A Proof-of-Concept for a Hypolipidemic Brown Trout Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
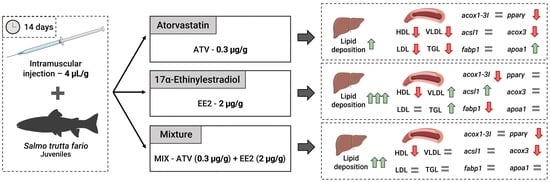
2.2. Exposure Procedures
2.3. Sampling
2.4. Blood Lipid Biochemistry
2.5. Liver Lipid Quantification
2.6. Liver RNA Expression
2.7. Statistical Analysis
3. Results
3.1. Mortality and Fish Biometry
3.2. Blood Lipid Biochemistry
3.3. Liver Lipid Quantification
3.4. Liver RNA Expression
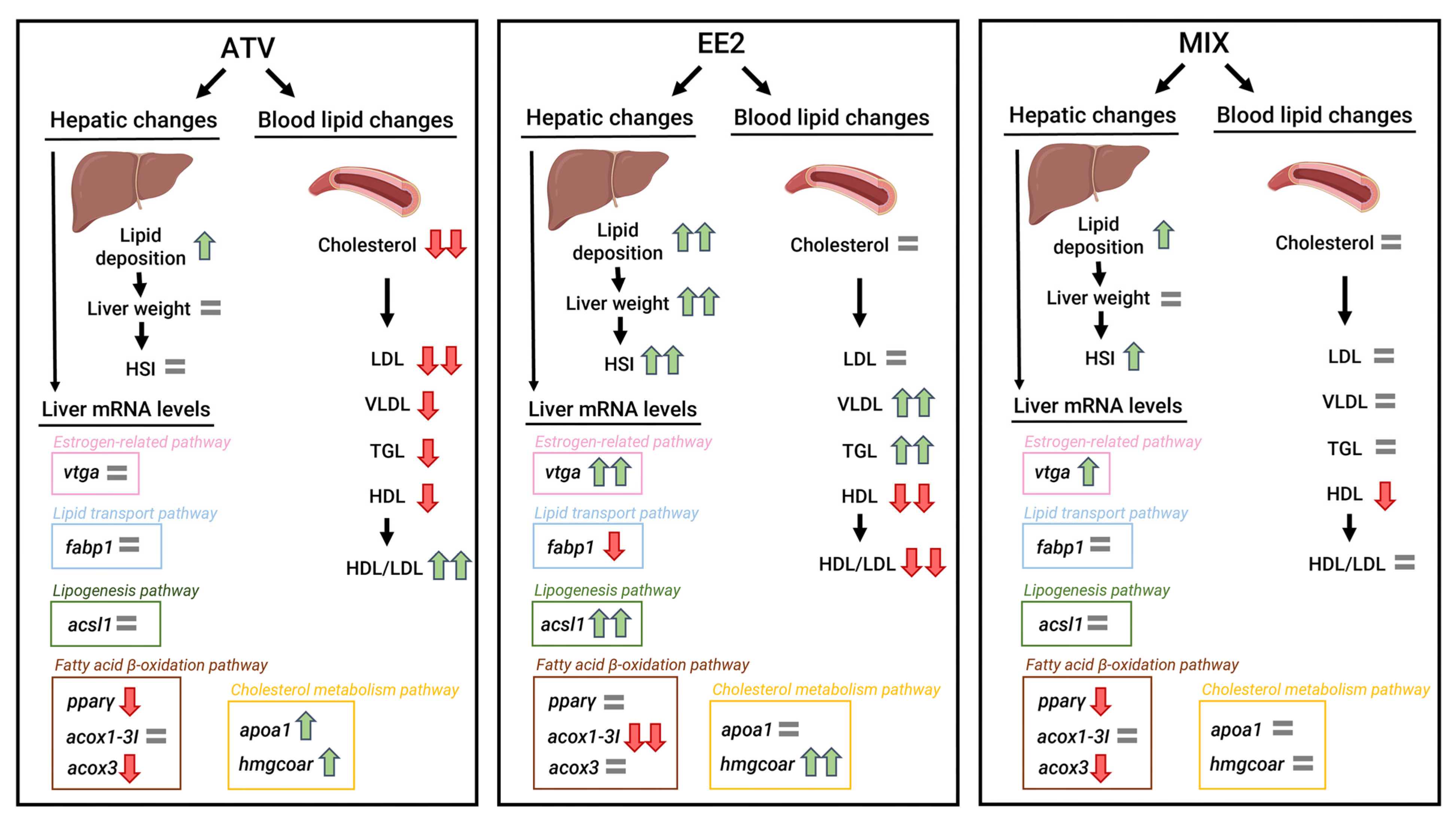
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tocher, D.R. Metabolism and functions of lipids and fatty acids in teleost fish. Rev. Fish. Sci. 2003, 11, 107–184. [Google Scholar] [CrossRef]
- Reading, B.J.; Andersen, L.K.; Ryu, Y.-W.; Mushirobira, Y.; Todo, T.; Hiramatsu, N. Oogenesis and egg quality in finfish: Yolk formation and other factors influencing female fertility. Fishes 2018, 3, 45. [Google Scholar] [CrossRef]
- Sheridan, M.A. Regulation of lipid metabolism in poikilothermic vertebrates. Comp. Biochem. Physiol. Part B Comp. Biochem. 1994, 107, 495–508. [Google Scholar] [CrossRef]
- Wallace, M.; Metallo, C.M. Tracing insights into de novo lipogenesis in liver and adipose tissues. Semin. Cell Dev. Biol. 2020, 108, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Olivares-Rubio, H.F.; Vega-López, A. Fatty acid metabolism in fish species as a biomarker for environmental monitoring. Environ. Pollut. 2016, 218, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Ayisi, C.L.; Yamei, C.; Zhao, J.-L. Genes, transcription factors and enzymes involved in lipid metabolism in fin fish. Agri Gene 2018, 7, 7–14. [Google Scholar] [CrossRef]
- Maradonna, F.; Carnevali, O. Lipid metabolism alteration by endocrine disruptors in animal models: An overview. Front. Endocrinol. 2018, 9, 654. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Xu, W.; Mai, K.; Zhou, H.; Zhang, Y.; Liufu, Z.; Zhang, K.; Ai, Q. Growth performance, lipid deposition and hepatic lipid metabolism related gene expression in juvenile turbot (Scophthalmus maximus L.) fed diets with various fish oil substitution levels by soybean oil. Aquaculture 2014, 433, 442–449. [Google Scholar] [CrossRef]
- Varga, T.; Czimmerer, Z.; Nagy, L. PPARs are a unique set of fatty acid regulated transcription factors controlling both lipid metabolism and inflammation. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2011, 1812, 1007–1022. [Google Scholar] [CrossRef]
- Karagianni, P.; Talianidis, I. Transcription factor networks regulating hepatic fatty acid metabolism. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2015, 1851, 2–8. [Google Scholar] [CrossRef]
- Jamalzadeh, H.; Hajirezaee, S.; Nazeri, S.; Khara, H.; Mirrasuli, E. Identification of follicular vitellogenesis stage by monitoring of plasma calcium and estradiol-17β concentrations in the cultured Caspian brown trout, Salmo trutta caspius Kessler, 1877. Biologia 2012, 67, 796–799. [Google Scholar] [CrossRef]
- Cleveland, B.; Weber, G. Effects of steroid treatment on growth, nutrient partitioning, and expression of genes related to growth and nutrient metabolism in adult triploid rainbow trout (Oncorhynchus mykiss). Domest. Anim. Endocrinol. 2016, 56, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Madureira, T.V.; Malhão, F.; Simões, T.; Pinheiro, I.; Lopes, C.; Gonçalves, J.F.; Urbatzka, R.; Castro, L.F.; Lemos, M.F.; Rocha, E. Sex-steroids and hypolipidemic chemicals impacts on brown trout lipid and peroxisome signaling—Molecular, biochemical and morphological insights. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2018, 212, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Kopin, L.; Lowenstein, C.J. Dyslipidemia. Ann. Intern. Med. 2017, 167, ITC81–ITC96. [Google Scholar] [CrossRef] [PubMed]
- Vancheri, F.; Backlund, L.; Strender, L.-E.; Godman, B.; Wettermark, B. Time trends in statin utilization and coronary mortality in Western European countries. BMJ Open 2016, 6, e010500. [Google Scholar] [CrossRef] [PubMed]
- Pirillo, A.; Casula, M.; Olmastroni, E.; Norata, G.D.; Catapano, A.L. Global epidemiology of dyslipidaemias. Nat. Rev. Cardiol. 2021, 18, 689–700. [Google Scholar] [CrossRef]
- Sulaiman, S.; Khamis, M.; Nir, S.; Lelario, F.; Scrano, L.; Bufo, S.A.; Mecca, G.; Karaman, R. Stability and removal of atorvastatin, rosuvastatin and simvastatin from wastewater. Environ. Technol. 2015, 36, 3232–3242. [Google Scholar] [CrossRef]
- Shattat, G.F. A review article on hyperlipidemia: Types, treatments and new drug targets. Biomed. Pharmacol. J. 2015, 7, 399–409. [Google Scholar] [CrossRef]
- Fent, K.; Weston, A.A.; Caminada, D. Ecotoxicology of human pharmaceuticals. Aquat. Toxicol. 2006, 76, 122–159. [Google Scholar] [CrossRef]
- Burg, J.S.; Espenshade, P.J. Regulation of HMG-CoA reductase in mammals and yeast. Prog. Lipid Res. 2011, 50, 403–410. [Google Scholar] [CrossRef]
- Al-Habsi, A.A.; Massarsky, A.; Moon, T.W. Exposure to gemfibrozil and atorvastatin affects cholesterol metabolism and steroid production in zebrafish (Danio rerio). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2016, 199, 87–96. [Google Scholar] [CrossRef]
- Barros, S.; Montes, R.; Quintana, J.B.; Rodil, R.; André, A.; Capitão, A.; Soares, J.; Santos, M.M.; Neuparth, T. Chronic environmentally relevant levels of simvastatin disrupt embryonic development, biochemical and molecular responses in zebrafish (Danio rerio). Aquat. Toxicol. 2018, 201, 47–57. [Google Scholar] [CrossRef]
- Lourenço, T.; Madureira, T.V.; Rocha, M.J.; Rocha, E. Fish as models to study liver and blood lipid-related effects of fibrates and statins and screen new hypolipidemic drugs. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2023, 274, 109756. [Google Scholar] [CrossRef]
- Mezzelani, M.; Regoli, F. The biological effects of pharmaceuticals in the marine environment. Annu. Rev. Mar. Sci. 2022, 14, 105–128. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Han, Z.; Zhong, H.; Yin, Q.; Xiao, J.; Wang, F.; Zhou, Y.; Luo, Y. Regulation of triglyceride synthesis by estradiol in the livers of hybrid tilapia (Oreochromis niloticus♀ × O. aureus♂). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2019, 238, 110335. [Google Scholar] [CrossRef]
- Madureira, T.V.; Costa, J.L.; Malhão, F.; Lopes, C.; Gonçalves, J.F.; Rocha, E. Design of a multi-parametric profile for assessing the acclimation period of juvenile brown trout after an acute transport to new housing environment. Appl. Anim. Behav. Sci. 2019, 219, 104835. [Google Scholar] [CrossRef]
- Cleveland, B.M.; Manor, M.L. Effects of phytoestrogens on growth-related and lipogenic genes in rainbow trout (Oncorhynchus mykiss). Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2015, 170, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Prindiville, J.S.; Mennigen, J.A.; Zamora, J.M.; Moon, T.W.; Weber, J.-M. The fibrate drug gemfibrozil disrupts lipoprotein metabolism in rainbow trout. Toxicol. Appl. Pharmacol. 2011, 251, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Mimeault, C.; Woodhouse, A.J.; Miao, X.-S.; Metcalfe, C.D.; Moon, T.W.; Trudeau, V.L. The human lipid regulator, gemfibrozil bioconcentrates and reduces testosterone in the goldfish, Carassius auratus. Aquat. Toxicol. 2005, 73, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Real, M.V.; Rocha, M.J.; Gonçalves, J.F.; Rocha, E. Histology and design-based estimation of hepatocellularity and volumes of hepatocytes in control and ethynylestradiol exposed males of platyfish (Xiphophorus maculatus). Tissue Cell 2020, 63, 101327. [Google Scholar] [CrossRef]
- Madsen, S.S.; Skovbølling, S.; Nielsen, C.; Korsgaard, B. 17-β Estradiol and 4-nonylphenol delay smolt development and downstream migration in Atlantic salmon, Salmo salar. Aquat. Toxicol. 2004, 68, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Zahl, I.H.; Samuelsen, O.; Kiessling, A. Anaesthesia of farmed fish: Implications for welfare. Fish Physiol. Biochem. 2012, 38, 201–218. [Google Scholar] [CrossRef] [PubMed]
- Sharpe, R.L.; MacLatchy, D.L. Lipid dynamics in goldfish (Carassius auratus) during a period of gonadal recrudescence: Effects of β-sitosterol and 17β-estradiol exposure. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2007, 145, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Gates, L.; Adler, R.R.; Elangbam, C.S. Osmium tetroxide post-fixation and periodic acid-Schiff dual-staining technique to demonstrate intracellular lipid and glycogen in the mouse liver section—A novel method for co-visualization of intracellular contents in paraffin-embedded tissue. J. Histotechnol. 2016, 39, 2–7. [Google Scholar] [CrossRef]
- Marcos, R.; Monteiro, R.A.; Rocha, E. The use of design-based stereology to evaluate volumes and numbers in the liver: A review with practical guidelines. J. Anat. 2012, 220, 303–317. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Urbatzka, R.; Galante-Oliveira, S.; Rocha, E.; Castro, L.F.; Cunha, I. Normalization strategies for gene expression studies by real-time PCR in a marine fish species, Scophthalmus maximus. Mar. Genom. 2013, 10, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Caballero-Solares, A.; Xue, X.; Parrish, C.C.; Foroutani, M.B.; Taylor, R.G.; Rise, M.L. Changes in the liver transcriptome of farmed Atlantic salmon (Salmo salar) fed experimental diets based on terrestrial alternatives to fish meal and fish oil. BMC Genom. 2018, 19, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Madureira, T.V.; Castro, L.F.; Rocha, E. Acyl-coenzyme A oxidases 1 and 3 in brown trout (Salmo trutta f. fario): Can peroxisomal fatty acid β-oxidation be regulated by estrogen signaling? Fish Physiol. Biochem. 2016, 42, 389–401. [Google Scholar] [CrossRef]
- Madureira, T.V.; Pinheiro, I.; Freire, R.d.P.; Rocha, E.; Castro, L.F.; Urbatzka, R. Genome specific PPARαB duplicates in salmonids and insights into estrogenic regulation in brown trout. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2017, 208–209, 94–101. [Google Scholar] [CrossRef]
- Körner, O.; Kohno, S.; Schönenberger, R.; Suter, M.J.-F.; Knauer, K.; Guillette, L.J.; Burkhardt-Holm, P. Water temperature and concomitant waterborne ethinylestradiol exposure affects the vitellogenin expression in juvenile brown trout (Salmo trutta). Aquat. Toxicol. 2008, 90, 188–196. [Google Scholar] [CrossRef]
- Minghetti, M.; Leaver, M.J.; Tocher, D.R. Transcriptional control mechanisms of genes of lipid and fatty acid metabolism in the Atlantic salmon (Salmo salar L.) established cell line, SHK-1. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2011, 1811, 194–202. [Google Scholar] [CrossRef]
- Todorčević, M.; Vegusdal, A.; Gjøen, T.; Sundvold, H.; Torstensen, B.E.; Kjær, M.A.; Ruyter, B. Changes in fatty acids metabolism during differentiation of Atlantic salmon preadipocytes; Effects of n-3 and n-9 fatty acids. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2008, 1781, 326–335. [Google Scholar] [CrossRef]
- Madureira, T.V.; Malhão, F.; Pinheiro, I.; Lopes, C.; Ferreira, N.; Urbatzka, R.; Castro, L.F.C.; Rocha, E. Estrogenic and anti-estrogenic influences in cultured brown trout hepatocytes: Focus on the expression of some estrogen and peroxisomal related genes and linked phenotypic anchors. Aquat. Toxicol. 2015, 169, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Lopes, C.; Malhão, F.; Guimarães, C.; Pinheiro, I.; Gonçalves, J.F.; Castro, L.F.; Rocha, E.; Madureira, T.V. Testosterone-induced modulation of peroxisomal morphology and peroxisome-related gene expression in brown trout (Salmo trutta f. fario) primary hepatocytes. Aquat. Toxicol. 2017, 193, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Vang, S.-H.; Kortner, T.M.; Arukwe, A. Steroidogenic acute regulatory (StAR) protein and cholesterol side-chain cleavage (p450scc) as molecular and cellular targets for 17α-ethynylestradiol in salmon previtellogenic oocytes. Chem. Res. Toxicol. 2007, 20, 1811–1819. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Ø.; Harper, D.A. Past: Paleontological statistics software package for educaton and data anlysis. Palaeontol. Electron. 2001, 4, 1. [Google Scholar]
- Zhou, J.; Xu, Y.-Q.; Guo, S.-Y.; Li, C.-Q. Rapid analysis of hypolipidemic drugs in a live zebrafish assay. J. Pharmacol. Toxicol. Methods 2015, 72, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Kenari, A.A.; Mozanzadeh, M.T.; Pourgholam, R. Effects of total fish oil replacement to vegetable oils at two dietary lipid levels on the growth, body composition, haemato-immunological and serum biochemical parameters in caspian brown trout (Salmo trutta caspius Kessler, 1877). Aquac. Res. 2011, 42, 1131–1144. [Google Scholar] [CrossRef]
- Guo, X.; Liang, X.-F.; Fang, L.; Yuan, X.; Zhou, Y.; He, S.; Shen, D. Effects of lipid-lowering pharmaceutical clofibrate on lipid and lipoprotein metabolism of grass carp (Ctenopharyngodon idellal Val.) fed with the high non-protein energy diets. Fish Physiol. Biochem. 2015, 41, 331–343. [Google Scholar] [CrossRef] [PubMed]
- Poli, A. Atorvastatin: Pharmacological characteristics and lipid-lowering effects. Drugs 2007, 67, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Rudling, M.; Angelin, B.; Ståhle, L.; Reihnér, E.; Sahlin, S.; Olivecrona, H.; Björkhem, I.; Einarsson, C. Regulation of hepatic low-density lipoprotein receptor, 3-hydroxy-3-methylglutaryl coenzyme A reductase, and cholesterol 7α-hydroxylase mRNAs in human liver. J. Clin. Endocrinol. Metab. 2002, 87, 4307–4313. [Google Scholar] [CrossRef] [PubMed]
- Al-Habsi, A.A.; Massarsky, A.; Moon, T.W. Atorvastatin alters gene expression and cholesterol synthesis in primary rainbow trout (Oncorhynchus mykiss) hepatocytes. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2018, 224, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Wallaert, C.; Babin, P.J. Effects of 17β-estradiol and starvation on trout plasma lipoproteins. Lipids 1992, 27, 1032–1041. [Google Scholar] [CrossRef] [PubMed]
- Robertson, J.C.; Bradley, T.M. Liver ultrastructure of juvenile Atlantic salmon (Salmo salar). J. Morphol. 1992, 211, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Tocher, D.R.; Glencross, B.D. Lipids and fatty acids. In Dietary Nutrients, Additives, and Fish Health; Lee, C.-S., Lim, C., Gatlin, D.M., III, Webster, C., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2015; pp. 47–94. [Google Scholar]
- Roglans, N.; Verd, J.C.; Peris, C.; Alegret, M.; Vázquez, M.; Adzet, T.; Diaz, C.; Hernández, G.; Laguna, J.C.; Sánchez, R.M. High doses of atorvastatin and simvastatin induce key enzymes involved in VLDL production. Lipids 2002, 37, 445–454. [Google Scholar] [CrossRef]
- Zhang, X.; Zhong, H.; Han, Z.; Tang, Z.; Xiao, J.; Guo, Z.; Wang, F.; Luo, Y.; Zhou, Y. Effects of waterborne exposure to 17β-estradiol on hepatic lipid metabolism genes in tilapia (Oreochromis niloticus). Aquac. Rep. 2020, 17, 100382. [Google Scholar] [CrossRef]
- Li, L.O.; Klett, E.L.; Coleman, R.A. Acyl-CoA synthesis, lipid metabolism and lipotoxicity. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2010, 1801, 246–251. [Google Scholar] [CrossRef]
- Singh, A.B.; Kan, C.F.; Dong, B.; Liu, J. SREBP2 activation induces hepatic long-chain acyl-CoA synthetase 1 (ACSL1) expression in vivo and in vitro through a sterol regulatory element (SRE) motif of the ACSL1 C-promoter. J. Biol. Chem. 2016, 291, 5373–5384. [Google Scholar] [CrossRef]
- Teles, M.; Fierro-Castro, C.; Na-Phatthalung, P.; Tvarijonaviciute, A.; Soares, A.; Tort, L.; Oliveira, M. Evaluation of gemfibrozil effects on a marine fish (Sparus aurata) combining gene expression with conventional endocrine and biochemical endpoints. J. Hazard. Mater. 2016, 318, 600–607. [Google Scholar] [CrossRef]
- Leaver, M.J.; Bautista, J.M.; Björnsson, B.T.; Jönsson, E.; Krey, G.; Tocher, D.R.; Torstensen, B.E. Towards fish lipid nutrigenomics: Current state and prospects for fin-fish aquaculture. Rev. Fish. Sci. 2008, 16, 73–94. [Google Scholar] [CrossRef]
- Venkatachalam, A.B.; Lall, S.P.; Denovan-Wright, E.M.; Wright, J.M. Tissue-specific differential induction of duplicated fatty acid-binding protein genes by the peroxisome proliferator, clofibrate, in zebrafish (Danio rerio). BMC Evol. Biol. 2012, 12, 112. [Google Scholar] [CrossRef] [PubMed]
- Landrier, J.-F.; Thomas, C.; Grober, J.; Duez, H.; Percevault, F.; Souidi, M.; Linard, C.; Staels, B.; Besnard, P. Statin induction of liver fatty acid-binding protein (L-FABP) gene expression is peroxisome proliferator-activated receptor-α-dependent. J. Biol. Chem. 2004, 279, 45512–45518. [Google Scholar] [CrossRef]
- Estey, C.; Chen, X.; Moon, T.W. 3-Hydroxy-3-methylglutaryl coenzyme A reductase in rainbow trout: Effects of fasting and statin drugs on activities and mRNA transcripts. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2008, 147, 386–398. [Google Scholar] [CrossRef] [PubMed]
- Dominiczak, M.H.; Caslake, M.J. Apolipoproteins: Metabolic role and clinical biochemistry applications. Ann. Clin. Biochem. Int. J. Biochem. Lab. Med. 2011, 48, 498–515. [Google Scholar] [CrossRef]
- Rigotti, A.; Miettinen, H.E.; Krieger, M. The role of the high-density lipoprotein receptor sr-bi in the lipid metabolism of endocrine and other tissues. Endocr. Rev. 2003, 24, 357–387. [Google Scholar] [CrossRef]
- Maejima, T.; Yamazaki, H.; Aoki, T.; Tamaki, T.; Sato, F.; Kitahara, M.; Saito, Y. Effect of pitavastatin on apolipoprotein A-I production in HepG2 cell. Biochem. Biophys. Res. Commun. 2004, 324, 835–839. [Google Scholar] [CrossRef]
- Qin, S.; Koga, T.; Ganji, S.H.; Kamanna, V.S.; Kashyap, M.L. Rosuvastatin selectively stimulates apolipoprotein A-I but not apolipoprotein A-II synthesis in Hep G2 cells. Metabolism 2008, 57, 973–979. [Google Scholar] [CrossRef]
- Cunha, V.; Santos, M.; Moradas-Ferreira, P.; Castro, L.; Ferreira, M. Simvastatin modulates gene expression of key receptors in zebrafish embryos. J. Toxicol. Environ. Health Part A 2017, 80, 465–476. [Google Scholar] [CrossRef]
- Rakhshandehroo, M.; Knoch, B.; Müller, M.; Kersten, S. Peroxisome proliferator-activated receptor alpha target genes. PPAR Res. 2010, 2010, 612089. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, J.; Winter, M.J.; Lange, A.; Cumming, R.; Owen, S.F.; Tyler, C.R. Effects of the lipid regulating drug clofibric acid on PPARα-regulated gene transcript levels in common carp (Cyprinus carpio) at pharmacological and environmental exposure levels. Aquat. Toxicol. 2015, 161, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Coimbra, A.M.; Peixoto, M.J.; Coelho, I.; Lacerda, R.; Carvalho, A.P.; Gesto, M.; Lyssimachou, A.; Lima, D.; Soares, J.; André, A.; et al. Chronic effects of clofibric acid in zebrafish (Danio rerio): A multigenerational study. Aquat. Toxicol. 2015, 160, 76–86. [Google Scholar] [CrossRef]
- Misra, P.; Reddy, J.K. Peroxisome proliferator-activated receptor-α activation and excess energy burning in hepatocarcinogenesis. Biochimie 2014, 98, 63–74. [Google Scholar] [CrossRef]
- Ruyter, B.; Andersen, Ø.; Dehli, A.; Farrants, A.K.; Gjøen, T.; Thomassen, M.S. Peroxisome proliferator activated receptors in Atlantic salmon (Salmo salar): Effects on PPAR transcription and acyl-CoA oxidase activity in hepatocytes by peroxisome proliferators and fatty acids. Biochim. Biophys. Acta (BBA)-Lipids Lipid Metab. 1997, 1348, 331–338. [Google Scholar] [CrossRef]
- Patel, O.; Muller, C.J.F.; Joubert, E.; Rosenkranz, B.; Louw, J.; Awortwe, C. Therapeutic effects of an aspalathin-rich green rooibos extract, pioglitazone and atorvastatin combination therapy in diabetic db/db mice. PLoS ONE 2021, 16, e0251069. [Google Scholar] [CrossRef] [PubMed]

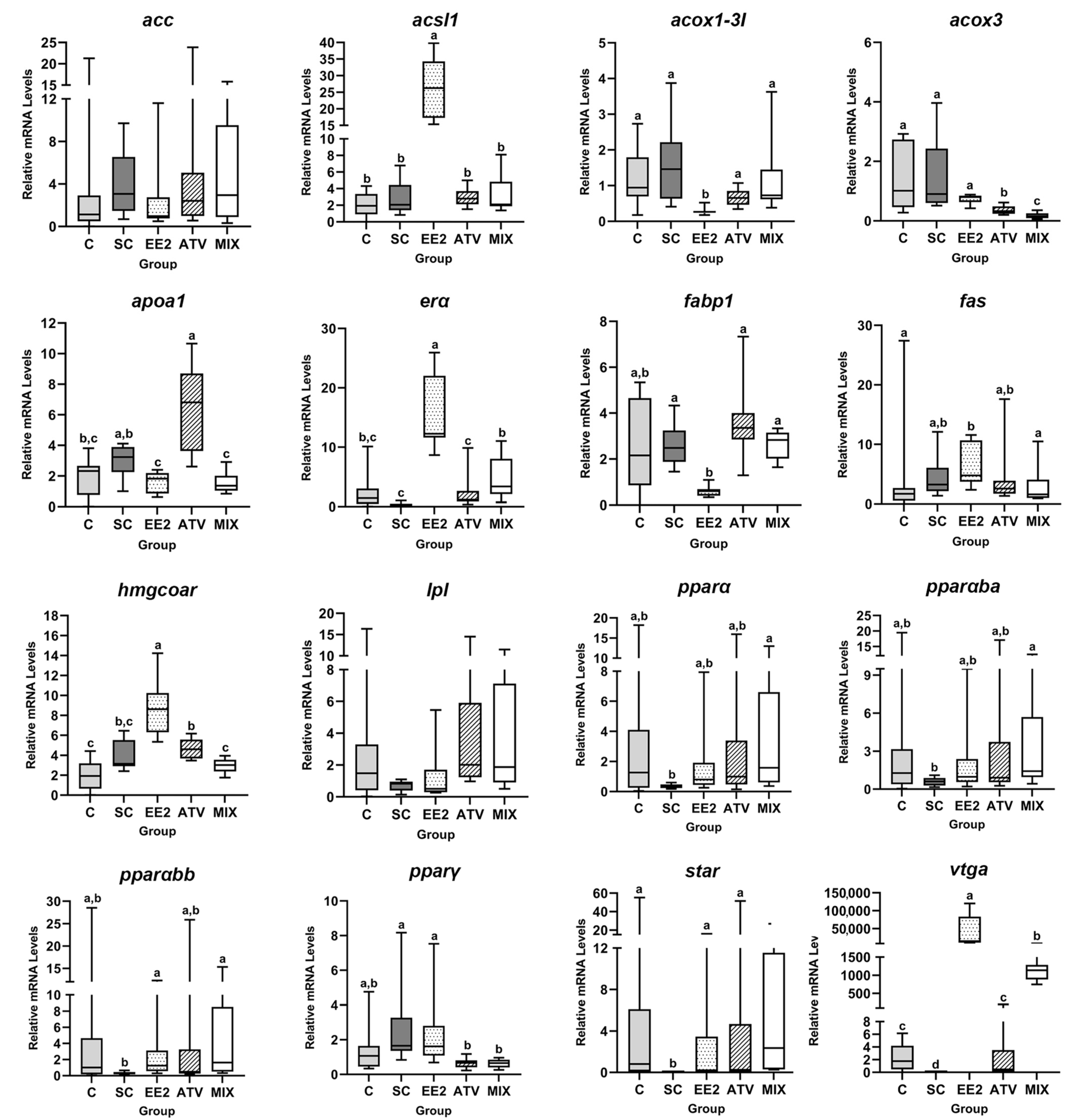
| Gene Name | Abbreviation | Primer Forward (5′-3′) | Primer Reverse (5′-3′) | Annealing Temperature (ᵒC) | Amplification Efficiency (%) | Reference |
|---|---|---|---|---|---|---|
| Acetyl-CoA carboxylase | acc | TTTTGATGGCGATCTTGACA | CATCACAATGCCTCGCTCTA | 60.0 | 102.2 # | [39] |
| Acyl-CoA long-chain synthetase 1 | acsl1 | CGACCAAGCCGCTATCTC | CCAACAGCCTCCACATCC | 55.0 | 97.8 # | [13] |
| Acyl-CoA oxidase 1 3I | acox1-3I | TGTAACAAGGAGCAGTTCG | TTGCCGTGGTTTCAAGCC | 56.0 | 96.9 * | [40] |
| Acyl-CoA oxidase 3 | acox3 | GGGAAGACGGCTACACACG | CAACAATTACTCCTGGCATCGC | 55.0 | 105.3 * | [40] |
| Apolipoprotein AI | apoa1 | ATGAAATTCCTGGCTCTTG | TACTCTTTGAACTCTGTGTC | 55.0 | 89.9 # | [41] |
| Estrogen receptor alpha | erα | GACATGCTCCTGGCCACTGT | TGGCTTTGAGGCACACAAAC | 61.6 | 91.2 # | [42] |
| Fatty acid binding protein 1 | fabp1 | GTCCGTCACCAACTCCTTC | GCGTCTCAACCATCTCTCC | 57.0 | 97.7 # | [41] |
| Fatty acid synthase | fas | ACCGCCAAGCTCAGTGTGC | CAGGCCCCAAAGGAGTAGC | 60.0 | 95.1 # | [43] |
| Glyceraldehyde-3-phosphate dehydrogenase | gapdh | CCACCTATGTAGTTGAGTC | ACCTTGAGGGAGTTATCG | 55.0 | 92.8 # or 100.6 * | [41] |
| Hydroxy-3-methylglutaryl Co-A reductase | hmgcoar | CCTTCAGCCATGAACTGGAT | TCCTGTCCACAGGCAATGTA | 58.0 | 94.2 # | [43] |
| Lipoprotein lipase | lpl | TGCTGGTAGCGGAGAAAGACAT | CTGACCACCAGGAAGACACCAT | 60.0 | 104.1 # | [44] |
| Peroxisome proliferator-activated receptor alpha | pparα | CGGGTGACAGGGAGGTGGAGGAC | GGTGAGGATGGTGCGGGCTTTGG | 59.0 | 100.6 # | [45] |
| Peroxisome proliferator-activated receptor alpha Ba | pparαba | ATCCACTACTCCCACAGG | GTCTAAACCCAGCCAAATAC | 55.0 | 106.7 # | [41] |
| Peroxisome proliferator-activated receptor alpha Bb | pparαbb | GAGTCTCCTGTCCTATCC | AGTTCTGCTGTTCTTCAC | 55.0 | 99.3 # | [41] |
| Peroxisome proliferator-activated receptor gamma | pparγ | CGGAATAAGTGCCAGTAC | GGGTCCACATCCATAAAC | 56.0 | 98.1 * | [46] |
| Ribosomal protein L8 | rpl8 | TCAGCTGAGCTTTCTTGCCAC | AGGACTGAGCTGTTCATTGCG | 59.0 | 93.8 # or 99.0 * | [42] |
| Steroidogenic acute regulatory protein | star | AGGATGGATGGACCACTGAG | GTCTCCCATCTGCTCCATGT | 63.0 | 104.5 # | [47] |
| Vitellogenin A | vtga | AACGGTGCTGAATGTCCATAG | ATTGAGATCCTTGCTCTTGGTC | 62.9 | 99.0 # | [42] |
| Group | Total Weight (g) | Total Length (cm) | Fulton’s Condition Factor (K) | Liver Weight (g) | Hepatosomatic Index (HSI) |
|---|---|---|---|---|---|
| C | 63.3 (41.3–83.3) | 18.0 (15.3–19.5) | 1.1 (1.0–1.3) | 0.91 (0.66–1.20) b | 1.44 (1.28–1.60) c |
| SC | 62.3 (40.7–91.3) | 17.4 (15.5–20.0) | 1.1 (0.9–1.3) | 0.93 (0.53–1.06) b | 1.37 (1.03–1.78) c |
| EE2 | 62.8 (48.0–114.2) | 18.3 (16.5–21.5) | 1.0 (0.9–1.3) | 2.13 (1.47–3.54) a | 3.30 (2.62–3.97) a |
| ATV | 52.8 (27.4–60.7) | 17.0 (14.0–18.0) | 1.0 (1.0–1.2) | 0.73 (0.34–0.99) b | 1.37 (1.03–1.83) c |
| MIX | 48.2 (36.8–74.7) | 16.5 (15.2–19.5) | 1.1 (0.9–1.1) | 0.87 (0.61–1.44) b | 1.84 (1.30–2.43) b |
| Blood Lipid Levels | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Group | Total Cholesterol (mg/dL) | HDL (mg/dL) | LDL (mg/dL) | VLDL (mg/dL) | Non-HDL (mg/dL) | TGLs (mg/dL) | Total Cholesterol/HDL | HDL/LDL | LDL/HDL | TGLs/HDL |
| C | 307.0 a (238.0–501.0) | 105.0 a (101.0–109.0) | 159.8 a (81.8–347.0) | 48.0 b,c (43.2–59.8) | 203.0 a (130.0–395.0) | 240.0 b,c (216.0–299.0) | 3.0 b (2.2–4.7) | 0.7 b (0.3–1.3) | 1.5 b (0.8–3.3) | 2.2 b (2.1–3.0) |
| SC | 376.0 a (247.0–434.0) | 105.0 a (101.0–110.0) | 215.2 a (92.2–281.8) | 50.2 b (37.4–68.4) | 269.0 a (138.0–332.0) | 251.0 b (187.0–342.0) | 3.5 b (2.3–4.3) | 0.5 b (0.4–1.2) | 2.1 b (0.9–2.8) | 2.5 a,b (1.8–3.4) |
| EE2 | 421.0 a (192.0–501.0) | 37.5 c (23.0–54.0) | 138.0 a (109.0–280.0) | 130.2 a (48.8–130.2) | 198.0 a (169.0–374.0) | 651.0 a (244.0–651.0) | 8.0 a (4.9–9.0) | 0.2 c (0.2–0.3) | 4.5 a (2.9–6.0) | 10.7 a (4.5–14.2) |
| ATV | 151.0 b (107.0–235.0) | 101.0 b (73.0–101.0) | 44.0 b (3.4–89.8) | 36.2 c (15.2–49.0) | 66.0 b (24.0–134.0) | 181.0 c (76.0–245.0) | 1.9 c (1.3–2.3) | 1.7 a (1.1–29.7) | 0.6 c (0.0–0.9) | 1.9 b,c (0.9–2.4) |
| MIX | 335.0 a (269.0–501.0) | 93.0 b (75.0–101.0) | 196.0 a (130.0–350.0) | 52.6 b (29.0–64.4) | 234.0 a (193.0–400.0) | 263.0 b (145.0–322.0) | 3.6 b (3.3–5.0) | 0.5 b (0.3–0.6) | 2.0 b (1.6–3.5) | 1.2 c (0.7–2.5) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lourenço, T.; Rocha, E.; Gonçalves, J.F.; Rocha, M.J.; Madureira, T.V. A Proof-of-Concept for a Hypolipidemic Brown Trout Model. Toxics 2024, 12, 219. https://doi.org/10.3390/toxics12030219
Lourenço T, Rocha E, Gonçalves JF, Rocha MJ, Madureira TV. A Proof-of-Concept for a Hypolipidemic Brown Trout Model. Toxics. 2024; 12(3):219. https://doi.org/10.3390/toxics12030219
Chicago/Turabian StyleLourenço, Tiago, Eduardo Rocha, José Fernando Gonçalves, Maria João Rocha, and Tânia Vieira Madureira. 2024. "A Proof-of-Concept for a Hypolipidemic Brown Trout Model" Toxics 12, no. 3: 219. https://doi.org/10.3390/toxics12030219
APA StyleLourenço, T., Rocha, E., Gonçalves, J. F., Rocha, M. J., & Madureira, T. V. (2024). A Proof-of-Concept for a Hypolipidemic Brown Trout Model. Toxics, 12(3), 219. https://doi.org/10.3390/toxics12030219