Modeling DNA Methylation Profiles and Epigenetic Analysis of Safflower (Carthamus tinctorius L.) Seedlings Exposed to Copper Heavy Metal
Abstract
1. Introduction
2. Materials and Methods
2.1. Hydroponic Growth of Safflower Seedlings and Cu Stress Treatments
2.2. DNA Isolation and PCR Amplification Protocols
2.3. CRED-RA Assay
2.4. Electrophoresis
2.5. Statistical Analysis
2.6. Detection of Methylation Patterns by CRED-RA Analysis
3. Results
3.1. Data Analysis of RAPD-PCR Results
3.2. CRED-RA Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Davis, P.H. Flora of Turkey and the East Aegeans Islands; The University Press: Edinburg, UK, 1975; Volume 5. [Google Scholar]
- Singh, R.J.; Jauhar, P.P. Genetic Resources, Chromosome Engineering, and Crop Improvement: Cereals; CRC Press: Boca Raton, FL, USA, 2006; Volume 2, pp. 167–194. [Google Scholar] [CrossRef]
- Babaoglu, M. Safflower and Its Cultivation; Trakya Agricultural Research Institute: Edirne, Türkiye, 2007. [Google Scholar]
- Delshad, E.; Yousefi, M.; Sasannezhad, P.; Rakhshandeh, H.; Ayati, Z. Medical uses of Carthamus tinctorius L. (Safflower): A comprehensive review from traditional medicine to modern medicine. Electron. Physician 2018, 10, 6672–6681. [Google Scholar] [CrossRef] [PubMed]
- Anonymous. Available online: https://www.agroscope.admin.ch/agroscope/en/home/topics/plantproduction/fieldcrops/kulturarten/alternativekulturpflanzen/saflor.html (accessed on 26 July 2021).
- Buyuk, I.; Bolukbasi, E.; Aras, E.S. Expression of CtFAD2 gene for early selection in safflower oleic linoleic oil content. J. Anim. Plant Sci. 2016, 26, 1383–1388. Available online: https://www.thejaps.org.pk/docs/v-26-05/27.pdf (accessed on 16 August 2022).
- Khurana, N.; Chatterjee, C. Influence of variable zinc on yield, oil content, and physiology of sunflower. Commun. Soil Sci. Plant Anal. 2001, 32, 3023–3030. [Google Scholar] [CrossRef]
- Yarsan, E.; Bilgili, A.; Türel, I. Heavy metal levels in mussels (Unio stevenianus Krynicki) obtained from Van Lake. Turk. J. Vet. Anim. Sci. 2000, 24, 93–96. Available online: https://www.journals.tubitak.gov.tr/veterinary/vol24/iss1/13 (accessed on 27 December 2021).
- Hu, H. Heavy metal poisoning. In Harrison’s Principles of Internal Medicine, 16th ed.; Kasper, D.L., Ed.; McGraw-Hill: New York, NY, USA, 2005; Volume 4, pp. 2577–2580. [Google Scholar]
- Kumar, N.; Soni, H.; Kumar, R.N.; Bhatt, I. Hyper accumulation and mobility of heavy metals in vegetable crops in India. J. Agric. Environ. 2009, 10, 29–38. [Google Scholar] [CrossRef]
- Bolukbasi, E.; Aras, E.S. Determination of DNA methylation levels with CRED-RA technique in the genome of sunflower seedlings (Helianthus annuus L.) subjected to zinc stress. Int. J. Environ. Agric. Biotechnol. 2016, 1, 438–444. [Google Scholar] [CrossRef]
- Yang, M.G.; Lin, X.Y.; Yang, X.E. Impact of Cd on growth and nutrient accumulation of different plant species. Chin. J. Appl. Ecol. 1998, 19, 89–94. Available online: https://www.cjae.net/EN/abstract/abstract5053.shtml (accessed on 25 March 2022).
- Kosnett, M.J. Heavy metal intoxication and chelators. In Basic and Clinical Pharmacology, 10th ed.; Katzung, B.G., Ed.; McGraw-Hill: New York, NY, USA, 2007; pp. 945–957. [Google Scholar]
- Waisberg, M.P.; Joseph, B.; Beyersmann, D. Molecular and cellular mechanisms of cadmium carcinogenesis. Toxicology 2003, 192, 95–117. [Google Scholar] [CrossRef]
- Jia, X.; Fu, T.; Hu, B.; Shi, Z.; Zhou, L.; Zhu., Y. Identification of the potential risk areas for soil heavy metal pollution based on the source-sink theory. J. Hazard. Mater. 2020, 393, 122424. [Google Scholar] [CrossRef]
- Dash, S.; Borah, S.S.; Kalamdhad, A.S. Heavy metal pollution and potential ecological risk assessment for surficial sediments of Deepor Beel, India. Ecol. Indic. 2021, 122, 107265. [Google Scholar] [CrossRef]
- Bolukbasi, E. Analysis of genetic and epigenetic effects of sunflower (Helianthus Annuus L.) seedlings in response to copper stress. Fresenius Environ. Bull. 2022, 31, 4596–4602. [Google Scholar]
- Yalcin, I.E.; Ozyigit, I.I.; Dogan, I.; Demir, G.; Yarci, C. Using the Turkish red pine tree to monitor heavy metal pollution. Pol. J. Environ. Stud. 2020, 29, 3881–3889. [Google Scholar] [CrossRef]
- Alirzayeva, E.G.; Shirvani, T.S.; Yazici, M.A.; Alverdiyeva, S.; Shukurov, E.S.; Ozturk, L.; Ali-Zade, V.M.; Cakmak, I. Heavy metal accumulation in Artemisia and foliaceous lichen species from the Azerbaijan flora. For. Snow Landsc. Res. 2006, 80, 339–348. Available online: https://:www.research.sabanciuniv.edu/id/eprint/114 (accessed on 5 September 2022).
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Kirbag, F.; Munzuroglu, O. Toxic effects of cadmium (Cd+2) on metabolism of sunflower (Helianthus annuus L.) seedlings. Acta Agric. Scand. Sect. B Soil Plant Sci. 2006, 56, 224–229. [Google Scholar] [CrossRef]
- Schützendübel, A.; Polle, A. Plant responses to abiotic stresses: Heavy metal induced oxidative stress and protection by mycorrhization. J. Exp. Bot. 2002, 53, 1351–1365. [Google Scholar] [CrossRef]
- Soydam, A.S.; Gokce, E.; Buyuk, I.; Aras, S. Characterization of stress induced by copper and zinc on cucumber (Cucumis sativus L.) seedlings by means of molecular and population parameters. Mutat. Res. 2012, 746, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Arnon, D.I.; Stout, P.R. The essentiality of certain elements in minute quantity for plants with special reference to copper. Plant Physiol. 1939, 14, 371–375. [Google Scholar] [CrossRef] [PubMed]
- Bolukbasi, E.; Aras, E.S. Determination of physiological biochemical and molecular effects of zinc stress on the growth of sunflower seedlings (Helianthus annuus L.). Int. J. Environ. Agric. Biotechnol. 2018, 3, 530–536. [Google Scholar] [CrossRef]
- Martin, C.; Zhang, Y. Mechanisms of epigenetic inheritance. Curr. Opin. Cell Biol. 2007, 19, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Zhu, J.K. Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 2009, 12, 133–139. [Google Scholar] [CrossRef]
- Suzuki, M.M.; Bird, A. DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev. Genet. 2008, 9, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Leljak-Levanic, D.; Bauer, N.; Mihaljevic, S.; Jelaska, S. Changes in DNA methylation during somatic embryogenesis in Cucurbita pepo L. Plant Cell Rep. 2004, 23, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Rein, T.; DePamphilis, M.L.; Zorbas, H. Identifying 5-methylcytosine and related modifications in DNA genomes. Nucleic Acids Res. 1998, 26, 2255–2264. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Li, P.; Qi, X.; Zhou, Q.; Zheng, L.; Sun, T.; Yang, Y. DNA changes in barley (Hordeum vulgare) seedlings induced by cadmium pollution using RAPD analysis. Chemosphere 2005, 61, 158–167. [Google Scholar] [CrossRef]
- Wang, W.S.; Pan, Y.J.; Zhao, X.Q.; Dwivedi, D.; Zhu, L.H.; Ali, J.; Fu, B.Y.; Li, Z.K. Drought-induced site-specific DNA methylation and its association with drought tolerance in rice (Oryza sativa L.). J. Exp. Bot. 2011, 62, 1951–1960. [Google Scholar] [CrossRef]
- Shams, M.; Yildirim, E.; Arslan, E.; Agar, G. Salinity induced alteration in DNA methylation pattern, enzyme activity, nutrient uptake and H2O2 content in pepper (Capsicum annuum L.) cultivars. Acta Physiol. Plant. 2020, 42, 1–12. [Google Scholar] [CrossRef]
- Grigg, G.; Clark, S. Genes and genomes: Sequencing 5-methylcytosine residues in genomic DNA. Bioessays 1994, 16, 431–436. [Google Scholar] [CrossRef]
- Altunkaynak, E.; Büyük, I.; Aydin, S.S.; Aras, S. New insight into evaluation of DNA methylation levels with CRED–RA technique in the genome of Lycopersicum esculentum subjected to NaCl and PEG. Biol. Divers. Conserv. 2016, 9, 163–171. [Google Scholar]
- Cai, Q.; Guy, C.L.; Moore, G.A. Detection of cytosine methylation and mapping of a gene influencing cytosine methylation in the genome of Citrus. Genome 1996, 39, 235–242. [Google Scholar] [CrossRef]
- Hoagland, D.R.; Arnon, D.I. The water-culture method for growing plants without soil. In Circular; California Agricultural Experiment Station: Davis, CA, USA, 1950; Volume 347, p. 32. [Google Scholar]
- Taspinar, M.S.; Aydin, M.; Sigmaz, B.; Yildirim, N.; Agar, G. Protective role of humic acids against picloram-induced genomic instability and DNA methylation in Phaseolus vulgaris. Environ. Sci. Pollut. Res. 2017, 24, 22948–22953. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Wang, W.; Zhao, X.; Zhu, L.; Fu, B.; Li, Z. DNA methylation alterations of rice in response to cold stress. Plant Omics J. 2011, 4, 364–369. Available online: https://:www.pomics.com/december2011.html (accessed on 5 January 2022).
- Karan, R.; DeLeon, T.; Biradar, H.; Subudhi, P.K. Salt stress induced variation in DNA methylation pattern and its influence on gene expression in contrasting rice genotypes. PLoS ONE 2012, 7, e40203. [Google Scholar] [CrossRef]
- Bolukbasi, E. Methylation modelling and epigenetic analysis of sunflower (Helianthus annuus L.) seedlings exposed to cadmium heavy metal stress. KSU J. Agric. Nat. 2021, 25, 467–475. [Google Scholar] [CrossRef]
- Liu, W.; Yang, Y.S.; Li, P.J.; Zhou, Q.X.; Xie, L.J.; Han, Y.P. Risk assessment of cadmium contaminated soil on plant DNA damage using RAPD and physiological indices. J. Hazard. Mater. 2009, 161, 878–883. [Google Scholar] [CrossRef]
- Mastin, B.J.; Rodgers, J.H. Toxicity and bioavailability of copper herbicides (Clearigate, Cutrine-Plus, and Copper Sulfate) to freshwater animals. Environ. Contam. Toxicol. 2014, 39, 445–451. [Google Scholar] [CrossRef]
- Tuteja, N.; Ahmad, P.; Panda, B.B.; Tuteja, R. Genotoxic stress in plants: Shedding light on DNA damage, repair and DNA repair helicases. Mutat Res. 2009, 681, 134–149. [Google Scholar] [CrossRef]
- Nagajyoti, P.C.; Lee, K.D.; Sreekanth, T.V.M. Heavy metals, occurrence and toxicity for plants: A review. Environ. Chem. Lett. 2010, 8, 199–216. [Google Scholar] [CrossRef]
- Liu, W.; Yang, Y.S.; Zhou, Q.X.; Xie, L.J.; Li, P.J.; Sun, T.H. Impact assessment of cadmium contamination on rice (Oryza sativa L.) seedlings at molecular and population levels using multiple biomarkers. Chemosphere 2007, 67, 1155–1163. [Google Scholar] [CrossRef]
- Taspinar, M.S.; Agar, G.; Alpsoy, L.; Yildirim, N.; Bozari, S. The protective role of zinc and calcium in Vicia faba seedlings subjected to cadmium stress. Toxicol. Ind. Health 2011, 27, 73–80. [Google Scholar] [CrossRef]
- Atienzar, F.A.; Cordi, B.; Donkin, M.E.; Evenden, A.J. Comparison of ultravioletinduced genotoxicity detected by random amplified polymorphic DNA with chlorophyll fluorescence and growth in a marine macroalgae. Palmaria Palmata. Aquat Toxicol 2000, 50, 1–12. [Google Scholar] [CrossRef]
- Gallo-Franco, J.J.; Sosa, C.C.; Ghneim-Herrera, T.; Quimbaya, M. Epigenetic control of plant response to heavy metal stress: A new view on aluminum tolerance. Front. Plant Sci. 2020, 11, 602625. [Google Scholar] [CrossRef] [PubMed]
- Harshitha, K.; Nair, R.A. Evaluation of DNA methylation changes by CRED-RA analysis following prednisone treatment of endophyte, Fusarium oxysporum. Indian J. Microbiol. 2020, 60, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Savva, D. The use of arbitrarily primed PCR (AP-PCR) fingerprinting to detect exposure to genotoxic chemicals. Ecotoxicology 2000, 9, 341–353. [Google Scholar] [CrossRef]
- Shi, D.; Zhuang, K.; Xia, Y.; Zhu, C.; Chen, C.; Hu, Z.; Shen, Z. Hydrilla verticillata employs two different ways to affect DNA methylation under excess copper stress. Aquat. Toxicol. 2017, 193, 97–104. [Google Scholar] [CrossRef]
- Greco, M.; Sáez, C.A.; Contreras, R.A.; Brown, M.T. Cadmium and/or copper excess induce interdependent metal accumulation, DNA methylation, induction of metal chelators and antioxidant defences in the seagrass Zostera marina. Chemosphere 2019, 224, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Boquete, M.T.; Schmid, M.W.; Wagemaker, N.C.; Carey, S.B.; Alonso, C. Molecular basis of intraspecific differentiation for heavy metal tolerance in the copper moss Scopelophila cataractae. Environ. Exp. Bot. 2022, 201, 104970. [Google Scholar] [CrossRef]
- Choi, C.S.; Sano, H. Abiotic-stress induces demethylation and transcriptional activation of a gene encoding a glycerophosphodiesterase-like protein in tobacco plants. Mol. Genet. Genom. 2007, 277, 589–600. [Google Scholar] [CrossRef]
- Ertürk, F.A.; Sunar, S.; Agar, G. Determination of cytogenetic and epigenetic effects of manganese and copper on Zea mays L. ISPEC J. Agric. Sci. 2021, 5, 529–543. [Google Scholar] [CrossRef]
- Ou, X.; Zhang, Y.; Xu, C.; Lin, X.; Zang, Q.; Zhuang, T.; Liu, B. Transgenerational inheritance of modified DNA methylation patterns and enhanced tolerance induced by heavy metal stress in rice (Oryza sativa L.). PLoS ONE 2012, 7, e41143. [Google Scholar] [CrossRef]
- McKergow, M.; Nkongolo, K.K. Gene regulation and global DNA methylation changes in white spruce (Picea glauca) in response to copper contaminations. Water Air Soil Pollut. 2023, 234, 1–23. [Google Scholar] [CrossRef]
- Yagci, S.; Yildirim, E.; Yildirim, N. Nitric oxide alleviates the effects of copper-induced DNA methylation, genomic instability, LTR retrotransposon polymorphism and enzyme activity in lettuce. Plant Physiol. Rep. 2019, 24, 289–295. [Google Scholar] [CrossRef]
- Colzi, I.; Arnetoli, M.; Gallo, A.; Doumett, S.; Gonnelli, C. Copper tolerance strategies involving the root cell wall pectins in Silene paradoxa L. Environ. Exp. Bot. 2012, 78, 91–98. [Google Scholar] [CrossRef]
- Cong, W.; Miao, Y.; Xu, L. Transgenerational memory of gene expression changes induced by heavy metal stress in rice (Oryza sativa L.). BMC Plant Biol. 2019, 19, 282. [Google Scholar] [CrossRef] [PubMed]

| Type | Methylation Status | HpaII | MspI | Score of Band Profile | ||||
|---|---|---|---|---|---|---|---|---|
| x | y | z | ||||||
| Type I | CCGG GGCC | digestion | digestion | −/1 | +/0 | +/0 | Non-methylation | |
| Type II | CCGG GGCC | CCGG GGCC | digestion | undigestion | −/1 | +/0 | −/1 | Semi-methylation |
| Type III | CCGG GGCC | undigestion | digestion | −/1 | −/1 | +/0 | Full-methylation | |
| Type IV | CCGG GGCC | undigestion | undigestion | −/1 | −/1 | −/1 | Full-methylation | |
| Primers’ Name | Polymorphism Rates (%) | Sequences (5′→3′) |
|---|---|---|
| OPC-01 * | 16.90 | TTCGAGCCAG |
| OPC-02 * | 14.40 | GTGAGGCGTC |
| OPC-04 * | 36.40 | CCGCATCTAC |
| OPC-06 | 52.90 | GAACGGACTC |
| OPC-07 * | 56.80 | GTCCCGACGA |
| OPC-08 | 53.00 | TGGACCGGTG |
| OPC-09 * | 59.70 | CTCACCGTCC |
| OPC-10 * | 54.10 | TGTCTGGGTG |
| OPC-11 | 55.90 | AAAGCTGCGG |
| OPA-08 | 32.50 | GTGACGTAGG |
| Samples | GTS Rate (%) |
|---|---|
| 20 mg L−1 | 82.75 |
| 40 mg L−1 | 88.90 |
| 80 mg L−1 | 86.26 |
| 160 mg L−1 | 83.90 |
| 320 mg L−1 | 82.25 |
| 640 mg L−1 | 84.60 |
| 1280 mg L−1 | 83.70 |
| Control | 20 mg L−1 | 40 mg L−1 | 80 mg L−1 | 160 mg L−1 | 320 mg L−1 | 640 mg L−1 | 1280 mg L−1 | |
|---|---|---|---|---|---|---|---|---|
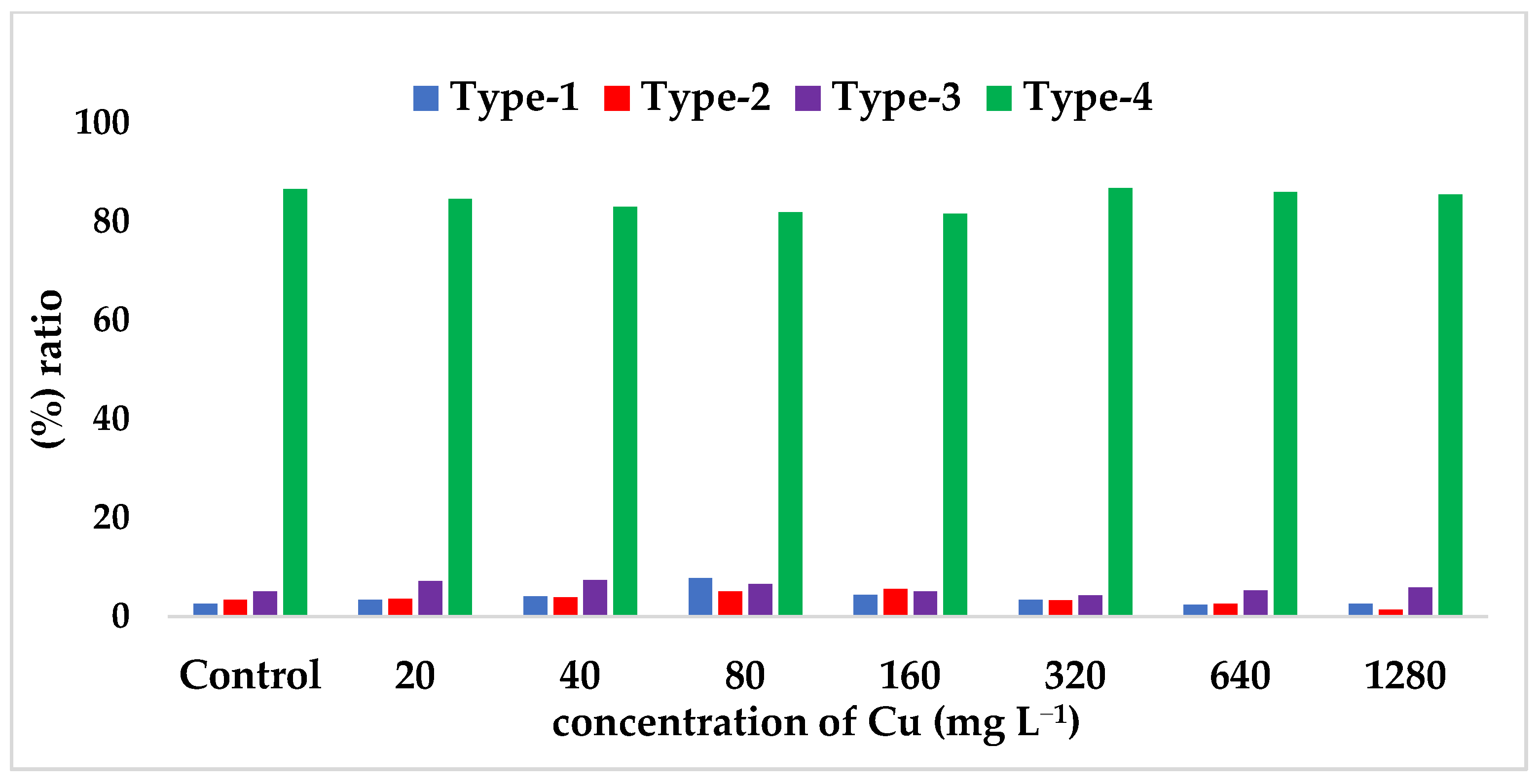
| Type-I (%) (Non-methylation) | 2.60 | 3.40 | 4.10 | 7.80 * | 4.40 | 3.40 | 2.40 * | 2.60 |
| Type-II (%) | 3.40 | 3.60 | 3.90 | 5.10 | 5.60 * | 3.30 | 2.60 | 1.40 * |
| Type-III (%) | 5.10 | 7.20 | 7.40 | 6.60 | 5.10 | 4.30 | 5.30 | 5.90 |
| Type-IV (%) | 86.60 * | 84.60 | 83.00 | 81.90 * | 81.60 * | 86.80 * | 86.00 | 85.50 |
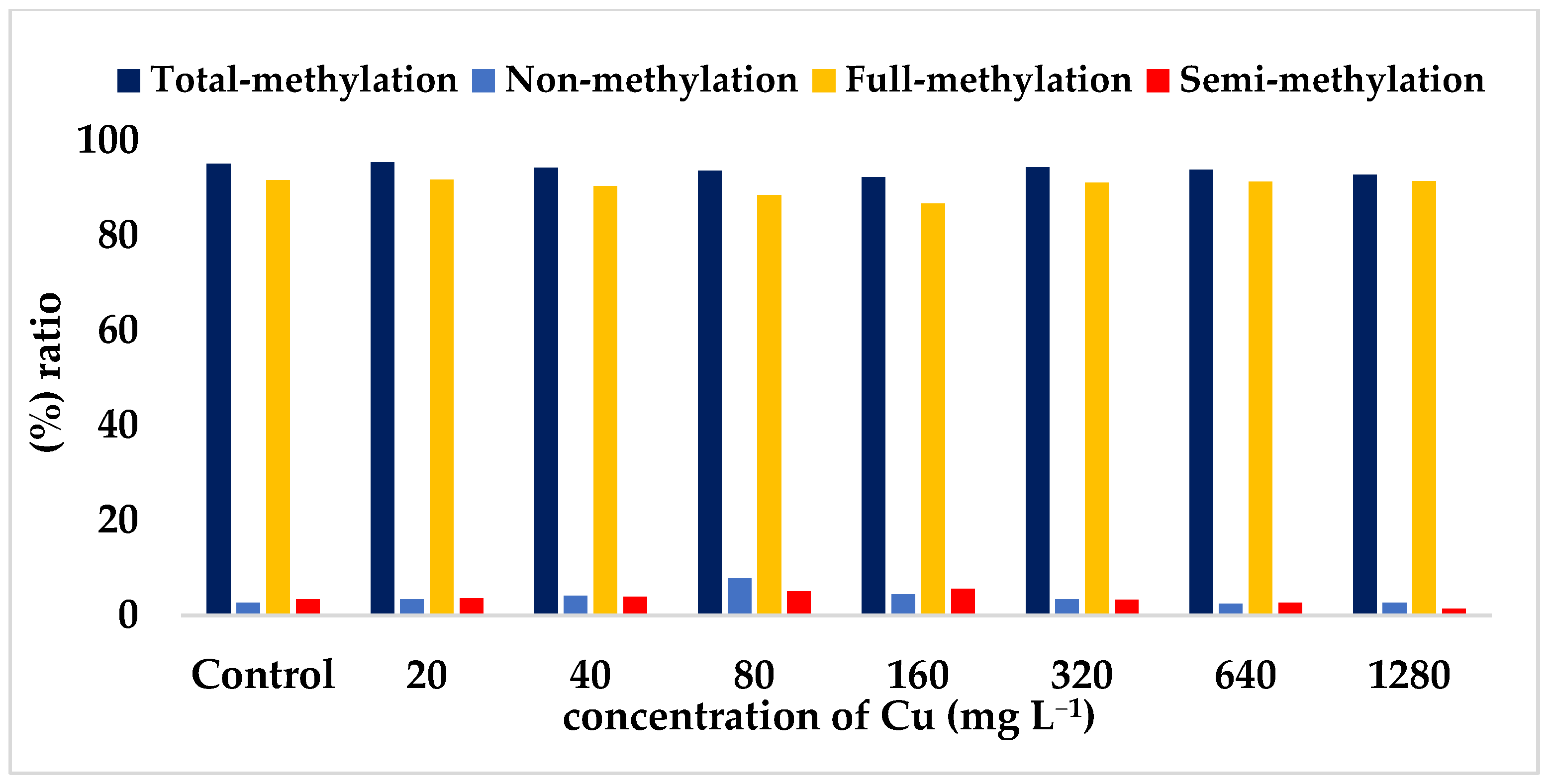
| Total methylated bands ratio (%) a | 95.10 | 95.40 * | 94.30 | 93.60 | 92.30 * | 94.40 | 93.90 | 92.80 |
| Full-methylated bands ratio (%) b | 91.70 | 91.80 * | 90.40 | 88.50 | 86.70 * | 91.10 | 91.30 | 91.40 |
| Semi-methylated bands ratio (%) c | 3.40 | 3.60 | 3.90 | 5.10 | 5.60 * | 3.30 | 2.60 * | 1.40 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bölükbaşı, E.; Karakaş, M. Modeling DNA Methylation Profiles and Epigenetic Analysis of Safflower (Carthamus tinctorius L.) Seedlings Exposed to Copper Heavy Metal. Toxics 2023, 11, 255. https://doi.org/10.3390/toxics11030255
Bölükbaşı E, Karakaş M. Modeling DNA Methylation Profiles and Epigenetic Analysis of Safflower (Carthamus tinctorius L.) Seedlings Exposed to Copper Heavy Metal. Toxics. 2023; 11(3):255. https://doi.org/10.3390/toxics11030255
Chicago/Turabian StyleBölükbaşı, Ekrem, and Mehmet Karakaş. 2023. "Modeling DNA Methylation Profiles and Epigenetic Analysis of Safflower (Carthamus tinctorius L.) Seedlings Exposed to Copper Heavy Metal" Toxics 11, no. 3: 255. https://doi.org/10.3390/toxics11030255
APA StyleBölükbaşı, E., & Karakaş, M. (2023). Modeling DNA Methylation Profiles and Epigenetic Analysis of Safflower (Carthamus tinctorius L.) Seedlings Exposed to Copper Heavy Metal. Toxics, 11(3), 255. https://doi.org/10.3390/toxics11030255