Exposure to Bisphenol S and Bisphenol F Alters Gene Networks Related to Protein Translation and Neuroinflammation in SH-SY5Y Human Neuroblastoma Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. SH-SY5Y Culturing, Differentiation, and Chemical Exposures
2.3. Cell Viability Assay
2.4. Caspase Activity Assay
2.5. Mitochondrial Membrane Potential
2.6. ATPase Activity
2.7. Mitochondrial Bioenergetics in Differentiated SH-SY5Y Cells
2.8. Intracellular Reactive Oxygen Species in Differentiated SH-SY5Y Cells
2.9. RNA-Sequencing
2.10. Bioinformatics of Pathways
2.11. Real-Time PCR Analysis
2.12. Statistical Analysis
3. Results
3.1. Cytotoxicity Based on Cell Viability
3.2. Caspase 3/7 Activity
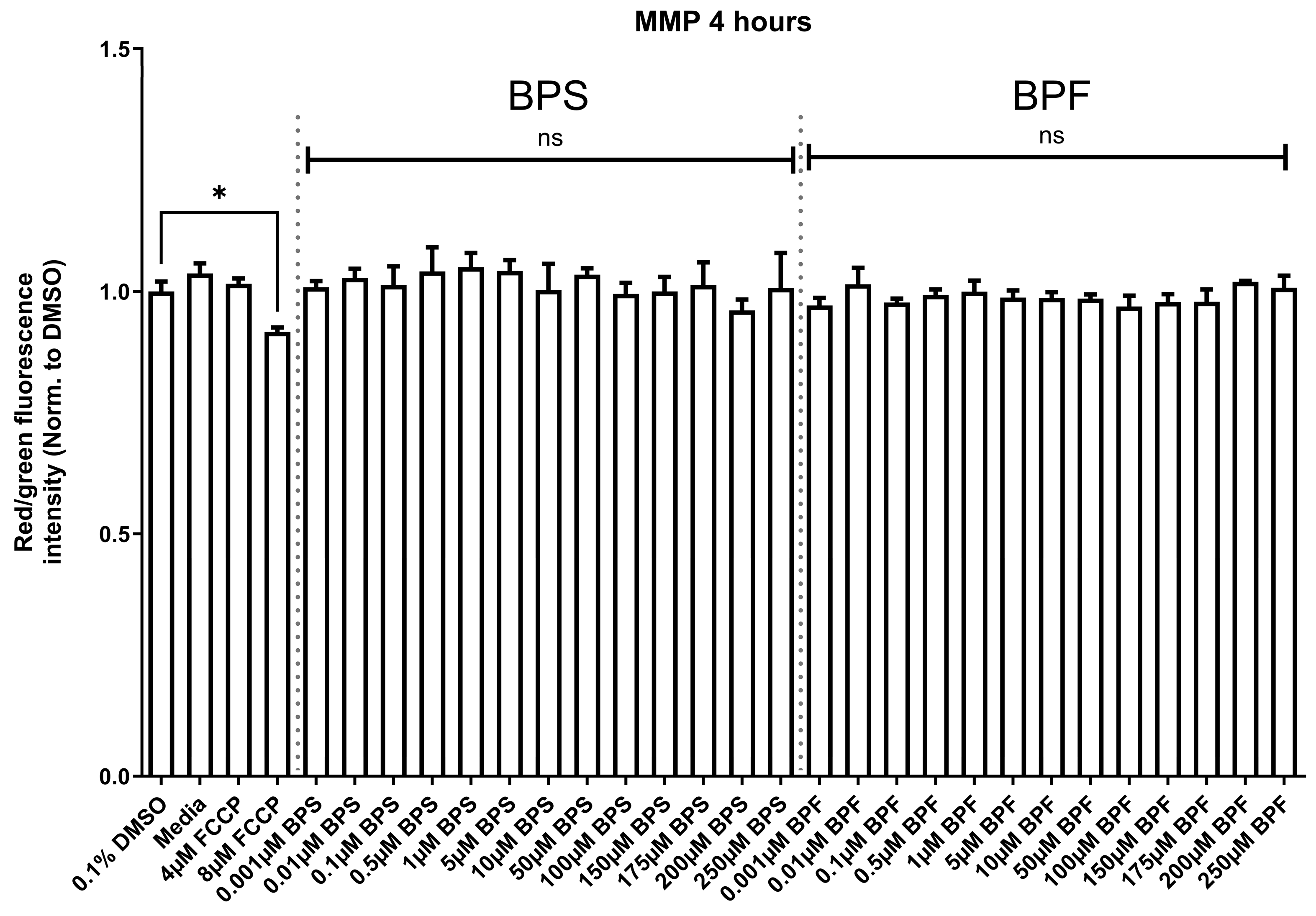
3.3. Mitochondrial Membrane Potential
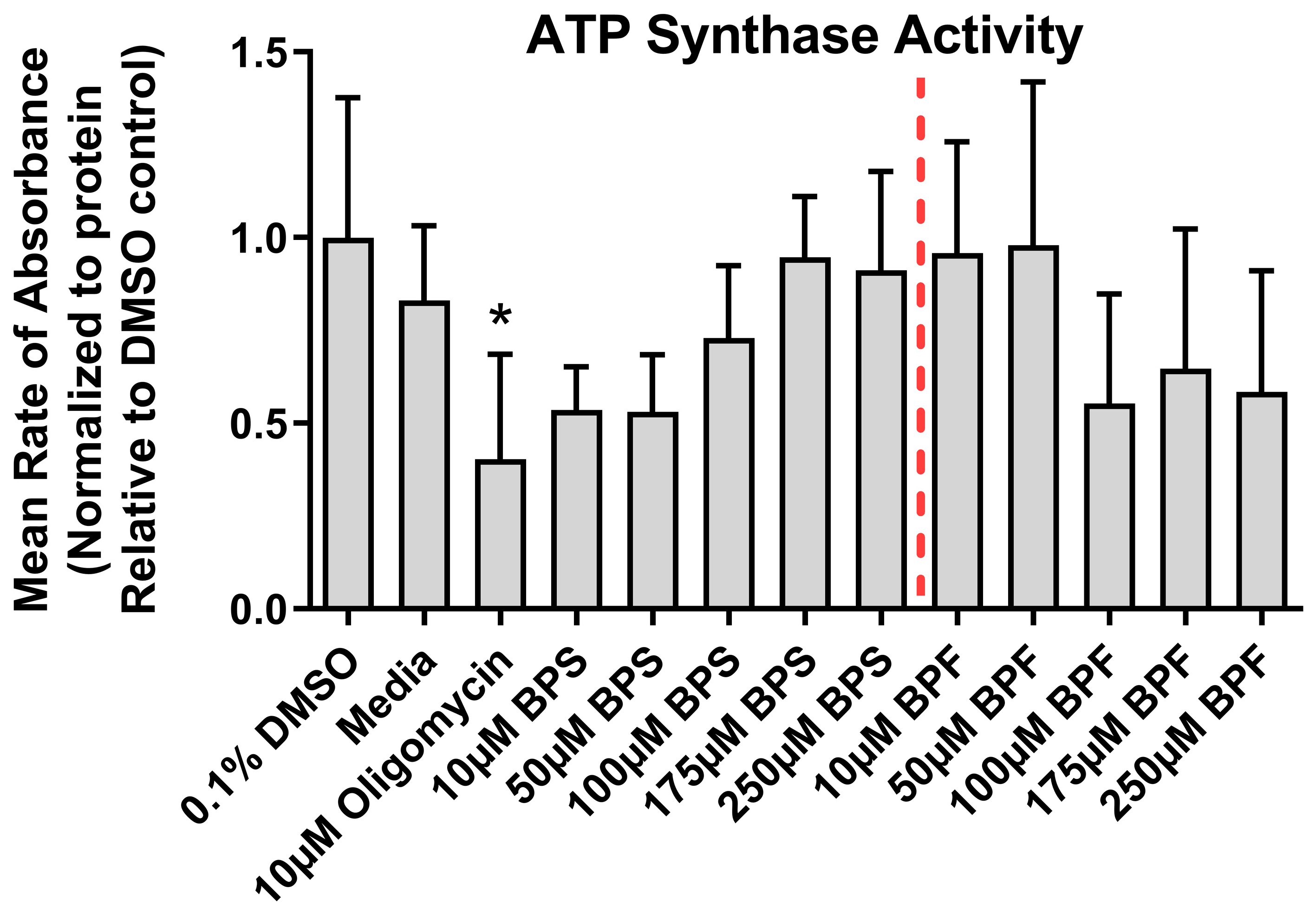
3.4. ATPase Activity
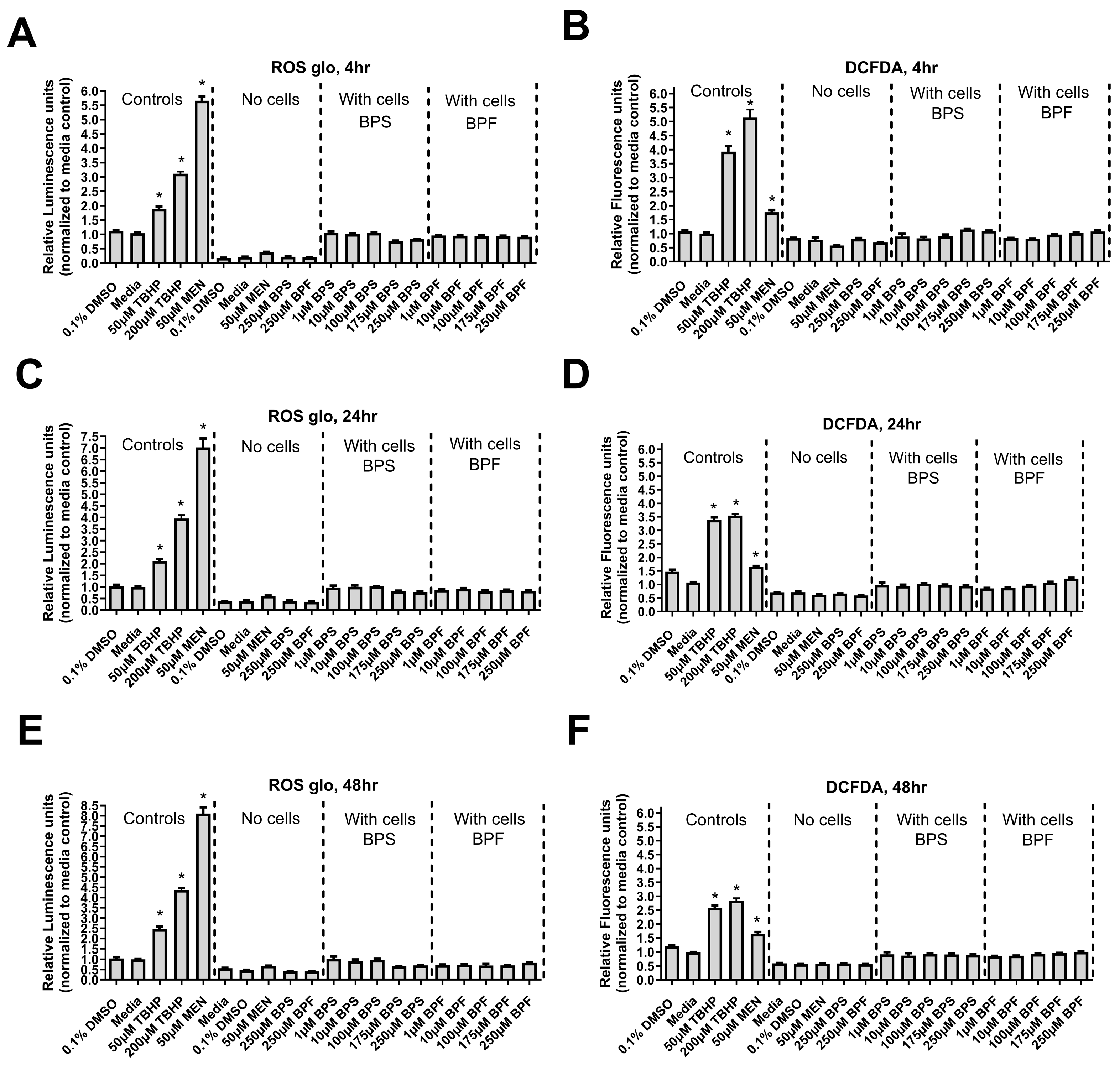
3.5. Reactive Oxygen Species
3.6. Mitochondrial Bioenergetics
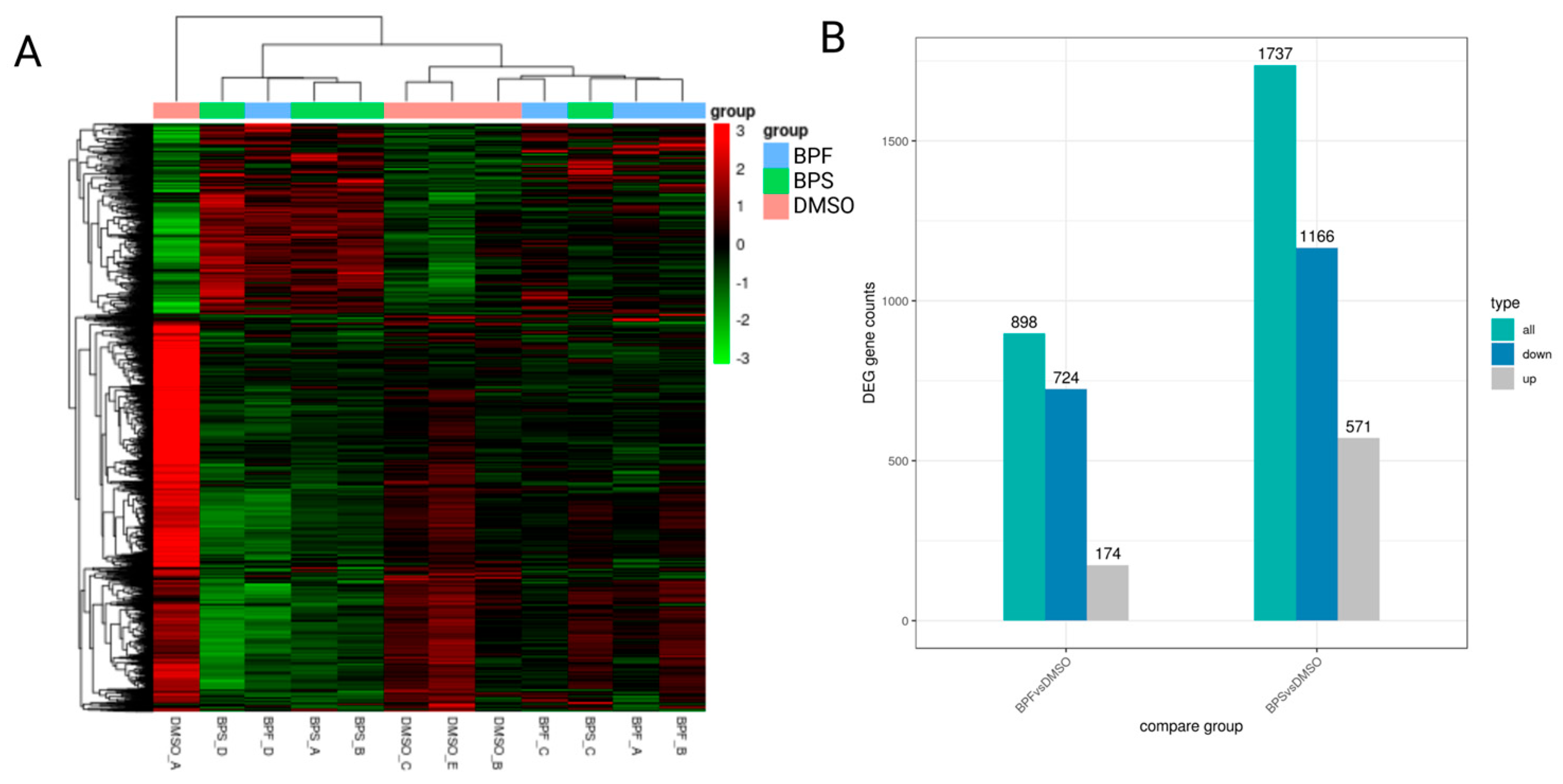
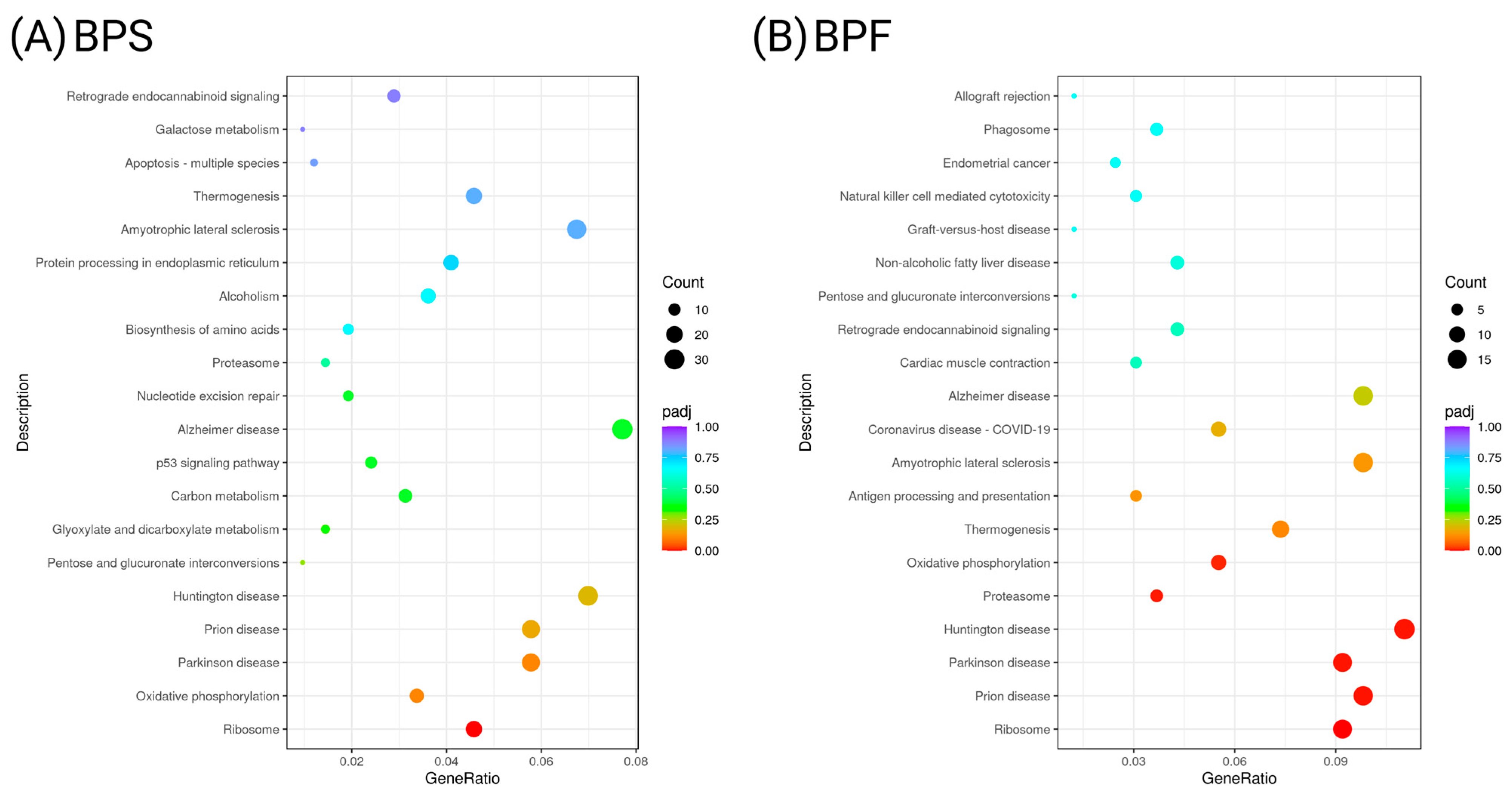
3.7. Transcriptome Profiling in SH-SY5Y Cells Following BPS and BPF Exposure
3.8. Real-Time PCR Analysis
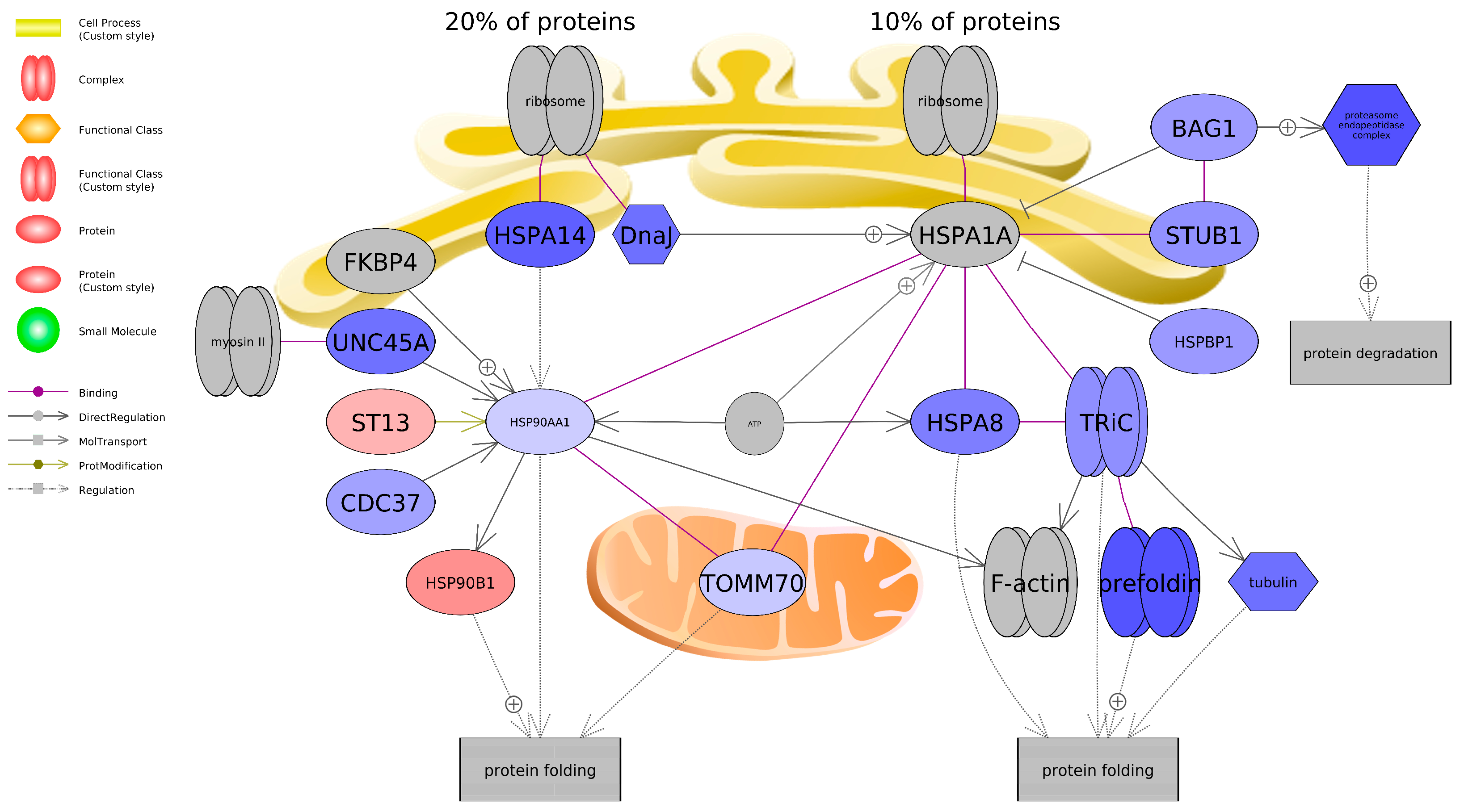
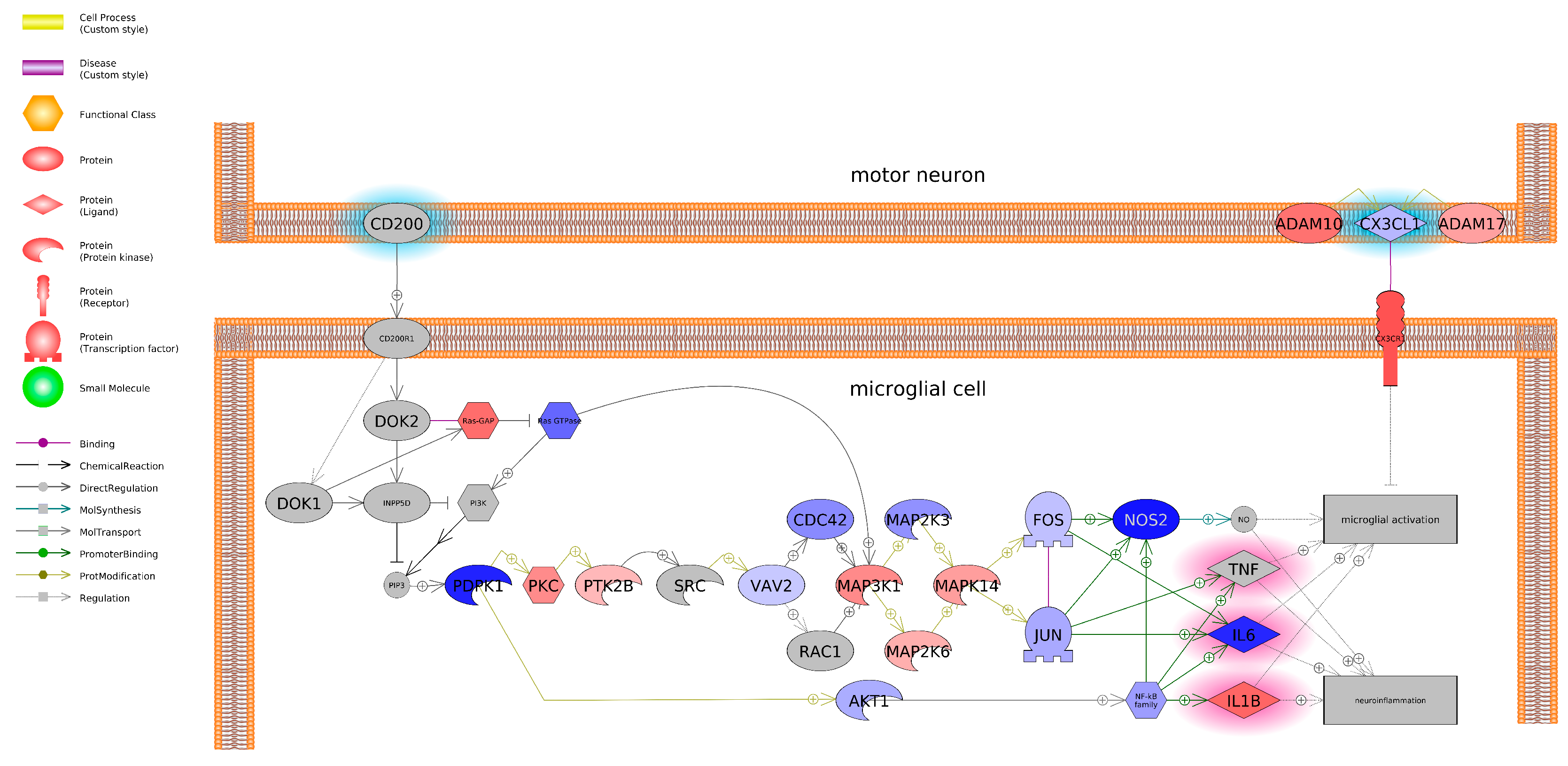
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AChE | Acetylcholinesterase |
| ADAM10 | A Disintegrin and Metalloproteinase Domain 10 |
| ADAM17 | A Disintegrin and Metalloproteinase Domain 17 |
| AM/Rot | Antimycin/Rotenone |
| BAK1 | Bcl-2 Antagonist/Killer 1 |
| BAX | Bcl-2-Associated X |
| BACE1 | Beta-secretase 1 |
| BLC-2 | B-Cell Leukemia/Lymphoma 2 |
| BPA | Bisphenol A |
| BPF | Bisphenol F |
| BPS | Bisphenol S |
| Casp3/7 | Caspase 3/7 |
| Caspase3 | Cysteinyl aspartate specific proteinase 3 |
| CYT C | Cytochrome c |
| DCFDA | 2′,7′-dichlorofluorescein diacetate |
| DIG | Digitonin |
| DMSO | Dimethyl sulfoxide |
| ER | Estrogen receptor |
| FCCP | Carbonyl cyanide-4-phenylhydrazone |
| FPKM | Fragments per kilobase of transcript per million |
| GEO | Gene Expression Omnibus |
| GO | Gene Ontology |
| GPCR | G-protein coupled receptors |
| GSEA | Gene set enrichment analysis |
| HSPA8 | Heat Shock Protein Family A Member 8 |
| HSPA14 | Heat Shock Protein Family A Member 14 |
| HSP90AA1 | Heat Shock Protein 90 Alpha Family A Member 1 |
| IL-1β | Interleukin-1β |
| IL2 | Interleukin 2 |
| LDH | Lactate dehydrogenase |
| MAO-A | Monoamine oxidase-A |
| MDA | Malondialdehyde |
| MEN | Menadione |
| MMP | Mitochondrial membrane potential |
| NF-κB | Nuclear factor-κB |
| OM | Oligomycin |
| POMC | Pro-opiomelanocortin |
| RA | Retinoic acid |
| rNSCs | Rat fetal neural stem cells |
| ROS | Reactive oxygen species |
| TBHP | Tert-Butyl hydroperoxide |
| TNF | Tumor necrosis factor |
| TOMM70 | Translocase of Outer Mitochondrial Membrane 70 |
References
- Faheem, M.; Bhandari, R.K. Detrimental effects of bisphenol compounds on physiology and reproduction in fish: A literature review. Environ. Toxicol. Pharmacol. 2021, 81, 103497. [Google Scholar] [CrossRef] [PubMed]
- Cariati, F.; D’Uonno, N.; Borrillo, F.; Iervolino, S.; Galdiero, G.; Tomaiuolo, R. Bisphenol a: An emerging threat to male fertility. Reprod. Biol. Endocrinol. 2019, 17, 6. [Google Scholar] [CrossRef]
- Pivonello, C.; Muscogiuri, G.; Nardone, A.; Garifalos, F.; Provvisiero, D.P.; Verde, N.; De Angelis, C.; Conforti, A.; Piscopo, M.; Auriemma, R.S. Bisphenol A: An emerging threat to female fertility. Reprod. Biol. Endocrinol. 2020, 18, 22. [Google Scholar] [CrossRef]
- Wang, X.H.; Souders II, C.L.; Zhao, Y.H.; Martyniuk, C.J. Paraquat affects mitochondrial bioenergetics, dopamine system expression, and locomotor activity in zebrafish (Danio rerio). Chemosphere 2018, 191, 106–117. [Google Scholar] [CrossRef]
- Górowska-Wójtowicz, E.; Duliban, M.; Kudrycka, M.; Dutka, P.; Pawlicki, P.; Milon, A.; Zarzycka, M.; Placha, W.; Kotula-Balak, M.; Ptak, A. Leydig cell tumorigenesis-implication of G-protein coupled membrane estrogen receptor, peroxisome proliferator-activated receptor and xenoestrogen exposure. In vivo and in vitro appraisal. Tissue Cell 2019, 61, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Wang, X.; Tan, H.; Shi, W.; Zhang, X.; Wei, S.; Giesy, J.P.; Yu, H. Molecular initiating events of bisphenols on androgen receptor-mediated pathways provide guidelines for in silico screening and design of substitute compounds. Environ. Sci. Technol. Lett. 2019, 6, 205–210. [Google Scholar] [CrossRef]
- Mu, X.; Liu, Z.; Zhao, X.; Yuan, L.; Li, Y.; Wang, C.; Xiao, G.; Mu, J.; Qiu, J.; Qian, Y. Bisphenol A Analogues Induce Neuroendocrine Disruption via Gut–Brain Regulation in Zebrafish. Environ. Sci. Technol. 2024, 58, 1022–1035. [Google Scholar] [CrossRef]
- Martínez, R.; Tu, W.; Eng, T.; Allaire-Leung, M.; Piña, B.; Navarro-Martín, L.; Mennigen, J.A. Acute and long-term metabolic consequences of early developmental Bisphenol A exposure in zebrafish (Danio rerio). Chemosphere 2020, 256, 127080. [Google Scholar] [CrossRef]
- Lehmler, H.-J.; Liu, B.; Gadogbe, M.; Bao, W. Exposure to bisphenol A, bisphenol F, and bisphenol S in US adults and children: The national health and nutrition examination survey 2013–2014. ACS Omega 2018, 3, 6523–6532. [Google Scholar] [CrossRef]
- Andújar, N.; Gálvez-Ontiveros, Y.; Zafra-Gómez, A.; Rodrigo, L.; Álvarez-Cubero, M.J.; Aguilera, M.; Monteagudo, C.; Rivas, A. Bisphenol A analogues in food and their hormonal and obesogenic effects: A review. Nutrients 2019, 11, 2136. [Google Scholar] [CrossRef]
- Jurek, A.; Leitner, E. Analytical determination of bisphenol A (BPA) and bisphenol analogues in paper products by GC-MS/MS. Food Addit. Contam. Part A 2017, 34, 1225–1238. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Zhao, J.-L.; Yang, Y.-Y.; Jia, Y.-W.; Zhang, Q.-Q.; Chen, C.-E.; Liu, Y.-S.; Yang, B.; Xie, L.; Ying, G.-G. Occurrence, mass loads and risks of bisphenol analogues in the Pearl River Delta region, South China: Urban rainfall runoff as a potential source for receiving rivers. Environ. Pollut. 2020, 263, 114361. [Google Scholar] [CrossRef]
- Zheng, C.; Liu, J.; Ren, J.; Shen, J.; Fan, J.; Xi, R.; Chen, W.; Chen, Q. Occurrence, distribution and ecological risk of bisphenol analogues in the surface water from a water diversion project in Nanjing, China. Int. J. Environ. Res. Public Health 2019, 16, 3296. [Google Scholar] [CrossRef]
- Long, F.; Ren, Y.; Bi, F.; Wu, Z.; Zhang, H.; Li, J.; Gao, R.; Liu, Z.; Li, H. Contamination Characterization, Toxicological Properties, and Health Risk Assessment of Bisphenols in Multiple Media: Current Research Status and Future Perspectives. Toxics 2025, 13, 109. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, M.H.; Woodward, M.; Bao, W.; Liu, B.; Trasande, L. Urinary bisphenols and obesity prevalence among US children and adolescents. J. Endocr. Soc. 2019, 3, 1715–1726. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Gao, R.; Liang, W.; Wei, S.; Zhou, Y.; Zeng, F. Assessment of BPA and BPS exposure in the general population in Guangzhou, China-Estimation of daily intakes based on urinary metabolites. Environ. Pollut. 2022, 315, 120375. [Google Scholar] [CrossRef]
- Ndaw, S.; Remy, A.; Denis, F.; Marsan, P.; Jargot, D.; Robert, A. Occupational exposure of cashiers to bisphenol S via thermal paper. Toxicol. Lett. 2018, 298, 106–111. [Google Scholar] [CrossRef]
- Kojima, H.; Takeuchi, S.; Sanoh, S.; Okuda, K.; Kitamura, S.; Uramaru, N.; Sugihara, K.; Yoshinari, K. Profiling of bisphenol A and eight of its analogues on transcriptional activity via human nuclear receptors. Toxicology 2019, 413, 48–55. [Google Scholar] [CrossRef]
- Řimnáčová, H.; Štiavnická, M.; Moravec, J.; Chemek, M.; Kolinko, Y.; García-Álvarez, O.; Mouton, P.R.; Trejo, A.M.C.; Fenclová, T.; Eretová, N. Low doses of Bisphenol S affect post-translational modifications of sperm proteins in male mice. Reprod. Biol. Endocrinol. 2020, 18, 56. [Google Scholar] [CrossRef]
- Malaisé, Y.; Lencina, C.; Cartier, C.; Olier, M.; Ménard, S.; Guzylack-Piriou, L. Perinatal oral exposure to low doses of bisphenol A, S or F impairs immune functions at intestinal and systemic levels in female offspring mice. Environ. Health 2020, 19, 93. [Google Scholar] [CrossRef]
- Ferguson, M.; Lorenzen-Schmidt, I.; Pyle, W.G. Bisphenol S rapidly depresses heart function through estrogen receptor-β and decreases phospholamban phosphorylation in a sex-dependent manner. Sci. Rep. 2019, 9, 15948. [Google Scholar] [CrossRef]
- Zhang, R.; Guo, J.; Wang, Y.; Sun, R.; Dong, G.; Wang, X.; Du, G. Prenatal bisphenol S exposure induces hepatic lipid deposition in male mice offspring through downregulation of adipose-derived exosomal miR-29a-3p. J. Hazard. Mater. 2023, 453, 131410. [Google Scholar] [CrossRef]
- Charisiadis, P.; Andrianou, X.D.; Van Der Meer, T.P.; Den Dunnen, W.F.; Swaab, D.F.; Wolffenbuttel, B.H.; Makris, K.C.; van Vliet-Ostaptchouk, J.V. Possible obesogenic effects of bisphenols accumulation in the human brain. Sci. Rep. 2018, 8, 8186. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, J.; Xu, S.; Zhou, Y.; Zhao, H.; Li, Y.; Xiong, C.; Sun, X.; Liu, H.; Liu, W. Prenatal exposure to bisphenol A and its alternatives and child neurodevelopment at 2 years. J. Hazard. Mater. 2020, 388, 121774. [Google Scholar] [CrossRef]
- Rebolledo-Solleiro, D.; Flores, L.Y.C.; Solleiro-Villavicencio, H. Impact of BPA on behavior, neurodevelopment and neurodegeneration. Front. Biosci.-Landmark 2020, 26, 363–400. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.; Kumara, V.R. Comparative neurodevelopment effects of bisphenol A and bisphenol F on rat fetal neural stem cell models. Cells 2021, 10, 793. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; He, J.; Xu, T.; Han, H.; Zhu, Z.; Meng, L.; Pang, Q.; Fan, R. Bisphenol A (BPA), BPS and BPB-induced oxidative stress and apoptosis mediated by mitochondria in human neuroblastoma cell lines. Ecotoxicol. Environ. Saf. 2021, 207, 111299. [Google Scholar] [CrossRef]
- Kim, S.S.; Kim, J.L.; Hwang, K.-S.; Park, H.-C.; Bae, M.A.; Kim, K.-T.; Cho, S.-H. Mechanism of action and neurotoxic effects of chronic exposure to bisphenol F in adult zebrafish. Sci. Total Environ. 2022, 851, 158258. [Google Scholar] [CrossRef] [PubMed]
- Gyimah, E.; Xu, H.; Dong, X.; Qiu, X.; Zhang, Z.; Bu, Y.; Akoto, O. Developmental neurotoxicity of low concentrations of bisphenol A and S exposure in zebrafish. Chemosphere 2021, 262, 128045. [Google Scholar] [CrossRef]
- Kinch, C.D.; Ibhazehiebo, K.; Jeong, J.-H.; Habibi, H.R.; Kurrasch, D.M. Low-dose exposure to bisphenol A and replacement bisphenol S induces precocious hypothalamic neurogenesis in embryonic zebrafish. Proc. Natl. Acad. Sci. USA 2015, 112, 1475–1480. [Google Scholar] [CrossRef]
- Sanchez, C.L.; Souders II, C.L.; Pena-Delgado, C.J.; Nguyen, K.T.; Kroyter, N.; El Ahmadie, N.; Aristizabal-Henao, J.J.; Bowden, J.A.; Martyniuk, C.J. Neurotoxicity assessment of triazole fungicides on mitochondrial oxidative respiration and lipids in differentiated human SH-SY5Y neuroblastoma cells. Neurotoxicology 2020, 80, 76–86. [Google Scholar] [CrossRef]
- Xie, H.-r.; Hu, L.-s.; Li, G.-y. SH-SY5Y human neuroblastoma cell line: In vitro cell model of dopaminergic neurons in Parkinson’s disease. Chin. Med. J. 2010, 123, 1086–1092. [Google Scholar] [PubMed]
- Kovalevich, J.; Langford, D. Considerations for the use of SH-SY5Y neuroblastoma cells in neurobiology. In Neuronal Cell Culture: Methods and Protocols; Humana Totowa: Totowa, NJ, USA, 2013; pp. 9–21. [Google Scholar]
- Korecka, J.A.; van Kesteren, R.E.; Blaas, E.; Spitzer, S.O.; Kamstra, J.H.; Smit, A.B.; Swaab, D.F.; Verhaagen, J.; Bossers, K. Phenotypic characterization of retinoic acid differentiated SH-SY5Y cells by transcriptional profiling. PLoS ONE 2013, 8, e63862. [Google Scholar] [CrossRef]
- Souders, C.L., II; Sanchez, C.L.; Malphurs, W.; Aristizabal-Henao, J.J.; Bowden, J.A.; Martyniuk, C.J. Metabolic profiling in human SH-SY5Y neuronal cells exposed to perfluorooctanoic acid (PFOA). Neurotoxicology 2021, 85, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Breton, T.S.; Murray, C.A.; Huff, S.R.; Phaneuf, A.M.; Tripp, B.M.; Patuel, S.J.; Martyniuk, C.J.; DiMaggio, M.A. Phoenixin-14 alters transcriptome and steroid profiles in female green-spotted puffer (Dichotomyctere nigroviridis). Sci. Rep. 2022, 12, 9454. [Google Scholar] [CrossRef]
- Patuel, S.J.; English, C.; Lopez-Scarim, V.; Konig, I.; Souders, C.L., II; Ivantsova, E.; Martyniuk, C.J. The novel insecticide broflanilide dysregulates transcriptional networks associated with ion channels and induces hyperactivity in zebrafish (Danio rerio) larvae. Sci. Total Environ. 2023, 904, 167072. [Google Scholar] [CrossRef] [PubMed]
- Souders, C.L., II; Rushin, A.; Sanchez, C.L.; Toth, D.; Adamovsky, O.; Martyniuk, C.J. Mitochondrial and transcriptome responses in rat dopaminergic neuronal cells following exposure to the insecticide fipronil. Neurotoxicology 2021, 85, 173–185. [Google Scholar] [CrossRef]
- Cicinnati, V.R.; Shen, Q.; Sotiropoulos, G.C.; Radtke, A.; Gerken, G.; Beckebaum, S. Validation of putative reference genes for gene expression studies in human hepatocellular carcinoma using real-time quantitative RT-PCR. BMC Cancer 2008, 8, 350. [Google Scholar] [CrossRef]
- Liang, X.; Yin, N.; Liang, S.; Yang, R.; Liu, S.; Lu, Y.; Jiang, L.; Zhou, Q.; Jiang, G.; Faiola, F. Bisphenol A and several derivatives exert neural toxicity in human neuron-like cells by decreasing neurite length. Food Chem. Toxicol. 2020, 135, 111015. [Google Scholar] [CrossRef]
- Pang, Q.; Li, Y.; Meng, L.; Li, G.; Luo, Z.; Fan, R. Neurotoxicity of BPA, BPS, and BPB for the hippocampal cell line (HT-22): An implication for the replacement of BPA in plastics. Chemosphere 2019, 226, 545–552. [Google Scholar] [CrossRef]
- Meng, L.; Liu, J.; Wang, C.; Ouyang, Z.; Kuang, J.; Pang, Q.; Fan, R. Sex-specific oxidative damage effects induced by BPA and its analogs on primary hippocampal neurons attenuated by EGCG. Chemosphere 2021, 264, 128450. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Liu, S.; Fu, L.; Jiang, X.; Yang, M. Bisphenol A and its analogues bisphenol S, bisphenol F and bisphenol AF induce oxidative stress and biomacromolecular damage in human granulosa KGN cells. Chemosphere 2020, 253, 126707. [Google Scholar] [CrossRef]
- Heusinkveld, H.J.; Westerink, R.H. Comparison of different in vitro cell models for the assessment of pesticide-induced dopaminergic neurotoxicity. Toxicol. Vitr. 2017, 45, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Miao, S.; Gao, Z.; Kou, Z.; Xu, G.; Su, C.; Liu, N. Influence of bisphenol a on developing rat estrogen receptors and some cytokines in rats: A two-generational study. J. Toxicol. Environ. Health Part A 2008, 71, 1000–1008. [Google Scholar] [CrossRef]
- Zhu, J.; Jiang, L.; Liu, Y.; Qian, W.; Liu, J.; Zhou, J.; Gao, R.; Xiao, H.; Wang, J. MAPK and NF-κB pathways are involved in bisphenol A-induced TNF-α and IL-6 production in BV2 microglial cells. Inflammation 2015, 38, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Sukjamnong, S.; Thongkorn, S.; Kanlayaprasit, S.; Saeliw, T.; Hussem, K.; Warayanon, W.; Hu, V.W.; Tencomnao, T.; Sarachana, T. Prenatal exposure to bisphenol A alters the transcriptome-interactome profiles of genes associated with Alzheimer’s disease in the offspring hippocampus. Sci. Rep. 2020, 10, 9487. [Google Scholar] [CrossRef]
- Takahashi, M.; Komada, M.; Miyazawa, K.; Goto, S.; Ikeda, Y. Bisphenol A exposure induces increased microglia and microglial related factors in the murine embryonic dorsal telencephalon and hypothalamus. Toxicol. Lett. 2018, 284, 113–119. [Google Scholar] [CrossRef]
- Akintunde, J.; Akintola, T.; Adenuga, G.; Odugbemi, Z.; Adetoye, R.; Akintunde, O. Naringin attenuates Bisphenol-A mediated neurotoxicity in hypertensive rats by abrogation of cerebral nucleotide depletion, oxidative damage and neuroinflammation. Neurotoxicology 2020, 81, 18–33. [Google Scholar] [CrossRef]
- García-Osta, A.; Cuadrado-Tejedor, M.; García-Barroso, C.; Oyarzabal, J.; Franco, R. Phosphodiesterases as therapeutic targets for Alzheimer’s disease. ACS Chem. Neurosci. 2012, 3, 832–844. [Google Scholar] [CrossRef]
- Luo, G.; Wang, S.; Li, Z.; Wei, R.; Zhang, L.; Liu, H.; Wang, C.; Niu, R.; Wang, J. Maternal bisphenol a diet induces anxiety-like behavior in female juvenile with neuroimmune activation. Toxicol. Sci. 2014, 140, 364–373. [Google Scholar] [CrossRef]
- Salehi, A.; Loganathan, N.; Belsham, D.D. Bisphenol A induces Pomc gene expression through neuroinflammatory and PPARγ nuclear receptor-mediated mechanisms in POMC-expressing hypothalamic neuronal models. Mol. Cell. Endocrinol. 2019, 479, 12–19. [Google Scholar] [CrossRef] [PubMed]



| Gene Name | Full Name | log2FC | p-Adj | p-Value |
|---|---|---|---|---|
| RPS2P55 | Ribosomal Protein S2 Pseudogene 55 | −1.455 | 0.001 | 4.4149 × 10−8 |
| MT-TT | Mitochondrially Encoded TRNA-Thr (ACN) | 1.587 | 0.006 | 6.6467 × 10−7 |
| HLA-C | Major Histocompatibility Complex, Class I, C | −2.063 | 0.006 | 1.4419 × 10−6 |
| AC016596.2 | Ribosomal protein L41 | −1.929 | 0.006 | 1.7420 × 10−6 |
| RPL35P5 | Ribosomal Protein L35 Pseudogene 5 | −2.069 | 0.006 | 1.7752 × 10−6 |
| AC010343.1 | Ribosomal protein S8 pseudogene | −1.794 | 0.006 | 2.1002 × 10−6 |
| RPS10P3 | Ribosomal Protein S10 Pseudogene 3 | −2.368 | 0.007 | 2.7400 × 10−6 |
| Unknown | Unknown | −2.907 | 0.007 | 3.1888 × 10−6 |
| DUSP8 | Dual Specificity Phosphatase 8 | −1.488 | 0.007 | 3.4916 × 10−6 |
| AC011005.1 | Unknown | −2.424 | 0.007 | 3.9371 × 10−6 |
| Chemical | Name | # of Entities | Expanded # of Entities | # of Measured Entities | Median Change | Normalized Score | p-Value |
|---|---|---|---|---|---|---|---|
| BPS | Humoral Immunity in Vitiligo | 94 | 161 | 63 | −1.06 | 1.71 | 0.0017 |
| Natural Killer T-Cell Roles in Diabetes Mellitus Type 1 | 82 | 132 | 56 | 1.03 | 1.78 | 0.0018 | |
| T-Cell Maturation (Hypothesis) | 77 | 436 | 52 | 1.04 | 1.66 | 0.0018 | |
| IL2 Expression Targets | 97 | 138 | 61 | −1.05 | 1.77 | 0.0018 | |
| Atopic Dermatitis Onset | 72 | 348 | 51 | −1.06 | 1.69 | 0.0018 | |
| Treg-Cell Activation in Diabetes Mellitus | 72 | 121 | 57 | 1.03 | 1.67 | 0.0018 | |
| CD8 Activation of NF-kB Expression Targets | 44 | 63 | 30 | 1.05 | 1.73 | 0.0019 | |
| IL15R Activation of NF-kB/NFATC Signaling | 15 | 22 | 10 | −1.21 | 1.80 | 0.0019 | |
| RAS/RAF/MAPK Signaling Activation by Blocking of Tumor Suppressors | 47 | 427 | 27 | −1.06 | 1.79 | 0.0019 | |
| CD80 Activation of AP-1 Expression Targets | 28 | 32 | 14 | 1.07 | 1.80 | 0.0019 | |
| BPF | Dectin-2 (CLEC6A)/Mincle (CLEC4E)/BDCA2 (CLEC4C) Signaling | 23 | 33 | 11 | −1.02 | 1.86 | 0.0019 |
| IgE Receptors Activation of Targets in Lymphoid System and Blood | 25 | 70 | 6 | −1.38 | 1.73 | 0.0020 | |
| GPCRs Family Activation of Expression Targets in Bone | 15 | 17 | 6 | 1.33 | −1.64 | 0.0021 | |
| S/G2 Phase Transition | 49 | 247 | 132 | −1.13 | 1.51 | 0.0032 | |
| TNF Activation of NF-kB Expression Targets | 127 | 136 | 62 | −1.05 | 1.72 | 0.0033 | |
| TLR9 Expression Targets | 42 | 47 | 24 | 1.03 | 1.82 | 0.0035 | |
| CNR Activation of Expression Targets in Nerve Tissue | 35 | 89 | 14 | 1.01 | 1.73 | 0.0036 | |
| Acute Phase in Atopic Dermatitis | 53 | 82 | 22 | 1.04 | 1.69 | 0.0038 | |
| Muscular Dystrophy, Facioscapulohumeral | 32 | 343 | 10 | 1.21 | 1.62 | 0.0039 | |
| Aminoglycosides and Cisplatin-Induced Ototoxicity (Mouse Model) | 25 | 237 | 9 | −1.13 | 1.70 | 0.0040 |
| Overlapping Pathways | # of Measured Entities BPF | Median Change BPF | p-Value PBF | # of Measured Entities BPS | Median Change BPS | p-Value PBS |
|---|---|---|---|---|---|---|
| CD4+ T-Cell Function Decline in HIV | 56 | −1.006 | 0.005 | 57 | 1.006 | 0.004 |
| Natural Killer T-Cell Roles in Diabetes Mellitus Type 1 | 57 | −1.008 | 0.047 | 56 | 1.030 | 0.002 |
| Eosinophil Activation | 58 | −1.047 | 0.007 | 59 | −1.111 | 0.026 |
| IL2 Expression Targets | 61 | −1.058 | 0.014 | 61 | −1.047 | 0.002 |
| TNF Activation of NF-kB Expression Targets | 62 | −1.048 | 0.003 | 63 | −1.043 | 0.005 |
| CD8+ T-Cell Response in Celiac Disease | 65 | −1.076 | 0.016 | 64 | −1.096 | 0.004 |
| Natural Killer Cell Activation | 65 | −1.076 | 0.032 | 64 | −1.079 | 0.012 |
| Protein Folding | 69 | −1.224 | 0.021 | 71 | −1.264 | 0.035 |
| IGF1 Activation of ELK/SRF/HIF1A/MYC/SREBF Expression Targets | 74 | −1.107 | 0.037 | 74 | −1.101 | 0.036 |
| Insulin Activation of MEF/MYOD Expression Targets | 77 | −1.056 | 0.029 | 79 | −1.125 | 0.046 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guzman, A.P.; Sanchez, C.L.; Ivantsova, E.; Watkins, J.; Sutton, S.E.; Souders, C.L., II; Martyniuk, C.J. Exposure to Bisphenol S and Bisphenol F Alters Gene Networks Related to Protein Translation and Neuroinflammation in SH-SY5Y Human Neuroblastoma Cells. Toxics 2025, 13, 772. https://doi.org/10.3390/toxics13090772
Guzman AP, Sanchez CL, Ivantsova E, Watkins J, Sutton SE, Souders CL II, Martyniuk CJ. Exposure to Bisphenol S and Bisphenol F Alters Gene Networks Related to Protein Translation and Neuroinflammation in SH-SY5Y Human Neuroblastoma Cells. Toxics. 2025; 13(9):772. https://doi.org/10.3390/toxics13090772
Chicago/Turabian StyleGuzman, Andrea P., Christina L. Sanchez, Emma Ivantsova, Jacqueline Watkins, Sara E. Sutton, Christopher L. Souders, II, and Christopher J. Martyniuk. 2025. "Exposure to Bisphenol S and Bisphenol F Alters Gene Networks Related to Protein Translation and Neuroinflammation in SH-SY5Y Human Neuroblastoma Cells" Toxics 13, no. 9: 772. https://doi.org/10.3390/toxics13090772
APA StyleGuzman, A. P., Sanchez, C. L., Ivantsova, E., Watkins, J., Sutton, S. E., Souders, C. L., II, & Martyniuk, C. J. (2025). Exposure to Bisphenol S and Bisphenol F Alters Gene Networks Related to Protein Translation and Neuroinflammation in SH-SY5Y Human Neuroblastoma Cells. Toxics, 13(9), 772. https://doi.org/10.3390/toxics13090772








