Transcriptomic Alterations in Water Flea (Daphnia magna) following Pravastatin Treatments: Insect Hormone Biosynthesis and Energy Metabolism
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemical Reagents
2.2. Biotests and Culture Conditions
2.3. D. magna Immobilization Study
2.4. Chronic Toxicity Test
2.5. RNA Sequencing
2.6. RNA-Seq Data Analyses
2.7. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Analysis
2.8. Statistical Analysis
3. Results
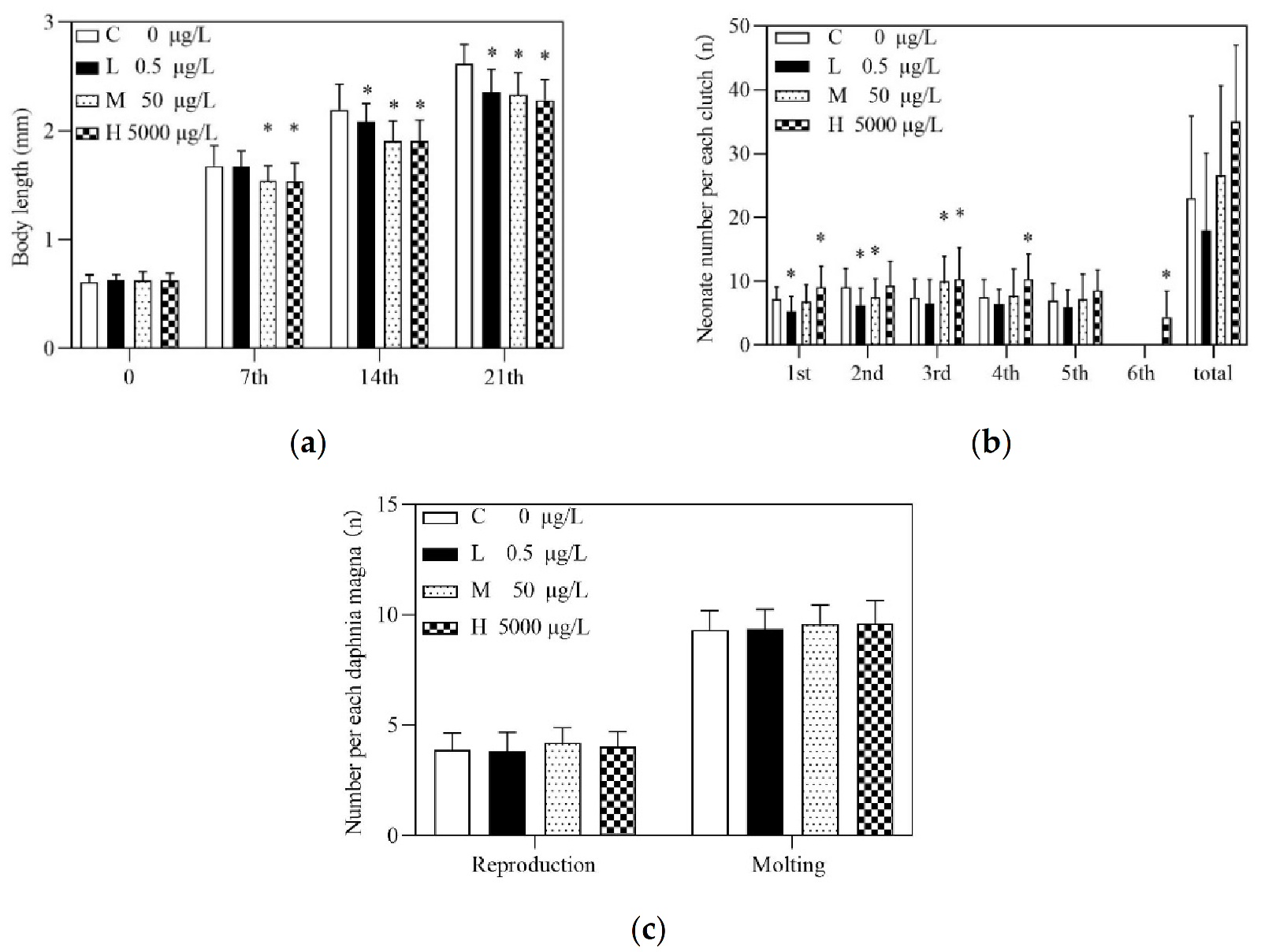
3.1. Effects of Acute and Chronic Exposures to PRA
3.2. Transcriptome Analysis
3.2.1. Sequence Assembly and Generation
3.2.2. RNA-Seq Reads and DEGs
3.2.3. Go Ontology and KEGG Pathway Analysis
4. Discussion
4.1. Genes Related to Xenobiotic Metabolism
4.2. Genes Related to Insect Hormone Biosynthesis
4.3. Genes Related to Energy Metabolism
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhou, Q.; Liao, J.K. Statins and cardiovascular diseases: From cholesterol lowering to pleiotropy. Curr. Pharm. Des. 2009, 15, 467–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamashita, S.; Tsubakio-Yamamoto, K.; Ohama, T.; Nakagawa-Toyama, Y.; Nishida, M. Molecular mechanisms of HDL-cholesterol elevation by statins and its effects on HDL functions. J. Atheroscler. Thromb. 2010, 17, 436–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hague, W.E.; Simes, J.; Kirby, A.; Keech, A.C.; White, H.D.; Hunt, D.; Nestel, P.J.; Colquhoun, D.M.; Pater, H.; Stewart, R.A. Long-term effectiveness and safety of pravastatin in patients with coronary heart disease: Sixteen years of follow-up of the LIPID study. Circulation 2016, 133, 1851–1860. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. Drugbank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006, 34, D668–D672. [Google Scholar] [CrossRef]
- Ellesat, K.S.; Tollefsen, K.-E.; Åsberg, A.; Thomas, K.V.; Hylland, K. Cytotoxicity of atorvastatin and simvastatin on primary rainbow trout (Oncorhynchus Mykiss) hepatocytes. Toxicol. In Vitro 2010, 24, 1610–1618. [Google Scholar] [CrossRef]
- Sulaiman, S.; Khamis, M.; Karaman, R. Stability and removal of several statins from wastewater. Environ. Technol. 2015, 36, 3232–3242. [Google Scholar] [CrossRef]
- Liu, Y.; Ding, R.; Pan, B.; Wang, L.; Liu, S.; Nie, X. Simvastatin affects the expression of detoxification-related genes and enzymes in Daphnia magna and alter its life history parameters. Ecotoxicol. Environ. Saf. 2019, 182, 109389. [Google Scholar] [CrossRef]
- Santos, M.M.; Ruivo, R.; Lopes-Marques, M.; Torres, T.; Carmen, B.; Castro, L.F.C.; Neuparth, T. Statins: An undesirable class of aquatic contaminants? Aquat. Toxicol. 2016, 174, 1–9. [Google Scholar] [CrossRef]
- Key, P.B.; Hoguet, J.; Reed, L.A.; Chung, K.W.; Fulton, M.H. Effects of the statin antihyperlipidemic agent simvastatin on grass shrimp, Palaemonetes pugio. Environ. Toxicol. Int. J. 2008, 23, 153–160. [Google Scholar] [CrossRef]
- Neuparth, T.; Martins, C.; Carmen, B.; Costa, M.H.; Martins, I.; Costa, P.M.; Santos, M.M. Hypocholesterolaemic pharmaceutical simvastatin disrupts reproduction and population growth of the amphipod Gammarus locusta at the ng/L range. Aquat. Toxicol. 2014, 155, 337–347. [Google Scholar] [CrossRef]
- Ribeiro, S.; Torres, T.; Martins, R.; Santos, M.M. Toxicity screening of diclofenac, propranolol, sertraline and simvastatin using Danio rerio and Paracentrotus lividus embryo bioassays. Ecotoxicol. Environ. Saf. 2015, 114, 67–74. [Google Scholar] [CrossRef] [Green Version]
- Dussault, È.B.; Balakrishnan, V.K.; Sverko, E.; Solomon, K.R.; Sibley, P.K. Toxicity of human pharmaceuticals and personal care products to benthic invertebrates. Environ. Toxicol. Chem. Int. J. 2008, 27, 425–432. [Google Scholar] [CrossRef]
- Costa, A.P.; Silva, D.A.; Rodrigues, A.C.; Marques, C.R.; Soares, A.M.; Rocha, R.J. Species-specific oxidative stress responses and cellular energy allocation after coral shipping. Aquac. Rep. 2021, 19, 100623. [Google Scholar] [CrossRef]
- Barata, C.; Baird, D.J. Determining the ecotoxicological mode of action of chemicals from measurements made on individuals: Results from instar-based tests with Daphnia magna Straus. Aquat. Toxicol. 2000, 48, 195–209. [Google Scholar] [CrossRef]
- OECD. Test No. 202: Daphnia sp. Acute Immobilisation Test, OECD Guidelines for the Testing of Chemicals; Section 2; OECD Publishing: Paris, France, 2004. [Google Scholar] [CrossRef]
- Pérez, S.; Beiras, R. The mysid Siriella Armata as a model organism in marine ecotoxicology: Comparative acute toxicity sensitivity with Daphnia magna. Ecotoxicology 2010, 19, 196–206. [Google Scholar] [CrossRef]
- Castro, B.B.; Freches, A.; Rodrigues, M.; Nunes, B.; Antunes, S. Transgenerational effects of toxicants: An extension of the daphnia 21-day chronic assay? Arch. Environ. Contam. Toxicol. 2018, 74, 616–626. [Google Scholar] [CrossRef]
- Kim, H.J.; Koedrith, P.; Seo, Y.R. Ecotoxicogenomic approaches for understanding molecular mechanisms of environmental chemical toxicity using aquatic invertebrate, Daphnia model organism. Int. J. Mol. Sci. 2015, 16, 12261–12287. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.-Y.; Choi, B.-S.; Kim, M.-S.; Park, J.C.; Jeong, C.-B.; Han, J.; Lee, J.-S. The genome of the freshwater water flea Daphnia magna: A potential use for freshwater molecular ecotoxicology. Aquat. Toxicol. 2019, 210, 69–84. [Google Scholar] [CrossRef]
- OECD. Test No. 211: Daphnia Magna Reproduction Test, OECD Guidelines for the Testing of Chemicals; Section 2; OECD Publishing: Paris, France, 2012. [Google Scholar] [CrossRef] [Green Version]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, S.; Li, H.; Dang, Y.; Liu, C. Effects of triphenyl phosphate on growth, reproduction and transcription of genes of Daphnia magna. Aquat. Toxicol. 2018, 195, 58–66. [Google Scholar] [CrossRef]
- Li, J.; Li, H.; Lin, D.; Li, M.; Wang, Q.; Xie, S.; Zhang, Y.; Liu, F. Effects of butyl benzyl phthalate exposure on Daphnia magna growth, reproduction, embryonic development and transcriptomic responses. J. Hazard. Mater. 2021, 404, 124030. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Li, C.Y.-T.; Kong, A.-N.T. Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch. Pharmacal Res. 2005, 28, 249–268. [Google Scholar] [CrossRef] [PubMed]
- Szakács, G.; Váradi, A.; Özvegy-Laczka, C.; Sarkadi, B. The role of ABC transporters in drug absorption, distribution, metabolism, excretion and toxicity (ADME–Tox). Drug Discov. Today 2008, 13, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B.; Gutteridge, J.M. Free Radicals in Biology and Medicine; Oxford University Press: New York, NY, USA, 2015. [Google Scholar] [CrossRef]
- Bhabak, K.P.; Mugesh, G. Functional mimics of glutathione peroxidase: Bioinspired synthetic antioxidants. Acc. Chem. Res. 2010, 43, 1408–1419. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.D.; Flanagan, J.U.; Jowsey, I.R. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 2005, 45, 51–88. [Google Scholar] [CrossRef]
- Hiruma, K.; Kaneko, K. Hormonal regulation of insect metamorphosis with special reference to juvenile hormone biosynthesis. Curr. Top. Dev. Biol. 2013, 103, 73–100. [Google Scholar] [CrossRef]
- Miyakawa, H.; Sato, T.; Song, Y.; Tollefsen, K.E.; Iguchi, T. Ecdysteroid and juvenile hormone biosynthesis, receptors and their signaling in the freshwater microcrustacean Daphnia. J. Steroid Biochem. Mol. Biol. 2018, 184, 62–68. [Google Scholar] [CrossRef] [Green Version]
- Riddiford, L.M. How does juvenile hormone control insect metamorphosis and reproduction? Gen. Comp. Endocrinol. 2012, 179, 477–484. [Google Scholar] [CrossRef]
- Liu, S.; Li, K.; Gao, Y.; Liu, X.; Chen, W.; Ge, W.; Feng, Q.; Palli, S.R.; Li, S. Antagonistic actions of juvenile hormone and 20-hydroxyecdysone within the ring gland determine developmental transitions in Drosophila. Proc. Natl. Acad. Sci. USA 2018, 115, 139–144. [Google Scholar] [CrossRef] [Green Version]
- Miyakawa, H.; Toyota, K.; Hirakawa, I.; Ogino, Y.; Miyagawa, S.; Oda, S.; Tatarazako, N.; Miura, T.; Colbourne, J.K.; Iguchi, T. A mutation in the receptor Methoprene-tolerant alters juvenile hormone response in insects and crustaceans. Nat. Commun. 2013, 4, 1856. [Google Scholar] [CrossRef] [Green Version]
- Laufer, H.; Biggers, W.J. Unifying concepts learned from methyl farnesoate for invertebrate reproduction and post-embryonic development. Am. Zool. 2001, 41, 442–457. [Google Scholar] [CrossRef] [Green Version]
- Reddy, P.R.; Nagaraju, G.P.C.; Reddy, P.S. Involvement of methyl farnesoate in the regulation of molting and reproduction in the freshwater crab Oziotelphusa senex. J. Crustacean Biol. 2004, 24, 511–515. [Google Scholar] [CrossRef]
- Mak, A.S.C.; Choi, C.L.; Tiu, S.H.K.; Hui, J.H.L.; He, J.G.; Tobe, S.S.; Chan, S.M. Vitellogenesis in the red crab Charybdis Feriatus: Hepatopancreas-specific expression and farnesoic acid stimulation of vitellogenin gene expression. Mol. Reprod. Dev. Inc. Gamete Res. 2005, 70, 288–300. [Google Scholar] [CrossRef]
- LeBlanc, G.A. Crustacean endocrine toxicology: A review. Ecotoxicology 2007, 16, 61–81. [Google Scholar] [CrossRef]
- Wang, P.; Ng, Q.X.; Zhang, H.; Zhang, B.; Ong, C.N.; He, Y. Metabolite changes behind faster growth and less reproduction of Daphnia similis exposed to low-dose silver nanoparticles. Ecotoxicol. Environ. Saf. 2018, 163, 266–273. [Google Scholar] [CrossRef]
- Schilling, O.; Biniossek, M.L.; Mayer, B.; Elsässer, B.; Brandstetter, H.; Goettig, P.; Stenman, U.-H.; Koistinen, H. Specificity profiling of human trypsin-isoenzymes. Biol. Chem. 2018, 399, 997–1007. [Google Scholar] [CrossRef]
- Esteghamat, F.; Broughton, J.S.; Smith, E.; Cardone, R.; Tyagi, T.; Guerra, M.; Szabó, A.; Ugwu, N.; Mani, M.V.; Azari, B. CELA2A mutations predispose to early-onset atherosclerosis and metabolic syndrome and affect plasma insulin and platelet activation. Nat. Genet. 2019, 51, 1233–1243. [Google Scholar] [CrossRef]
- Ratcliffe, D.R.; Iqbal, J.; Hussain, M.M.; Cramer, E.B. Fibrillar collagen type I stimulation of apolipoprotein B secretion in Caco-2 cells is mediated by β1 integrin. Biochim. et Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2009, 1791, 1144–1154. [Google Scholar] [CrossRef] [Green Version]
- Basson, M.D. Invited research review: Cell-matrix interactions in the gut epithelium. Surgery 2003, 133, 263–267. [Google Scholar] [CrossRef]


| Category | Pathway | p Value | Up-Gene | Down-Gene |
|---|---|---|---|---|
| C-vs-L | ||||
| Metabolism | Glutathione metabolism | 1.6 × 10−3 | gclc, gss, gst | - |
| Metabolism | Insect hormone biosynthesis | 4.4 × 10−3 | cyp15a1_c1, jhamt | - |
| Metabolism | Drug metabolism—cytochrome P450 | 4.0 × 10−4 | hpgds, ugt, | - |
| Metabolism | Drug metabolism—other enzymes | 3.4 × 10−2 | ugt | - |
| Metabolism | Metabolism of xenobiotics by cytochrome P450 | 4.9 × 10−4 | gst, ugt | - |
| C-vs-M | ||||
| Metabolism | Insect hormone biosynthesis | 8.5 × 10−3 | cyp15a1_c1, jhamt | - |
| C-vs-H | ||||
| Metabolism | Glutathione metabolism | 5.8 × 10−3 | gclc, ggt1_5, gshb, gst | anpep |
| Metabolism | Insect hormone biosynthesis | 5.2 × 10−4 | cyp15a1_c1, jhamt | cyp18a1 |
| Organismal Systems | Pancreatic secretion | 2.7 × 10−3 | - | prss1_2_3, cela2, clca2 |
| Organismal Systems | Protein digestion and absorption | 6.36 × 10−7 | - | cela2, col1a, col4a, col5a,prss1_2_3, |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lei, Y.; Guo, J.; Chen, Q.; Mo, J.; Tian, Y.; Iwata, H.; Song, J. Transcriptomic Alterations in Water Flea (Daphnia magna) following Pravastatin Treatments: Insect Hormone Biosynthesis and Energy Metabolism. Toxics 2022, 10, 110. https://doi.org/10.3390/toxics10030110
Lei Y, Guo J, Chen Q, Mo J, Tian Y, Iwata H, Song J. Transcriptomic Alterations in Water Flea (Daphnia magna) following Pravastatin Treatments: Insect Hormone Biosynthesis and Energy Metabolism. Toxics. 2022; 10(3):110. https://doi.org/10.3390/toxics10030110
Chicago/Turabian StyleLei, Yuan, Jiahua Guo, Qiqi Chen, Jiezhang Mo, Yulu Tian, Hisato Iwata, and Jinxi Song. 2022. "Transcriptomic Alterations in Water Flea (Daphnia magna) following Pravastatin Treatments: Insect Hormone Biosynthesis and Energy Metabolism" Toxics 10, no. 3: 110. https://doi.org/10.3390/toxics10030110
APA StyleLei, Y., Guo, J., Chen, Q., Mo, J., Tian, Y., Iwata, H., & Song, J. (2022). Transcriptomic Alterations in Water Flea (Daphnia magna) following Pravastatin Treatments: Insect Hormone Biosynthesis and Energy Metabolism. Toxics, 10(3), 110. https://doi.org/10.3390/toxics10030110








