Environmental Epigenetics in Soil Ecosystems: Earthworms as Model Organisms
Abstract
:1. Introduction
1.1. Epigenetics
1.2. Environmental Epigenetics
1.3. Earthworms Role in Soil Ecosystems
2. Earthworms as Models for Environmental Stress Assessment
3. Earthworms as Models in Epigenetic Research
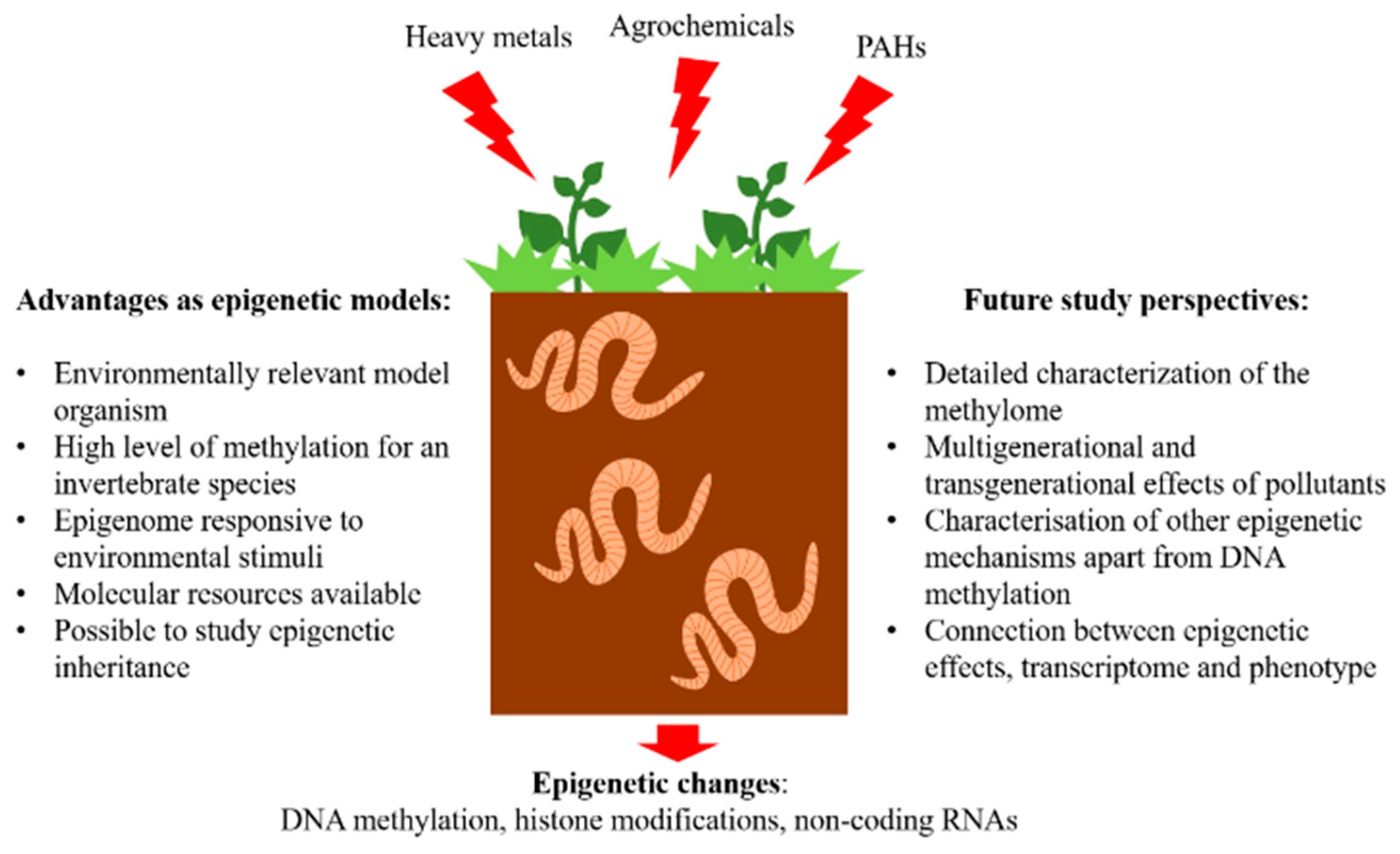
4. Advantages of Earthworm Models in Environmental Epigenetics
- Environmentally relevant sentinel species. Earthworms are important and often used species in ecotoxicological research. A range of effects of acute and chronic exposure to pollutants both in laboratory settings as well as in situ have been described. Earthworms are key soil organisms, providing important ecosystem services such as improving soil fertility and structure. Therefore, they represent a relevant model organism to study the effects of soil pollution.
- High level of genome methylation for an invertebrate species. In earthworm species, L. rubellus and A. caliginosa around 13% of methylated cytosine content was determined [51,59]. Therefore, earthworm epigenome has an important role in adaptation to environmental challenges. For comparison, the commonly used invertebrate model D. magna has only around 0.85% of methylated cytosines [8].
- Epigenome is responsive to environmental stimuli. As reviewed in the last paragraph, earthworm methylome responds to exposure to environmental pollution and shows changes even in the case of low, environmentally relevant concentrations of pollutants.
- Availability of molecular resources. To assess the genome methylation at high resolution it is of paramount importance to have genomic data available. So far, the genome is sequenced in two earthworm species E. fetida [60] and L. rubellus (unpublished). Additionally, there is available transcriptome for E. fetida [61], E. andrei [62], and L. terrestris (unpublished).
- Possibility to study epigenetic inheritance. Generational studies in a laboratory setting can easily be performed on earthworms. For instance, the full generational time of E. andrei and E. fetida is between 63 and 83 days and they reach sexual maturity in 40 to 60 days allowing studies on longer-lived species in comparison to commonly used model organisms such as D. pulex (5–10 days generational time) and C. elegans (3–4 days generational time) [53].
5. Future Study Perspectives
- Detailed characterization of the DNA methylation landscape. All of the studies performed so far on earthworms used low-resolution methods such as MSAP. Although this technique might be a good approach to cost-effectively screen a large number of samples, it has a very limited resolution, as only small number of loci are screened. WGBS is recommended to obtain an overview of methylation in the entire genome, however, due to its high cost, the method can not be used on a large number of samples. To overcome this issue, it is possible to use reduced representation approach methods or targeted approaches, however, in this way risking the possibility to miss out on some important alterations. In any case, the use of the NGS approaches will significantly improve our understanding of the DNA methylation landscape in the earthworm genome and its perturbations when challenged with environmental pollution.
- Multigenerational and transgenerational effects of pollutants. Long-term exposures covering several generations of earthworms to environmental pollutants have been seldom performed. It has been observed that earthworms originating from contaminated sites tend to have lower reproductive fitness in laboratory settings and it is therefore difficult and time-consuming to perform experiments on F1 and F2 generations [64]. A long-term multigenerational and transgenerational study on earthworms E. fetida and E. andrei exposed to arsenic, cadmium, and imidacloprid was performed to assess the phenotypic effects caused by pollution on three exposed and two unexposed generations [53]. However, such long studies exploring the earthworm epigenome have so far not been performed. The assessment of multigenerational and transgenerational effects of pollutants is of paramount importance as it can inform on long-lasting impacts that can also be inheritable. For example, in an annelid worm Enchytraeus crypticus exposed to nanomaterials (CuO and nanostructured tungsten carbide cobalt (WCCo NMs)) over several generations, an increase in global DNA methylation was associated with phenotypic effects (reproduction) [65,66,67]. Additionally, the authors noticed transgenerational effects as well as reported both global and tissue-specific changes in DNA methylation patterns in F6 and F7 generations whose ancestors were exposed to Cu nanomaterials and Cu salt [65,67].
- Characterisation of other epigenetic mechanisms apart from DNA methylation. All epigenetic mechanisms including DNA methylation, histone modifications, and micro RNA are important to study topics. However, for earthworms, only DNA methylation has been, so far, studied in more detail.
- Connection between epigenetic effects, transcriptome, and phenotype. The connection between epigenetic changes, gene expression, and phenotypic endpoints such as growth, development, reproduction, etc., is of paramount importance as it can show adaptive responses triggered by epigenetic alterations. This connection is crucial when considering the inheritance potential of epigenetic changes and transgenerational effects. These kinds of studies present a critical research challenge and can aid in our understanding of the adaptability and plasticity potential of organisms under stressful environments.
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Feil, R.; Fraga, M.F. Epigenetics and the environment: Emerging patterns and implications. Nat. Rev. Genet. 2012, 13, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Horemans, N.; Spurgeon, D.J.; Lecomte-Pradines, C.; Saenen, E.; Bradshaw, C.; Oughton, D.; Rasnaca, I.; Kamstra, J.H.; Adam-Guillermin, C. Current evidence for a role of epigenetic mechanisms in response to ionizing radiation in an ecotoxicological context. Environ. Pollut. 2019, 251, 469–483. [Google Scholar] [CrossRef] [PubMed]
- Norouzitallab, P.; Baruah, K.; Bossier, P.; Vanrompay, D. Nonmammalian model organisms in epigenetic research. In Transgenerational Epigenetics; Elsevier: Amsterdam, The Netherlands, 2019; pp. 251–261. [Google Scholar]
- De Mendoza, A.; Hatleberg, W.L.; Pang, K.; Leininger, S.; Bogdanovic, O.; Pflueger, J.; Buckberry, S.; Technau, U.; Hejnol, A.; Adamska, M.; et al. Convergent evolution of a vertebrate-like methylome in a marine sponge. Nat. Ecol. Evol. 2019, 3, 1464–1473. [Google Scholar] [CrossRef] [PubMed]
- Schübeler, D. Function and information content of DNA methylation. Nature 2015, 517, 321–326. [Google Scholar] [CrossRef]
- Noordhoek, J.W.; Koning, J.T.; Mariën, J.; Kamstra, J.H.; Amorim, M.J.B.; van Gestel, C.A.M.; van Straalen, N.M.; Roelofs, D. Exploring DNA methylation patterns in copper exposed Folsomia candida and Enchytraeus crypticus. Pedobiologia 2018, 66, 52–57. [Google Scholar] [CrossRef]
- Kurdyukov, S.; Bullock, M. DNA methylation analysis: Choosing the right method. Biology 2016, 5, 3. [Google Scholar] [CrossRef]
- Trijau, M.; Asselman, J.; Armant, O.; Adam-Guillermin, C.; De Schamphelaere, K.A.; Alonzo, F. Transgenerational DNA Methylation Changes in Daphnia magna Exposed to Chronic γ Irradiation. Environ. Sci. Technol. 2018, 52, 4331–4339. [Google Scholar] [CrossRef]
- Asselman, J.; De Coninck, D.I.; Beert, E.; Janssen, C.R.; Orsini, L.; Pfrender, M.E.; Decaestecker, E.; De Schamphelaere, K.A. Bisulfite sequencing with daphnia highlights a role for epigenetics in regulating stress response to microcystis through preferential differential methylation of serine and threonine amino acids. Environ. Sci. Technol. 2017, 51, 924–931. [Google Scholar] [CrossRef] [Green Version]
- Guan, D.L.; Ding, R.R.; Hu, X.Y.; Yang, X.R.; Xu, S.Q.; Gu, W.; Zhang, M. Cadmium-induced genome-wide DNA methylation changes in growth and oxidative metabolism in Drosophila melanogaster. BMC Genom. 2019, 20, 356. [Google Scholar] [CrossRef] [Green Version]
- Rondon, R.; Grunau, C.; Fallet, M.; Charlemagne, N.; Sussarellu, R.; Chaparro, C.; Montagnani, C.; Mitta, G.; Bachère, E.; Akcha, F. Effects of a parental exposure to diuron on Pacific oyster spat methylome. Environ. Epigenet. 2017, 3, dvx004. [Google Scholar] [CrossRef] [Green Version]
- Schield, D.R.; Walsh, M.R.; Card, D.C.; Andrew, A.L.; Adams, R.H.; Castoe, T.A. EpiRADseq: Scalable analysis of genomewide patterns of methylation using next-generation sequencing. Methods Ecol. Evol. 2016, 7, 60–69. [Google Scholar] [CrossRef]
- Van Gurp, T.P.; Wagemaker, N.C.A.M.; Wouters, B.; Vergeer, P.; Ouborg, J.N.J.; Verhoeven, K.J.F. EpiGBS: Reference-free reduced representation bisulfite sequencing. Nat. Methods 2016, 13, 322–324. [Google Scholar] [CrossRef]
- Trucchi, E.; Mazzarella, A.B.; Gilfillan, G.D.; Romero, M.L.; Schönswetter, P.; Paun, O. BsRADseq: Screening DNA methylation in natural populations of non-model species. Mol. Ecol. 2016, 8, 1697–1713. [Google Scholar] [CrossRef] [Green Version]
- Dixon, G.; Matz, M. Benchmarking DNA methylation assays in a reef-building coral. Mol. Ecol. Resour. 2021, 21, 464–477. [Google Scholar] [CrossRef]
- Novo, M.; Verdú, I.; Trigo, D.; Martínez-guitarte, J.L. Endocrine disruptors in soil: Effects of bisphenol A on gene expression of the earthworm Eisenia fetida. Ecotoxicol. Environ. Saf. 2018, 150, 159–167. [Google Scholar] [CrossRef]
- Aigner, G.P.; Pittl, V.; Fiechtner, B.; Egger, B.; Šrut, M.; Höckner, M. Common mechanisms cannot explain time- and dose-dependent DNA methylation changes in earthworms exposed to cadmium. Sci. Total Environ. 2022, 812, 151468. [Google Scholar] [CrossRef]
- Planques, A.; Kerner, P.; Ferry, L.; Grunau, C.; Gazave, E.; Vervoort, M. DNA methylation atlas and machinery in the developing and regenerating annelid Platynereis dumerilii. BMC Biol. 2021, 19, 148. [Google Scholar] [CrossRef]
- Dimond, J.L.; Roberts, S.B. Germline DNA methylation in reef corals: Patterns and potential roles in response to environmental change. Mol. Ecol. 2016, 25, 1895–1904. [Google Scholar] [CrossRef] [Green Version]
- Lyko, F.; Foret, S.; Kucharski, R.; Wolf, S.; Falckenhayn, C.; Maleszka, R. The honey bee epigenomes: Differential methylation of brain DNA in queens and workers. PLoS Biol. 2010, 8, e1000506. [Google Scholar] [CrossRef] [Green Version]
- Riviere, G.; Wu, G.-C.; Fellous, A.; Goux, D.; Sourdaine, P.; Favrel, P. DNA methylation is crucial for the early development in the Oyster C. gigas. Mar. Biotechnol. 2013, 15, 739–753. [Google Scholar] [CrossRef]
- Suzuki, M.M.; Bird, A. DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev. Genet. 2008, 9, 465–476. [Google Scholar] [CrossRef]
- Keller, T.E.; Han, P.; Yi, S.V. Evolutionary transition of promoter and gene body DNA methylation across invertebrate-vertebrate boundary. Mol. Biol. Evol. 2016, 33, 1019–1028. [Google Scholar] [CrossRef]
- Dixon, G.; Bay, L.K.; Matz, M.V. Evolutionary consequences of DNA methylation in a basal metazoan. Mol. Biol. Evol. 2016, 33, 2285–2293. [Google Scholar] [CrossRef] [Green Version]
- Dixon, G.B.; Bay, L.K.; Matz, M.V. Bimodal signatures of germline methylation are linked with gene expression plasticity in the coral Acropora millepora. BMC Genom. 2014, 15, 2–11. [Google Scholar] [CrossRef] [Green Version]
- Gatzmann, F.; Falckenhayn, C.; Gutekunst, J.; Hanna, K.; Raddatz, G.; Carneiro, V.C.; Lyko, F. The methylome of the marbled crayfish links gene body methylation to stable expression of poorly accessible genes. Epigenet. Chromatin 2018, 11, 57. [Google Scholar] [CrossRef]
- Vogt, G. Investigating the genetic and epigenetic basis of big biological questions with the parthenogenetic marbled crayfish: A review and perspectives. J. Biosci. 2018, 43, 189–223. [Google Scholar] [CrossRef]
- Gavery, M.R.; Roberts, S.B. Epigenetic considerations in aquaculture. PeerJ 2017, 5, e4147. [Google Scholar] [CrossRef] [Green Version]
- Olson, C.E.; Roberts, S.B. Genome-wide profiling of DNA methylation and gene expression in Crassostrea gigas male gametes. Front. Physiol. 2014, 5, 224. [Google Scholar] [CrossRef] [Green Version]
- Song, K.; Li, L.; Zhang, G. The association between DNA methylation and exon expression in the Pacific oyster Crassostrea gigas. PLoS ONE 2017, 12, e0185224. [Google Scholar] [CrossRef]
- Mirbahai, L.; Chipman, J.K. Epigenetic memory of environmental organisms: A reflection of lifetime stressor exposures. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2014, 764–765, 10–17. [Google Scholar] [CrossRef]
- Jeremias, G.; Gonçalves, F.J.M.; Pereira, J.L.; Asselman, J. Prospects for incorporation of epigenetic biomarkers in human health and environmental risk assessment of chemicals. Biol. Rev. 2020, 95, 822–846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suarez-Ulloa, V.; Gonzalez-Romero, R.; Eirin-Lopez, J.M. Environmental epigenetics: A promising venue for developing next-generation pollution biomonitoring tools in marine invertebrates. Mar. Pollut. Bull. 2015, 98, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Eirin-Lopez, J.M.; Putnam, H.M. Marine Environmental Epigenetics. Ann. Rev. Mar. Sci. 2018, 11, 335–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Šrut, M. Ecotoxicological epigenetics in invertebrates: Emerging tool for the evaluation of present and past pollution burden. Chemosphere 2021, 282, 131026. [Google Scholar] [CrossRef]
- Liew, Y.J.; Zoccola, D.; Li, Y.; Tambutté, E.; Venn, A.A.; Michell, C.T.; Deutekom, E.S.; Kaandorp, J.A.; Voolstra, C.R. Epigenome-associated phenotypic acclimatization to ocean acidification in a reef-building coral. Sci. Adv. 2018, 4, eaar8028. [Google Scholar] [CrossRef] [Green Version]
- Baderna, D.; Lomazzi, E.; Pogliaghi, A.; Ciaccia, G.; Lodi, M.; Benfenati, E. Acute phytotoxicity of seven metals alone and in mixture: Are Italian soil threshold concentrations suitable for plant protection? Environ. Res. 2015, 140, 102–111. [Google Scholar] [CrossRef]
- Bartlett, M.D.; Briones, M.J.; Neilson, R.; Schmidt, O.; Spurgeon, D.; Creamer, R.E. A critical review of current methods in earthworm ecology: From individuals to populations. Eur. J. Soil Biol. 2010, 46, 67–73. [Google Scholar] [CrossRef]
- Velki, M.; Ečimović, S. Important Issues in Ecotoxicological Investigations Using Earthworms. Rev. Environ. Contam. Toxicol. 2016, 239, 157–184. [Google Scholar] [CrossRef]
- ISO. Soil Quality—Effects of Pollutants on Earthworms (Eisenia fetida)—Part 1: Determination of Acute Toxicity Using Artificial Soil Substrate; ISO: Geneva, Switzerland, 2012. [Google Scholar]
- Fox, C.J.S. The effects of five herbicides on the numbers of certain invertebrate animals in grassland soil. Can. J. Plant Sci. 1964, 44, 405–409. [Google Scholar] [CrossRef] [Green Version]
- Edwards, C.A. Soil pollutants and soil animals. Sci. Am. 1969, 220, 88–99. [Google Scholar] [CrossRef]
- OECD. Test No. 207: Earthworm, Acute Toxicity Tests; OECD: Paris, France, 1984. [Google Scholar]
- OECD. Guideline for the Testing of Chemicals Nr. 222 “Earthworm, Reproduction Test”; OECD: Paris, France, 2004. [Google Scholar]
- ISO. Soil Quality—Effects of Pollutants on Earthworm (Eisenia fetida)—Part 2: Determination of Effects on Reproduction; ISO: Geneva, Switzerland, 2012. [Google Scholar]
- Vasseur, P.; Bonnard, M. Ecogenotoxicology in earthworms: A review. Curr. Zool. 2014, 60, 255–272. [Google Scholar] [CrossRef]
- Gong, P.; Perkins, E.J. Earthworm toxicogenomics: A renewed genome-wide quest for novel biomarkers and mechanistic insights. Appl. Soil Ecol. 2016, 104, 12–24. [Google Scholar] [CrossRef]
- Spurgeon, D.J.; Weeks, J.M.; van Gestel, C.A.M. A summary of eleven years progress in earthworm ecotoxicology. Pedobiologia 2003, 47, 588–606. [Google Scholar] [CrossRef]
- Klobučar, G.I.; Štambuk, A.; Šrut, M.; Husnjak, I.; Merkaš, M.; Traven, L.; Cvetković, Ž. Aporrectodea caliginosa, a suitable earthworm species for field based genotoxicity assessment? Environ. Pollut. 2011, 159, 841–849. [Google Scholar] [CrossRef]
- Yadav, A.; Vishwakarma, S.; Krishna, N.; Katara, P. Integrative Omics: Current Status and Future Directions. In Recent Trends in ‘Computational Omics: Concepts and Methodology’; Nova Science Publishers Inc.: New York, NY, USA, 2020; pp. 1–46. [Google Scholar]
- Kille, P.; Andre, J.; Anderson, C.; Ang, H.N.; Bruford, M.W.; Bundy, J.; Donnelly, R.; Hodson, M.E.; Juma, G.; Lahive, E.; et al. DNA sequence variation and methylation in an arsenic tolerant earthworm population. Soil Biol. Biochem. 2013, 57, 524–532. [Google Scholar] [CrossRef] [Green Version]
- Rasnaca, I.; Kille, P.; Newbold, L.K.; Spurgeon, D.J. Impacts of Life-Time Exposure of Arsenic, Cadmium and Fluoranthene on the Earthworms’ L. rubellus Global DNA Methylation as Detected by msAFLP. Genes 2022, 13, 770. [Google Scholar] [CrossRef]
- Rasnaca, I. Tracking the Ghost of the Genome: The Epigenetics of Pollution Adaptation in an Environmental Sentinel. Ph.D. Thesis, Cardiff University, Cardiff, UK, 2019. [Google Scholar]
- Šrut, M.; Drechsel, V.; Höckner, M. Low levels of Cd induce persisting epigenetic modifications and acclimation mechanisms in the earthworm Lumbricus terrestris. PLoS ONE 2017, 12, e0176047. [Google Scholar] [CrossRef] [Green Version]
- Drechsel, V.; Schauer, K.; Šrut, M.; Höckner, M. Regulatory plasticity of earthworm wMT-2 gene expression. Int. J. Mol. Sci. 2017, 18, 1113. [Google Scholar] [CrossRef] [Green Version]
- Aigner, G.P.; Nenning, P.; Fiechtner, B.; Šrut, M.; Höckner, M. Lumbricus terrestris Exposed to Cadmium and the DNA Demethylation Agent 5-aza-2-deoxycytidine. Toxics 2022, 10, 100. [Google Scholar] [CrossRef]
- Newbold, L.K.; Robinson, A.; Rasnaca, I.; Lahive, E.; Soon, G.H.; Lapied, E.; Oughton, D.; Gashchak, S.; Beresford, N.A.; Spurgeon, D.J. Genetic, epigenetic and microbiome characterisation of an earthworm species (Octolasion lacteum) along a radiation exposure gradient at Chernobyl. Environ. Pollut. 2019, 255, 113238. [Google Scholar] [CrossRef]
- Santoyo, M.M.; Flores, C.R.; Torres, A.L.; Wrobel, K.; Wrobel, K. Global DNA methylation in earthworms: A candidate biomarker of epigenetic risks related to the presence of metals/metalloids in terrestrial environments. Environ. Pollut. 2011, 159, 2387–2392. [Google Scholar] [CrossRef]
- Regev, A.; Lamb, M.J.; Jablonka, E. The Role of DNA Methylation in Invertebrates: Developmental Regulation or Genome Defense? Mol. Biol. Evol. 1998, 15, 880–891. [Google Scholar] [CrossRef]
- Zwarycz, A.S.; Nossa, C.W.; Putnam, N.H.; Ryan, J.F. Timing and Scope of Genomic Expansion within Annelida: Evidence from Homeoboxes in the Genome of the Earthworm Eisenia fetida. Genome Biol. Evol. 2015, 8, 271–281. [Google Scholar] [CrossRef] [Green Version]
- Gong, P.; Pirooznia, M.; Guan, X.; Perkins, E.J. Design, Validation and Annotation of Transcriptome-Wide Oligonucleotide Probes for the Oligochaete Annelid Eisenia fetida. PLoS ONE 2010, 5, e14266. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.S.; Cho, S.J.; Tak, E.S.; Lee, J.A.; Cho, H.J.; Park, B.J.; Shin, C.; Kim, D.K.; Park, S.C. Transcriptome analysis in the midgut of the earthworm (Eisenia andrei) using expressed sequence tags. Biochem. Biophys. Res. Commun. 2005, 328, 1196–1204. [Google Scholar] [CrossRef]
- Lloyd, V.K.; Brisson, J.A.; Fitzpatrick, K.A.; McEachern, L.A.; Verhulst, E.C. The Epigenetics of Emerging and Nonmodel Organisms. Genet. Res. Int. 2012, 2012, 491204. [Google Scholar] [CrossRef]
- Langdon, C.J.; Piearce, T.G.; Meharg, A.A.; Semple, K.T. Interactions between earthworms and arsenic in the soil environment: A review. Environ. Pollut. 2003, 124, 361–373. [Google Scholar] [CrossRef]
- Bicho, R.C.; Roelofs, D.; Mariën, J.; Scott-Fordsmand, J.J.; Amorim, M.J.B. Epigenetic effects of (nano)materials in environmental species—Cu case study in Enchytraeus crypticus. Environ. Int. 2020, 136, 105447. [Google Scholar] [CrossRef]
- Bicho, R.C.; Scott-Fordsmand, J.J.; Amorim, M.J.B. Multigenerational Exposure to WCCo Nanomaterials—Epigenetics in the Soil Invertebrate Enchytraeus crypticus. Nanomaterials 2020, 10, 836. [Google Scholar] [CrossRef] [PubMed]
- Bicho, R.C.; Faustino, A.M.R.; Rêma, A.; Scott-Fordsmand, J.J.; Amorim, M.J.B. Confirmatory assays for transient changes of omics in soil invertebrates—Copper materials in a multigenerational exposure. J. Hazard. Mater. 2021, 402, 123500. [Google Scholar] [CrossRef]
| Toxicant | Model Earthworm | Epigenetic Endpoint | Method | Main Finding | Reference |
|---|---|---|---|---|---|
| Arsenic (As) and copper (Cu) mine | L. rubellus | Genome-wide DNA methylation | meAFLP * | Association of methylation patterns with soil As concentrations in one earthworm lineage | [51] |
| As, cadmium (Cd), fluoranthene | L. rubellus | Genome-wide DNA methylation | MSAP * | No effects of As and Cd, fluoranthene changed DNA methylation patterns | [52] |
| Zinc (Zn), lead (Pb), and Cd smelter | L. rubellus | Genome-wide DNA methylation | MSAP * | No methylation changes | [53] |
| Cd | L. terrestris | Genome-wide DNA methylation | MSAP * | Hypermethylation | [54] |
| Cd | L. terrestris | Global and gene-specific DNA methylation; DNMT1, DNMT3, TET gene expression and activity | Dot blot; bisulfite conversion and sequencing of the MT2 gene body region; qPCR | Time and dose dependant changes in DNA methylation patterns; no significant changes in MT2 gene body methylation, no changes in DNMT and TET gene expression and activity | [17] |
| Cd | L. terrestris | Gene-specific DNA methylation | Bisulfite conversion and sequencing of the MT2 promoter region | No methylation in the MT2 promoter region | [55] |
| Cd | L. terrestris | Global DNA methylation | Dot blot | No methylation changes | [56] |
| Bisphenol A (BPA) | Eisenia fetida | DNMT1 and DNMT3b gene expression | qPCR | Lower expression at higher BPA concentrations | [16] |
| Ionizing radiation within the Chernobyl exclusion zone | Octolasion lacteum | Genome-wide DNA methylation | meAFLP * | No methylation changes | [57] |
| Silver and gold mine | Earthworms | Global DNA methylation | HPLC * | Inverse correlation between the percentage of methylated DNA and total tissue As, As + Hg, As + Hg + Se + Sb, and inorganic As + Hg | [58] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Šrut, M. Environmental Epigenetics in Soil Ecosystems: Earthworms as Model Organisms. Toxics 2022, 10, 406. https://doi.org/10.3390/toxics10070406
Šrut M. Environmental Epigenetics in Soil Ecosystems: Earthworms as Model Organisms. Toxics. 2022; 10(7):406. https://doi.org/10.3390/toxics10070406
Chicago/Turabian StyleŠrut, Maja. 2022. "Environmental Epigenetics in Soil Ecosystems: Earthworms as Model Organisms" Toxics 10, no. 7: 406. https://doi.org/10.3390/toxics10070406
APA StyleŠrut, M. (2022). Environmental Epigenetics in Soil Ecosystems: Earthworms as Model Organisms. Toxics, 10(7), 406. https://doi.org/10.3390/toxics10070406





