDM), Leadscope, Inc., (2016): http://www.leadscope.com/ | ||
| TBG | [52] | ID_1 | PBBs | Molecular Weight (MW); Critical temperature (CT); Critical pressure (CP); Topological diameter (TD) | PaDEL [180]; Gaussian (Gaussian 09 (Gaussian Inc., Wallingford, CT, USA); ChemDraw 12.0 |
| ID_2 | PBBs and OH-PBBs | Quadrupole moment Qyy (Qyy); Most negative Mulliken charge number (q−); Frequency (Freq); TD | PaDEL [180]; Gaussian (Gaussian 09 (Gaussian Inc., Wallingford, CT, USA); ChemDraw 12.0 | ||
| ID_3 | PBBs and 2OH-PBBs | q−; CP; TD; Topological Shape (TS) | PaDEL [180]; Gaussian (Gaussian 09 (Gaussian Inc., Wallingford, CT, USA); ChemDraw 12.0 | ||
| ID_4 | PBBs, OH-PBBs, and 2OH-PBBs | q−; CP; TD; CT | PaDEL [180]; Gaussian (Gaussian 09 (Gaussian Inc., Wallingford, CT, USA); ChemDraw 12.0 | ||
| NIS | [51] | ID_25 | Heterogeneous | Calculation of 119 RDKit chemical descriptors | RDKit: Open-source cheminformatics. http://www.rdkit.org |
| [104] | ID_30 | Heterogeneous | Calculation of extended fingerprints with a KNIME implementation of the CDK toolkit | CDK toolkit: https://cdk.github.io/ | |
| [119] | ID_59 | Heterogeneous | Calculation of count-based Morgan fingerprints with a radius of 2 bonds and a length of 2048 bits, and of all 119 one-dimensional and two-dimensional RDKit chemical descriptors | RDKit: Open-source cheminformatics. http://www.rdkit.org | |
| Albumin | [93] | ID_12 | PFAS | PDI; GATS8v; MATS8m; QED | AlvaDesc 2.0.16 [183] |
| ID_13 | PFAS | Eig12_AEA(bo); DECC; X4A | AlvaDesc 2.0.16 [183] | ||
| ID_14 | PFAS | QED; PDI; GATS8v; MATS8m | AlvaDesc 2.0.16 [183] | ||
| DIO1 | [51] | ID_22 | Heterogeneous | Calculation of 119 RDKit chemical descriptors | RDKit: Open-source cheminformatics. http://www.rdkit.org |
| [119] | ID_56 | Heterogeneous | Calculation of count-based Morgan fingerprints with a radius of 2 bonds and a length of 2048 bits, and of all 119 one-dimensional and two-dimensional RDKit chemical descriptors | RDKit: Open-source cheminformatics. http://www.rdkit.org | |
| DIO2 | [51] | ID_23 | Heterogeneous | Calculation of 119 RDKit chemical descriptors | RDKit: Open-source cheminformatics. http://www.rdkit.org |
| [119] | ID_57 | Heterogeneous | Calculation of count-based Morgan fingerprints with a radius of 2 bonds and a length of 2048 bits, and of all 119 one-dimensional and two-dimensional RDKit chemical descriptors | RDKit: Open-source cheminformatics. http://www.rdkit.org | |
| DIO3 | [51] | ID_24 | Heterogeneous | Calculation of 119 RDKit chemical descriptors | RDKit: Open-source cheminformatics. http://www.rdkit.org |
| [119] | ID_58 | Heterogeneous | Calculation of count-based Morgan fingerprints with a radius of 2 bonds and a length of 2048 bits, and of all 119 one-dimensional and two-dimensional RDKit chemical descriptors | RDKit: Open-source cheminformatics. http://www.rdkit.org | |
| TRHR | [51] | ID_21 | Heterogeneous | Calculation of 119 RDKit chemical descriptors | RDKit: Open-source cheminformatics. http://www.rdkit.org |
| [119] | ID_55 | Heterogeneous | Calculation of count-based Morgan fingerprints with a radius of 2 bonds and a length of 2048 bits, and of all 119 one-dimensional and two-dimensional RDKit chemical descriptors | RDKit: Open-source cheminformatics. http://www.rdkit.org |
| Model ID | Reference | Year | MIE | Algorithm | C or R | Chemical Class | Data Source Type | Data Source Literature Reference(s) |
|---|---|---|---|---|---|---|---|---|
| ID_2025_1 | [186] | 2025 | Albumin | MLR | R | Phenoxyacetic acid-derived congeners | Primary | [186] |
| ID_2025_2 | [187] | 2025 | TTR | RF | C | Heterogenous | Secondary | [188] |
| ID_2025_3 | [189] | 2025 | TTR | LDA | C | PFAS | Secondary | [190] |
| ID_2025_4 | [189] | 2025 | TTR | MLR | R | PFAS | Secondary | [190] |
| ID_2025_5 | [191] | 2025 | TTR | DTC | C | PFAS | Primary | [191] |
| ID_2025_6 | [191] | 2025 | TTR | MLR | R | PFAS | Primary | [191] |
| MIE | Ref. | Model ID | Chemical class | Descriptors | Software |
|---|---|---|---|---|---|
| TTR | [187] | ID_2025_2 | Heterogenous | Thirty-one descriptors sorted by permutation importance: CrippenLogP; ATSC3c; ATSC5c; C1SP3; ETA_BetaP_s; naAromAtom; ZMIC1; ATSC4m; ZMIC5; ATSC4c; hmin; hmax; ATSC2m; ATSC5m; ETA_Beta_ns_d; ATSC0m; VE1_DzZ; C1SP2; ZMIC2; ATSC1m; nHBAcc; ZMIC3; ATSC3m; ATSC2c; ETA_dAlpha_A; ETA_Shape_Y; ATSC0c; maxdssC; ZMIC4; nHBDon; ATSC1c | PaDEL descriptors from OPERA software v2.9 [192] |
| [189] | ID_2025_3 | PFAS | GATS3e; ATSC6p; GATS8m; MIC2 | PaDEL [180] | |
| [189] | ID_2025_4 | PFAS | piPC5; GGI9; AATSC0e | PaDEL [180] | |
| [191] | ID_2025_5 | PFAS | SM4_D; GATS3m | AlvaDesc [182] | |
| [191] | ID_2025_6 | PFAS | AMW; GATS7p; B10[F-F] | AlvaDesc [182] | |
| Albumin | [186] | ID_2025_1 | Phenoxyacetic acid-derived congeners | logkBMC; α; sum of HBD and HBA | ACD/Percepta software, version 1994–2012 (ACD/Labs, Advanced Chemistry Development, Inc., Toronto, ON, Canada) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Evangelista, M.; Papa, E. A Review of Quantitative Structure–Activity Relationship (QSAR) Models to Predict Thyroid Hormone System Disruption by Chemical Substances. Toxics 2025, 13, 799. https://doi.org/10.3390/toxics13090799
Evangelista M, Papa E. A Review of Quantitative Structure–Activity Relationship (QSAR) Models to Predict Thyroid Hormone System Disruption by Chemical Substances. Toxics. 2025; 13(9):799. https://doi.org/10.3390/toxics13090799
Chicago/Turabian StyleEvangelista, Marco, and Ester Papa. 2025. "A Review of Quantitative Structure–Activity Relationship (QSAR) Models to Predict Thyroid Hormone System Disruption by Chemical Substances" Toxics 13, no. 9: 799. https://doi.org/10.3390/toxics13090799
APA StyleEvangelista, M., & Papa, E. (2025). A Review of Quantitative Structure–Activity Relationship (QSAR) Models to Predict Thyroid Hormone System Disruption by Chemical Substances. Toxics, 13(9), 799. https://doi.org/10.3390/toxics13090799






