Comprehensive Analysis of lncRNA–mRNA Expression Profiles in Depression-like Responses of Mice Related to Polystyrene Nanoparticle Exposure
Abstract
1. Introduction
2. Materials and Methods
2.1. Characterization of PS NPs
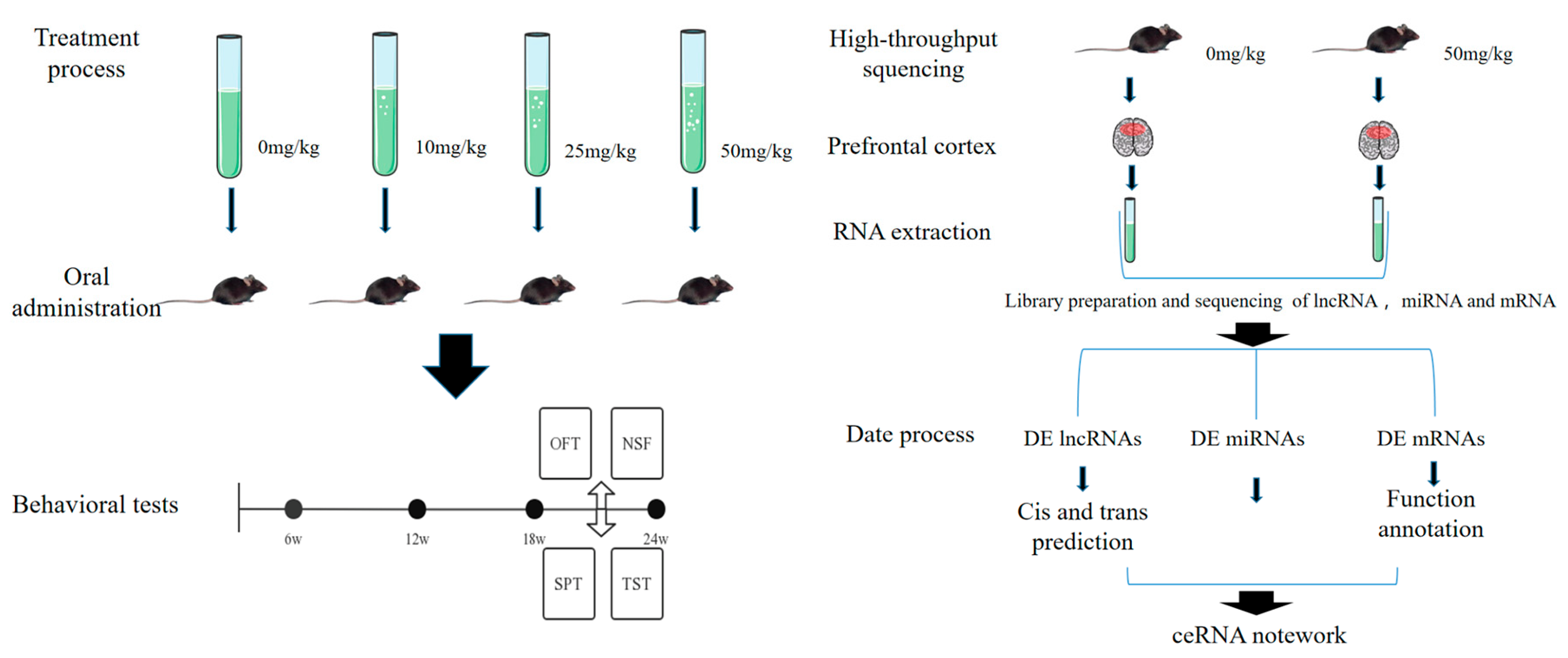
2.2. Animals and Treatment
2.3. Behavioral Tests
2.3.1. Open Field Test (OFT)
2.3.2. Novelty-Suppressed Feeding Test (NSF)
2.3.3. Sucrose Preference Test (SPT)
2.3.4. Mouse Tail Suspension Test (TST)
2.4. RNA Sequencing
2.5. Target Gene Prediction of lncRNAs
2.6. Construction of the Competing Endogenous RNA (ceRNA) Regulatory Network
2.7. Construction of the Protein–Protein Interaction (PPI) Network
2.8. Bioinformatic Analyses
2.9. Real-Time Quantitative PCR (qRT-PCR) Validation
2.10. Statistical Analysis
3. Results
3.1. Characterization of PS NPs
3.2. PS NPs Exposure Induced Depression-like Responses in Mice
3.3. LncRNA, miRNA, and mRNA Expression Profiles
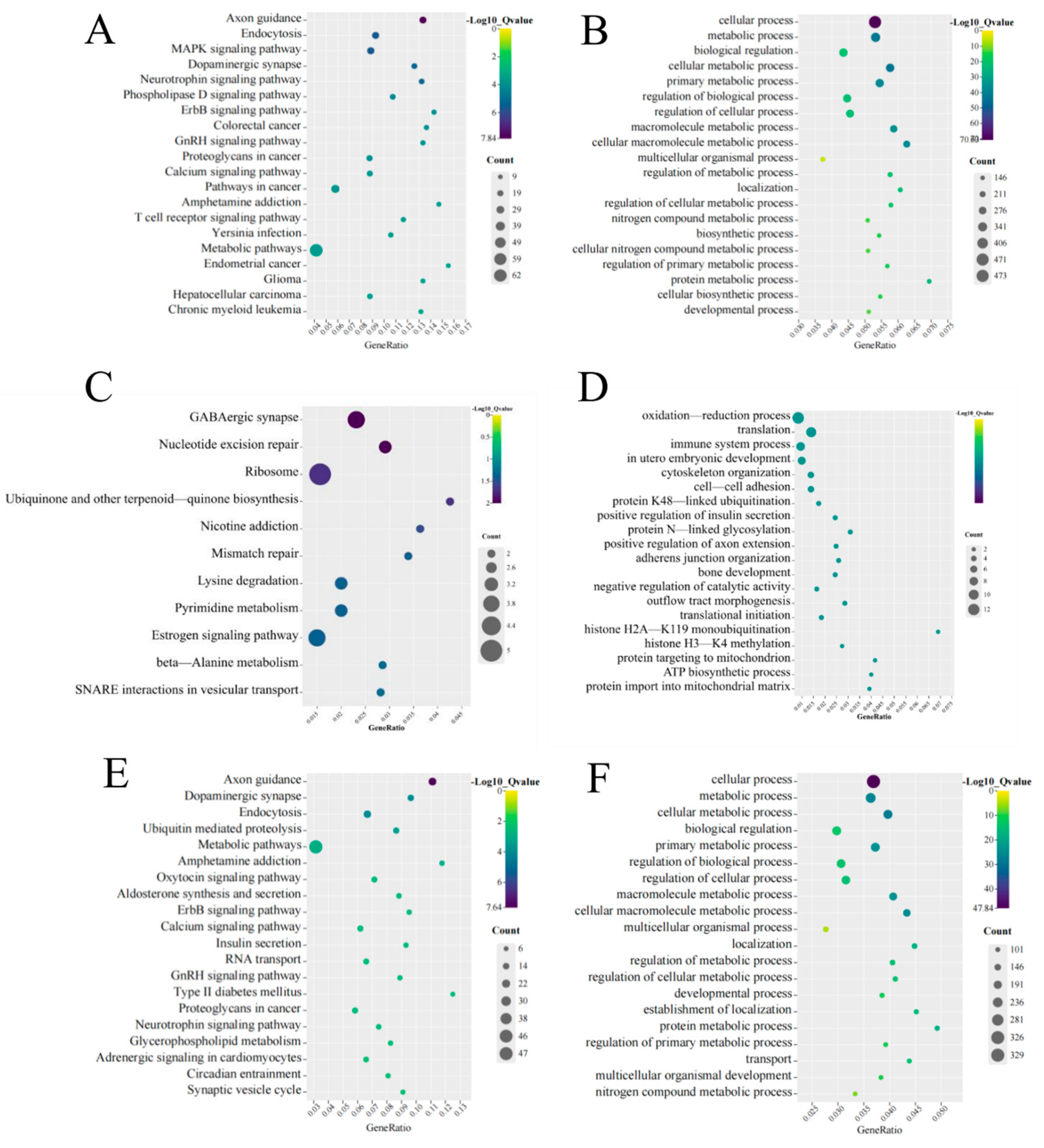
3.4. Function and Pathway Analysis of the Identified DE mRNAs and DE lncRNAs
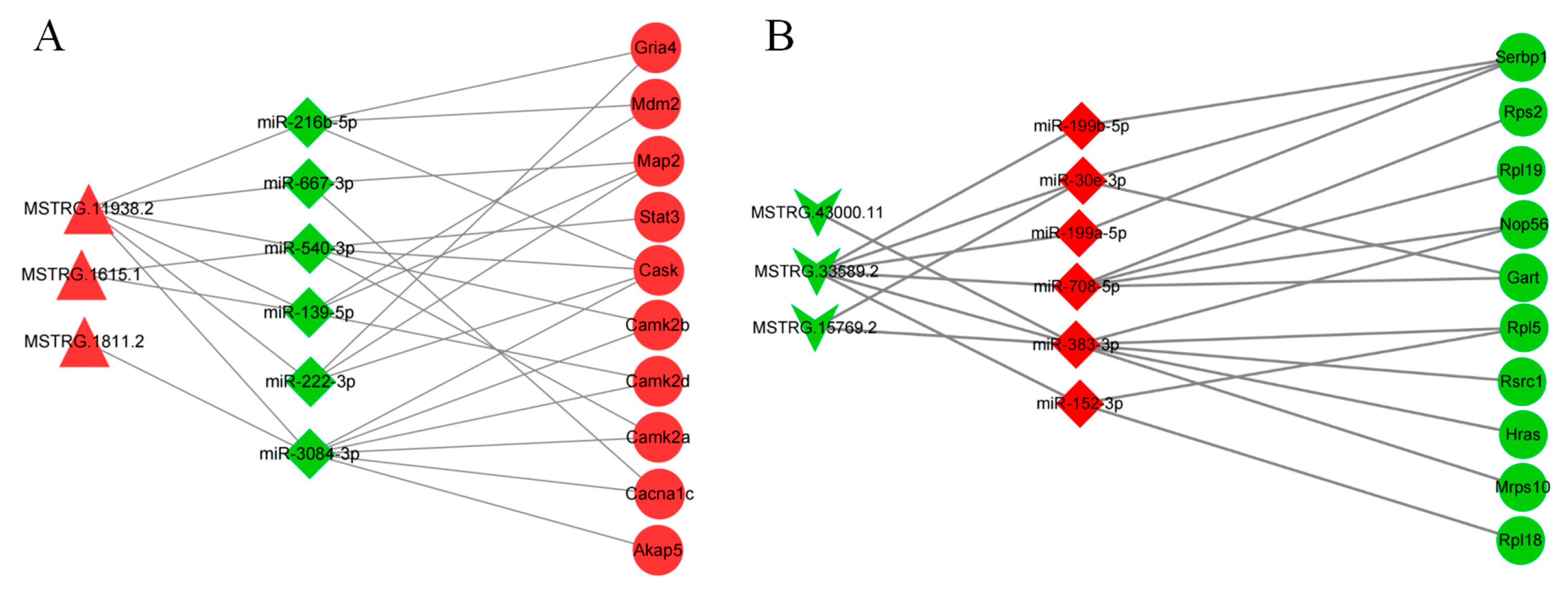
3.5. Construction of the LncRNA–miRNA-mRNA ceRNA Regulatory Network
3.6. Function Analysis of mRNAs in ceRNA Network
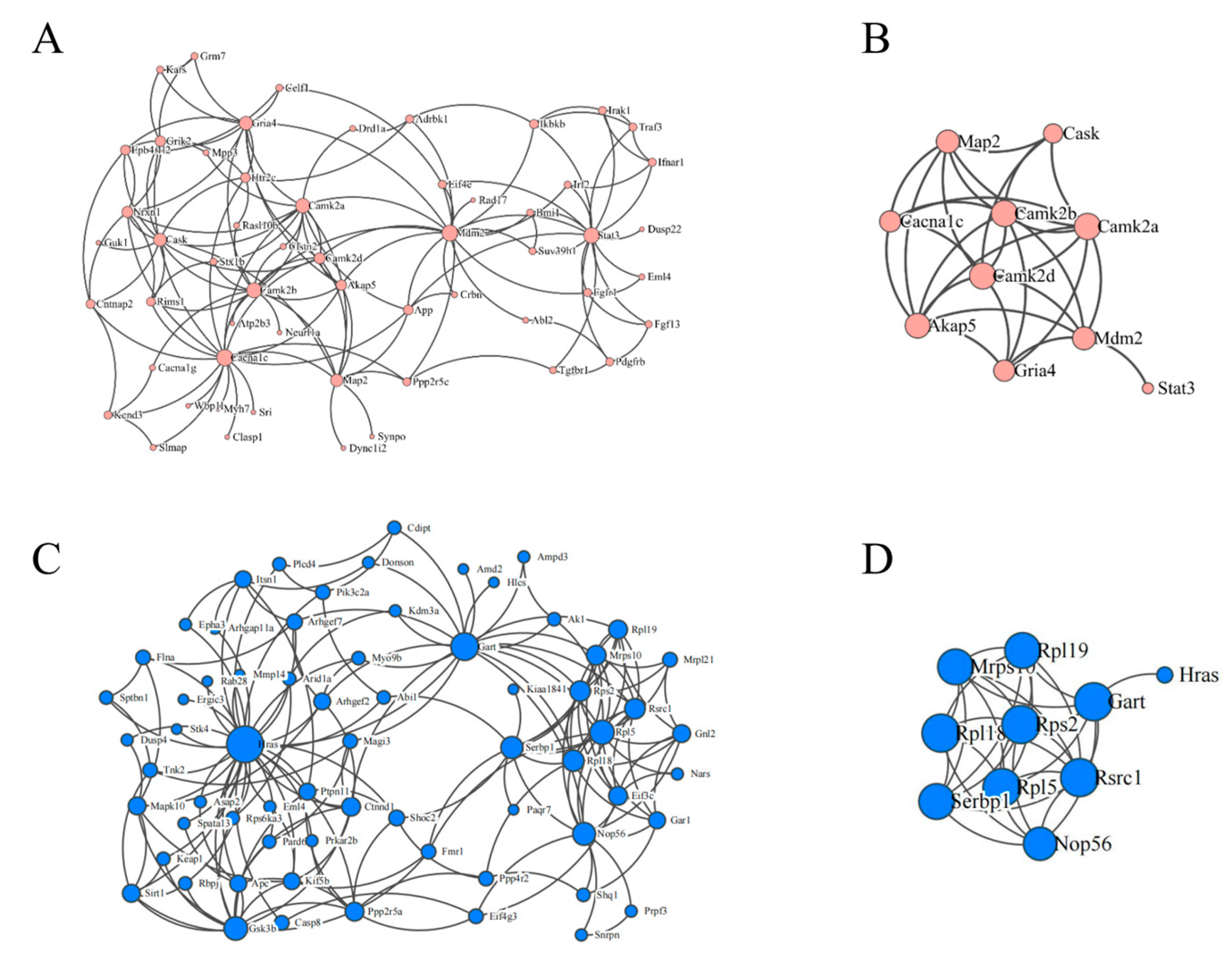
3.7. PPI Network Analysis of mRNAs
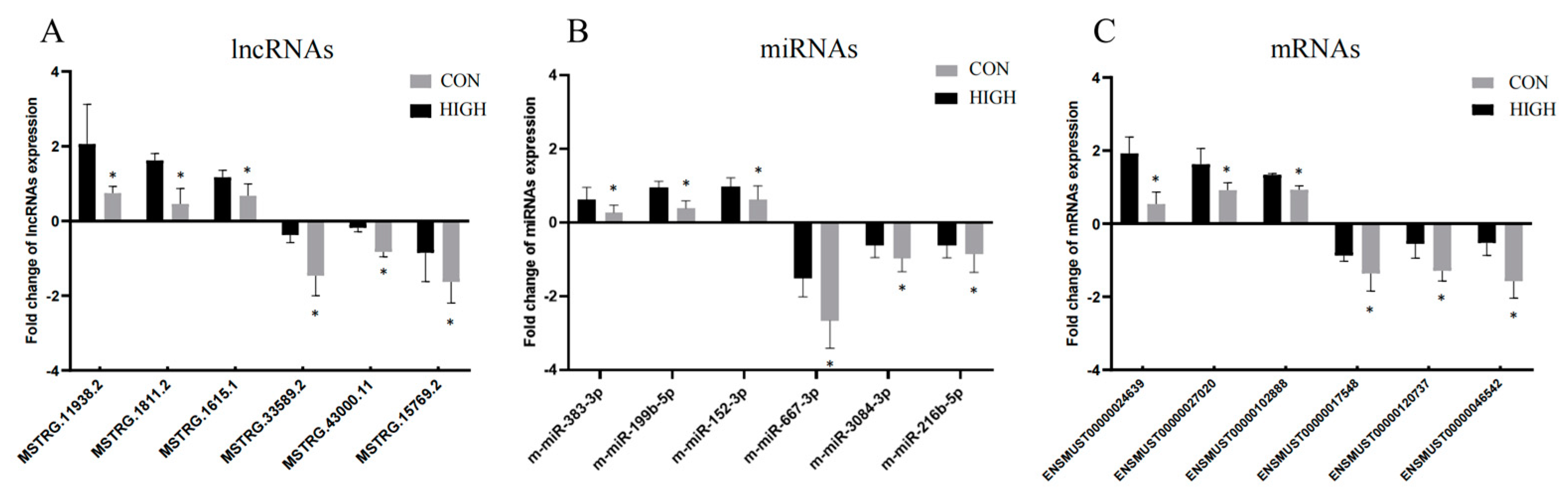
3.8. qRT-PCR Experiments
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Thompson, R.C.; Moore, C.J.; vom Saal, F.S.; Swan, S.H. Plastics, the environment and human health: Current consensus and future trends. Philos. Trans. R. Soc. 2009, 364, 2153–2166. [Google Scholar] [CrossRef]
- Morrison, M.; Trevisan, R.; Ranasinghe, P.; Merrill, G.B.; Santos, J.; Hong, A.; Edward, W.C.; Jayasundara, N.; Somarelli, J.A. A growing crisis for One Health: Impacts of plastic pollution across layers of biological function. Front. Mar. Sci. 2022, 9, 980705. [Google Scholar] [CrossRef]
- Available online: https://www.statista.com/statistics/282732/global-production-of-plastics-since-1950/ (accessed on 1 July 2023).
- Borrelle, S.B.; Ringma, J.; Law, K.L.; Monnahan, C.C.; Lebreton, L.; McGivern, A.; Murphy, E.; Jambeck, J.; Leonard, G.H.; Hilleary, M.A.; et al. Predicted growth in plastic waste exceeds efforts to mitigate plastic pollution. Science 2022, 369, 1515–1518. [Google Scholar] [CrossRef]
- Bergmann, M.; Almroth, B.C.; Brander, S.M.; Dey, T.; Green, D.S.; Gundogdu, S.; Krieger, A.; Wagner, M.; Walker, T.R. A global plastic treaty must cap production. Science 2022, 376, 469–470. [Google Scholar] [CrossRef] [PubMed]
- Ragusa, A.; Notarstefano, V.; Svelato, A.; Belloni, A.; Gioacchini, G.; Blondeel, C.; Zucchelli, E.; De Luca, C.; D’avino, S.; Gulotta, A.; et al. Raman microspectroscopy detection and characterisation of microplastics in human breastmilk. Polymers 2022, 14, 2700. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Shelver, W.L. Micro- and nanoplastic induced cellular toxicity in mammals: A review. Sci. Total Environ. 2021, 755 Pt 2, 142518. [Google Scholar] [CrossRef]
- Shan, S.; Zhang, Y.; Zhao, H.; Zeng, T.; Zhao, X. Polystyrene nanoplastics penetrate across the blood-brain barrier and induce activation of microglia in the brain of mice. Chemosphere 2022, 298, 134261. [Google Scholar] [CrossRef] [PubMed]
- Vagner, M.; Boudry, G.; Courcot, L.; Vincent, D.; Dehaut, A.; Duflos, G.; Huvet, A.; Tallec, K.; Zambonino-Infante, J.-L. Experimental evidence that polystyrene nanoplastics cross the intestinal barrier of European seabass. Environ. Int. 2022, 166, 107340. [Google Scholar] [CrossRef]
- Sun, H.; Jiang, C.; Wu, L.; Bai, X.; Zhai, S. Cytotoxicity-related bioeffects induced by nanoparticles: The role of surface chemistry. Front. Bioeng. Biotechnol. 2019, 7, 414. [Google Scholar] [CrossRef]
- Maafa, I.M. Pyrolysis of polystyrene waste: A review. Polymers 2021, 13, 225. [Google Scholar] [CrossRef]
- Prüst, M.; Meijer, J.; Westerink, R.H.S. The plastic brain: Neurotoxicity of micro- and nanoplastics. Part. Fibre Toxicol. 2020, 17, 24. [Google Scholar] [CrossRef]
- Jung, B.-K.; Han, S.-W.; Park, S.-H.; Bae, J.-S.; Choi, J.; Ryu, K.-Y. Neurotoxic potential of polystyrene nanoplastics in primary cells originating from mouse brain. Neurotoxicology 2020, 81, 189–196. [Google Scholar] [CrossRef]
- Yang, D.; Zhu, J.; Zhou, X.; Pan, D.; Nan, S.; Yin, R.; Lei, Q.; Ma, N.; Zhu, H.; Chen, J.; et al. Polystyrene micro- and nano-particle coexposure injures fetal thalamus by inducing ROS-mediated cell apoptosis. Environ. Int. 2022, 166, 107362. [Google Scholar] [CrossRef]
- Teng, M.; Zhao, X.; Wu, F.; Wang, C.; Wang, C.; White, J.C.; Zhao, W.; Zhou, L.; Yan, S.; Tian, S. Charge-specific adverse effects of polystyrene nanoplastics on zebrafish (Danio rerio) development and behavior. Environ. Int. 2022, 163, 107154. [Google Scholar] [CrossRef]
- Sarasamma, S.; Audira, G.; Siregar, P.; Malhotra, N.; Lai, Y.-H.; Liang, S.-T.; Chen, J.-R.; Chen, K.H.-C.; Hsiao, C.-D. Nanoplastics cause neurobehavioral impairments, reproductive and oxidative damages, and biomarker responses in zebrafish: Throwing up alarms of wide spread health risk of exposure. Int. J. Mol. Sci. 2020, 21, 1410. [Google Scholar] [CrossRef]
- Available online: https://www.who.int/news-room/fact-sheets/detail/depression (accessed on 1 July 2023).
- COVID-19 Mental Disorders Collaborators. Global prevalence and burden of depressive and anxiety disorders in 204 countries and territories in 2020 due to the COVID-19 pandemic. Lancet 2021, 398, 1700–1712. [Google Scholar] [CrossRef]
- Malhi, G.S.; Mann, J.J. Depression. Lancet 2018, 392, 2299–2312. [Google Scholar] [CrossRef]
- Lin, C.; Liu, G.; Huang, Y.; Liu, S.; Tang, B. Rare-earth nanoparticles induce depression, anxiety-like behavior, and memory impairment in mice. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2021, 156, 112442. [Google Scholar] [CrossRef]
- Lima, V.S.; Guimarães, A.T.B.; Araújo, A.P.D.C.; Estrela, F.N.; da Silva, I.C.; de Melo, N.F.S.; Fraceto, L.F.; Malafaia, G. Depression, anxiety-like behavior, and memory impairment in mice exposed to chitosan-coated zein nanoparticles. Environ. Sci. Pollut. Res. Int. 2019, 26, 10641–10650. [Google Scholar] [CrossRef]
- Ehsanifar, M.; Tameh, A.A.; Farzadkia, M.; Kalantari, R.R.; Zavareh, M.S.; Nikzaad, H.; Jafari, A.J. Exposure to nanoscale diesel exhaust particles: Oxidative stress, neuroinflammation, anxiety and depression on adult male mice. Ecotoxicol. Environ. Saf. 2019, 168, 338–347. [Google Scholar] [CrossRef]
- Estrela, F.N.; Guimarães, A.T.B.; Araújo, A.P.D.C.; Silva, F.G.; da Luz, T.M.; Silva, A.M.; Pereira, P.S.; Malafaia, G. Toxicity of polystyrene nanoplastics and zinc oxide to mice. Chemosphere 2021, 271, 129476. [Google Scholar] [CrossRef]
- Chu, C.; Zhang, Y.; Liu, Q.; Pang, Y.; Niu, Y.; Zhang, R. Identification of ceRNA network to explain the mechanism of cognitive dysfunctions induced by PS NPs in mice. Ecotoxicol. Environ. Saf. 2022, 241, 113785. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Luo, Y.-L.; Mao, Y.-S.; Ji, J.-L. The link between long noncoding RNAs and depression. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2017, 73, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, E.; Bagheri-Hosseinabadi, Z.; De Toma, I.; Jafarisani, M.; Sadeghi, I. The importance of long non-coding RNAs in neuropsychiatric disorders. Mol. Asp. Med. 2019, 70, 127–140. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Wang, Q.; Song, R.; Kong, Y.; Zhang, Z. Non-coding RNAs in depression: Promising diagnostic and therapeutic biomarkers. EBioMedicine 2021, 71, 103569. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Qu, M.; Zhao, Y.; Zhao, Y.; Rui, Q.; Kong, Y.; Wang, D. Identification of long non-coding RNAs in response to nanopolystyrene in Caenorhabditis elegans after long-term and low-dose exposure. Environ. Pollut. 2019, 255 Pt 1, 113137. [Google Scholar] [CrossRef]
- Fan, Z.; Xiao, T.; Luo, H.; Chen, D.; Lu, K.; Shi, W.; Sun, C.; Bian, Q. A study on the roles of long non-coding RNA and circular RNA in the pulmonary injuries induced by polystyrene microplastics. Environ. Int. 2022, 163, 107223. [Google Scholar] [CrossRef]
- Chu, C.; Zhang, H.; Cui, S.; Han, B.; Zhou, L.; Zhang, N.; Su, X.; Niu, Y.; Chen, W.; Chen, R.; et al. Ambient PM2.5 caused depressive-like responses through Nrf2/NLRP3 signaling pathway modulating inflammation. J. Hazard. Mater. 2019, 369, 180–190. [Google Scholar] [CrossRef] [PubMed]
- Jung, E.-M.; Moffat, J.J.; Liu, J.; Dravid, S.M.; Gurumurthy, C.B.; Kim, W.-Y. Arid1b haploinsufficiency disrupts cortical interneuron development and mouse behavior. Nat. Neurosci. 2017, 20, 1694–1707. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, Y.; Ye, Z.-Q.; Liu, X.-Q.; Zhao, S.-Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef]
- Senathirajah, K.; Attwood, S.; Bhagwat, G.; Carbery, M.; Wilson, S.; Palanisami, T. Estimation of the mass of microplastics ingested—A pivotal first step towards human health risk assessment. J. Hazard. Mater. 2021, 404 Pt B, 124004. [Google Scholar] [CrossRef]
- Travis, C.C.; White, R.K. Interspecific scaling of toxicity data. Risk Anal. 1988, 8, 119–125. [Google Scholar] [CrossRef]
- Lee, Y.-S.; Ehninger, D.; Zhou, M.; Oh, J.-Y.; Kang, M.; Kwak, C.; Ryu, H.-H.; Butz, D.; Araki, T.; Cai, Y.; et al. Mechanism and treatment for learning and memory deficits in mouse models of Noonan syndrome. Nat. Neurosci. 2014, 17, 1736–1743. [Google Scholar] [CrossRef]
- Cattaneo, A.; Suderman, M.; Cattane, N.; Mazzelli, M.; Begni, V.; Maj, C.; D’Aprile, I.; Pariante, C.M.; Luoni, A.; Berry, A.; et al. Long-term effects of stress early in life on microRNA-30a and its network: Preventive effects of lurasidone and potential implications for depression vulnerability. Neurobiol. Stress 2020, 13, 100271. [Google Scholar] [CrossRef]
- Ochs, S.M.; Dorostkar, M.M.; Aramuni, G.; Schön, C.; Filser, S.; Pöschl, J.; Kremer, A.; Van Leuven, F.; Ovsepian, S.V.; Herms, J. Loss of neuronal GSK3β reduces dendritic spine stability and attenuates excitatory synaptic transmission via β-catenin. Mol. Psychiatry 2015, 20, 482–489. [Google Scholar] [CrossRef]
- Uellendahl-Werth, F.; Maj, C.; Borisov, O.; Juzenas, S.; Wacker, E.M.; Jørgensen, I.F.; Steiert, T.A.; Bej, S.; Krawitz, P.; Hoffmann, P.; et al. Cross-tissue transcriptome-wide association studies identify susceptibility genes shared between schizophrenia and inflammatory bowel disease. Commun. Biol. 2022, 5, 80. [Google Scholar] [CrossRef]
- Chen, C.; Jiang, X.; Li, Y.; Yu, H.; Li, S.; Zhang, Z.; Xu, H.; Yang, Y.; Liu, G.; Zhu, F.; et al. Low-dose oral copper treatment changes the hippocampal phosphoproteomic profile and perturbs mitochondrial function in a mouse model of Alzheimer’s disease. Free Radic. Biol. Med. 2019, 135, 144–156. [Google Scholar] [CrossRef]
- Li, Y.; Fan, C.; Wang, L.; Lan, T.; Gao, R.; Wang, W.; Yu, S.Y. MicroRNA-26a-3p rescues depression-like behaviors in male rats via preventing hippocampal neuronal anomalies. J. Clin. Investig. 2021, 131, e148853. [Google Scholar] [CrossRef] [PubMed]
- Im, H.-I.; Kenny, P.J. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 2012, 35, 325–334. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Wu, Z.; Xu, J.; Zhou, L.; Di Wang, D.; Yang, L.; Bin Zhu, B.; Chen, G.; Liu, C.; et al. miR-98-5p plays a critical role in depression and antidepressant effect of ketamine. Transl. Psychiatry 2021, 11, 454. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Yang, J.; Fang, Q.; Shao, H.; Yang, D.; Sun, J.; Gao, L. MiRNA-199a-5p targets WNT2 to regulate depression through the CREB/BDNF signaling in hippocampal neuron. Brain Behav. 2021, 11, e02107. [Google Scholar] [CrossRef] [PubMed]
- Liang, F.; Zhang, Y.; Wang, X.; Yang, S.; Fang, T.; Zheng, S.; Zeng, L. Integrative mRNA and Long Noncoding RNA Analysis Reveals the Regulatory Network of Floral Bud Induction in Longan (Dimocarpus longan Lour.). Front. Plant Sci. 2022, 13, 923183. [Google Scholar] [CrossRef]
- Vardar, G.; Chang, S.; Arancillo, M.; Wu, Y.-J.; Trimbuch, T.; Rosenmund, C. Distinct Functions of Syntaxin-1 in Neuronal Maintenance, Synaptic Vesicle Docking, and Fusion in Mouse Neurons. J. Neurosci. Off. J. Soc. Neurosci. 2016, 36, 7911–7924. [Google Scholar] [CrossRef]
- Park, J.-C.; Jeong, W.-J.; Kim, M.-Y.; Min, D.; Choi, K.-Y. Retinoic-acid-mediated HRas stabilization induces neuronal differentiation of neural stem cells during brain development. J. Cell Sci. 2016, 129, 2997–3007. [Google Scholar] [CrossRef]
- Gao, Y.; Xu, T.; Zhao, Y.-X.; Ling-Hu, T.; Liu, S.-B.; Tian, J.-S.; Qin, X.-M. A Novel Network Pharmacology Strategy to Decode Metabolic Biomarkers and Targets Interactions for Depression. Front. Psychiatry 2020, 11, 667. [Google Scholar] [CrossRef]
- Chen, M.; Lai, X.; Wang, X.; Ying, J.; Zhang, L.; Zhou, B.; Liu, X.; Zhang, J.; Wei, G.; Hua, F. Long Non-coding RNAs and Circular RNAs: Insights Into Microglia and Astrocyte Mediated Neurological Diseases. Front. Mol. Neurosci. 2021, 14, 745066. [Google Scholar] [CrossRef]
- Robison, A.J.; Vialou, V.; Sun, H.-S.; Labonte, B.; Golden, S.A.; Dias, C.; Turecki, G.; Tamminga, C.; Russo, S.; Mazei-Robison, M.; et al. Fluoxetine epigenetically alters the CaMKIIα promoter in nucleus accumbens to regulate ΔFosB binding and antidepressant effects. Neuropsychopharmacology 2014, 39, 1178–1186. [Google Scholar] [CrossRef]
- Rahman, A.; Weber, J.; Labin, E.; Lai, C.; Prieto, A.L. Developmental expression of Neuregulin-3 in the rat central nervous system. J. Comp. Neurol. 2019, 527, 797–817. [Google Scholar] [CrossRef]
- Kao, W.-T.; Wang, Y.; Kleinman, J.E.; Lipska, B.K.; Hyde, T.M.; Weinberger, D.R.; Law, A.J. Common genetic variation in Neuregulin 3 (NRG3) influences risk for schizophrenia and impacts NRG3 expression in human brain. Proc. Natl. Acad. Sci. USA 2010, 107, 15619–15624. [Google Scholar] [CrossRef]
- Rahman-Enyart, A.; Lai, C.; Prieto, A.L. Neuregulins 1, 2, and 3 Promote Early Neurite Outgrowth in ErbB4-Expressing Cortical GABAergic Interneurons. Mol. Neurobiol. 2020, 57, 3568–3588. [Google Scholar] [CrossRef]
- Zhou, C.; Wu, Y.; Ding, X.; Shi, N.; Cai, Y.; Pan, Z.Z. SIRT1 Decreases Emotional Pain Vulnerability with Associated CaMKIIα Deacetylation in Central Amygdala. J. Neurosci. 2020, 40, 2332–2342. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Q.; Hu, W.; Zhang, Y.; Ning, J.; Pang, Y.; Hu, H.; Chen, M.; Wu, M.; Wang, M.; Yang, P.; et al. Comprehensive Analysis of lncRNA–mRNA Expression Profiles in Depression-like Responses of Mice Related to Polystyrene Nanoparticle Exposure. Toxics 2023, 11, 600. https://doi.org/10.3390/toxics11070600
Liu Q, Hu W, Zhang Y, Ning J, Pang Y, Hu H, Chen M, Wu M, Wang M, Yang P, et al. Comprehensive Analysis of lncRNA–mRNA Expression Profiles in Depression-like Responses of Mice Related to Polystyrene Nanoparticle Exposure. Toxics. 2023; 11(7):600. https://doi.org/10.3390/toxics11070600
Chicago/Turabian StyleLiu, Qingping, Wentao Hu, Yaling Zhang, Jie Ning, Yaxian Pang, Huaifang Hu, Meiyu Chen, Mengqi Wu, Mengruo Wang, Peihao Yang, and et al. 2023. "Comprehensive Analysis of lncRNA–mRNA Expression Profiles in Depression-like Responses of Mice Related to Polystyrene Nanoparticle Exposure" Toxics 11, no. 7: 600. https://doi.org/10.3390/toxics11070600
APA StyleLiu, Q., Hu, W., Zhang, Y., Ning, J., Pang, Y., Hu, H., Chen, M., Wu, M., Wang, M., Yang, P., Bao, L., Niu, Y., & Zhang, R. (2023). Comprehensive Analysis of lncRNA–mRNA Expression Profiles in Depression-like Responses of Mice Related to Polystyrene Nanoparticle Exposure. Toxics, 11(7), 600. https://doi.org/10.3390/toxics11070600





