Purification and Characterization of Pathogenesis Related Class 10 Panallergens
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Sera
2.2. Cloning
2.3. Expression and Purification of Recombinant PR-10 Proteins
2.4. Liquid Chromatography-Mass Spectrometry (LC-MSMS)
2.5. IgE ELISA
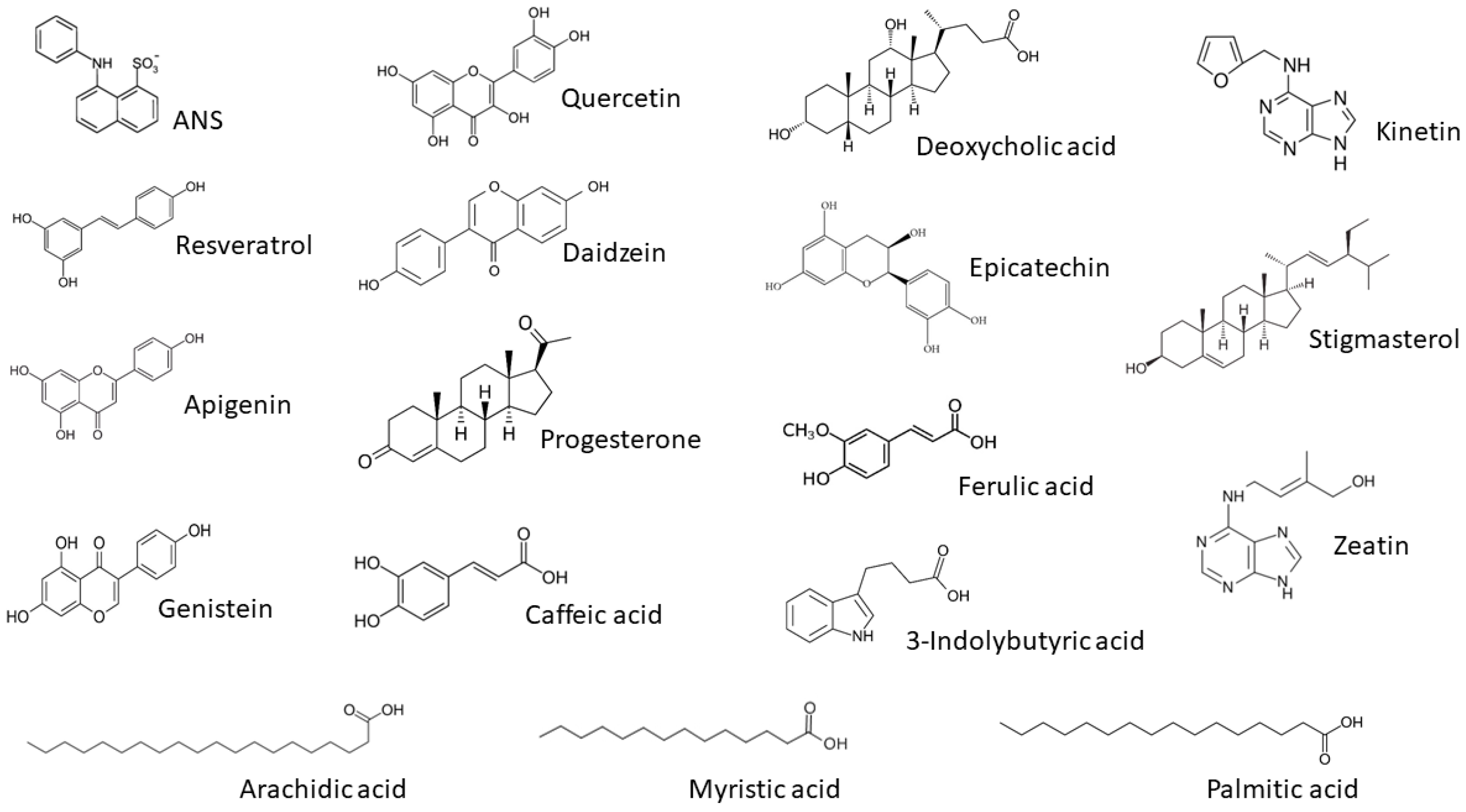
2.6. ANS Displacement Assay
2.7. t-Resveratrol Binding
2.8. Far UV Circular Dichroism
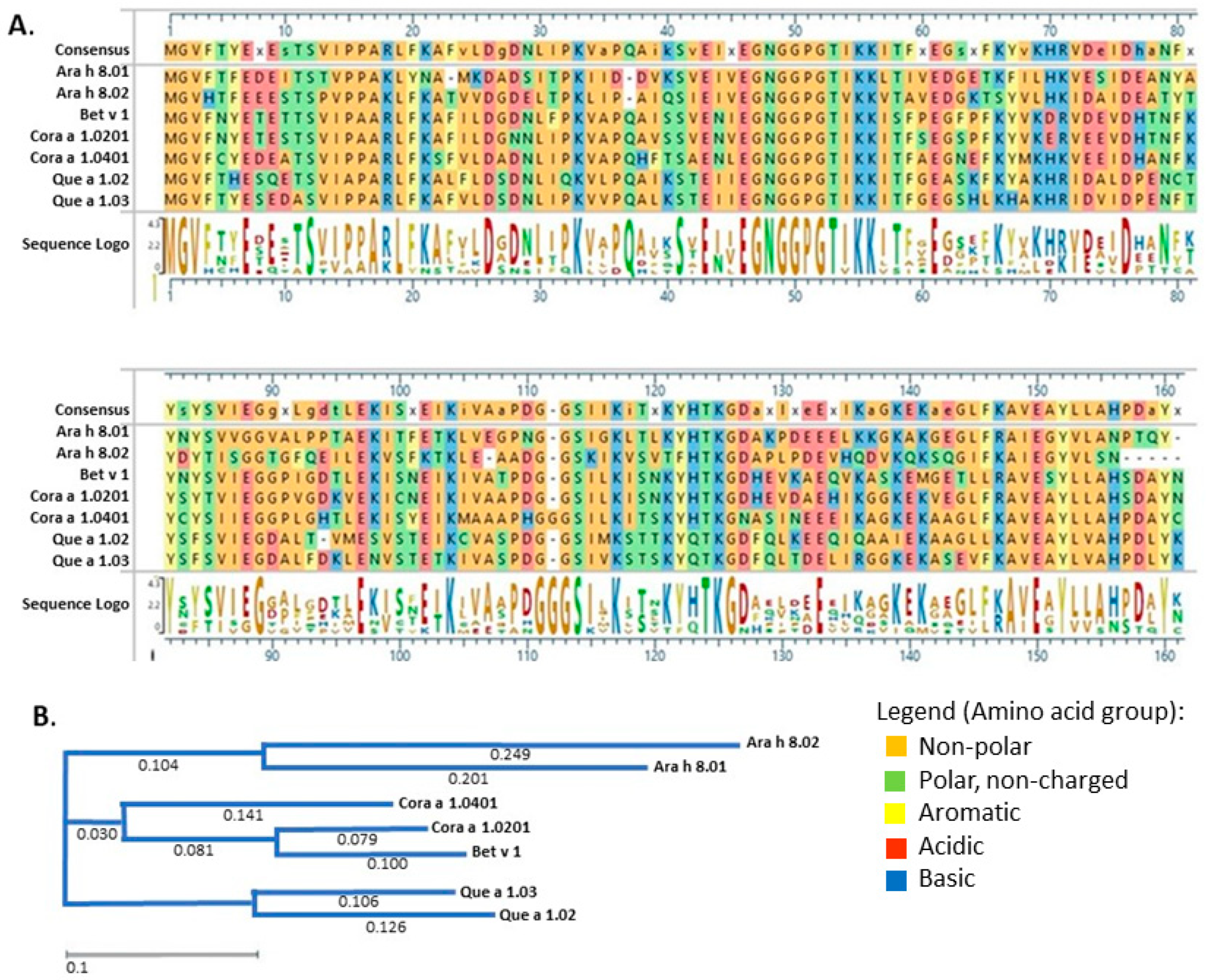
3. Results
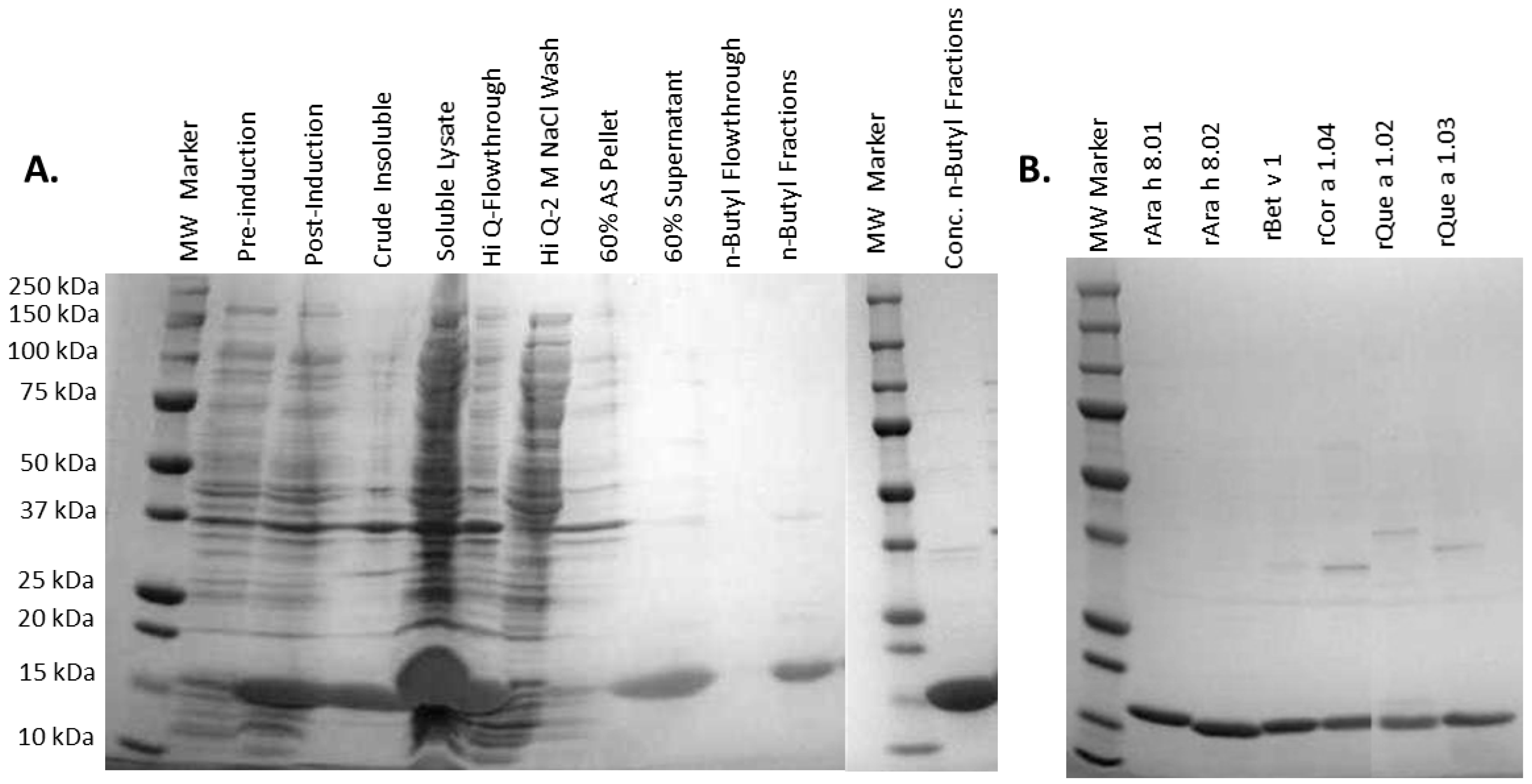
3.1. Recombinant Protein Expression and Purification
3.2. IgE Binding of PR-10 Proteins
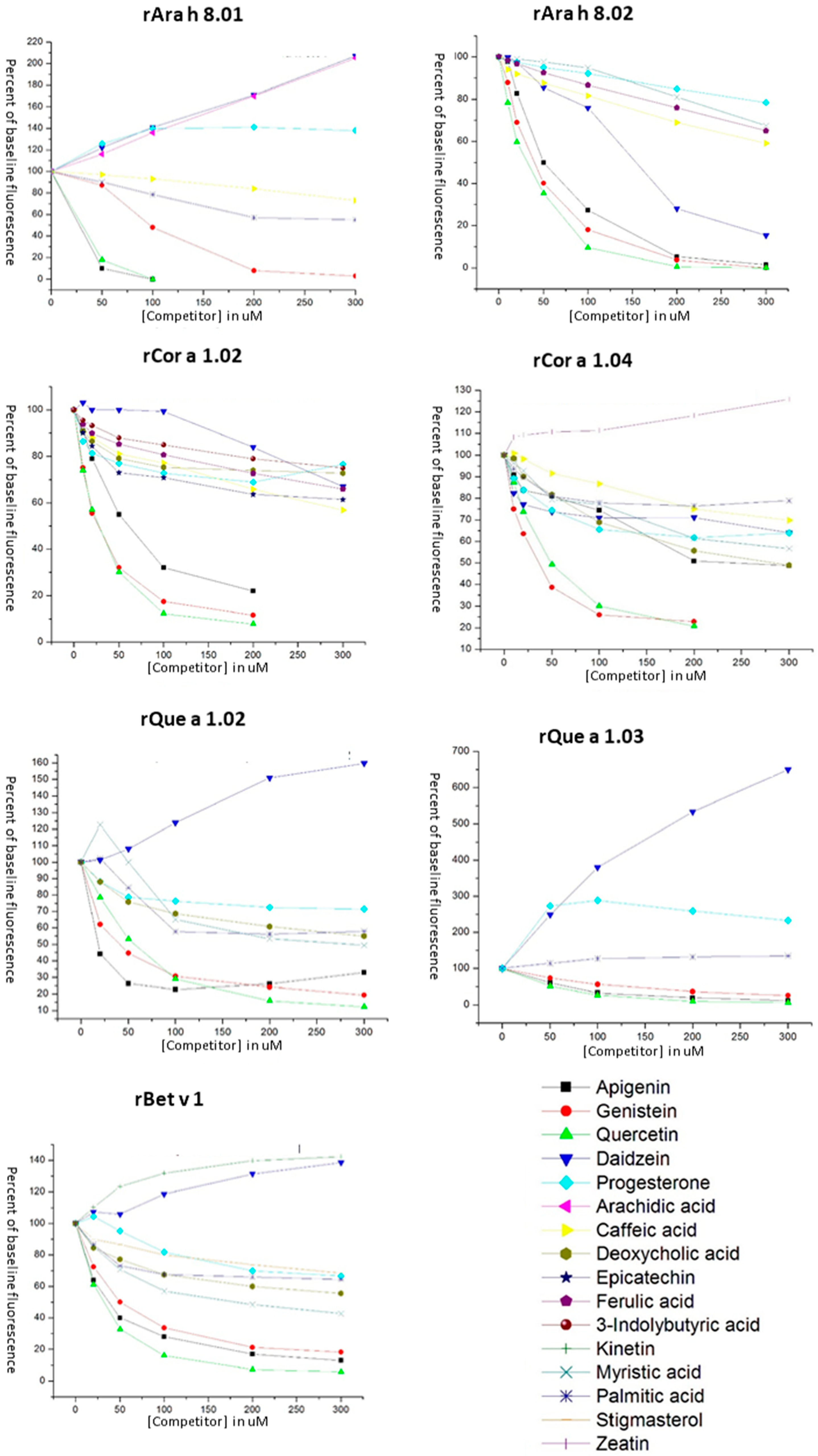
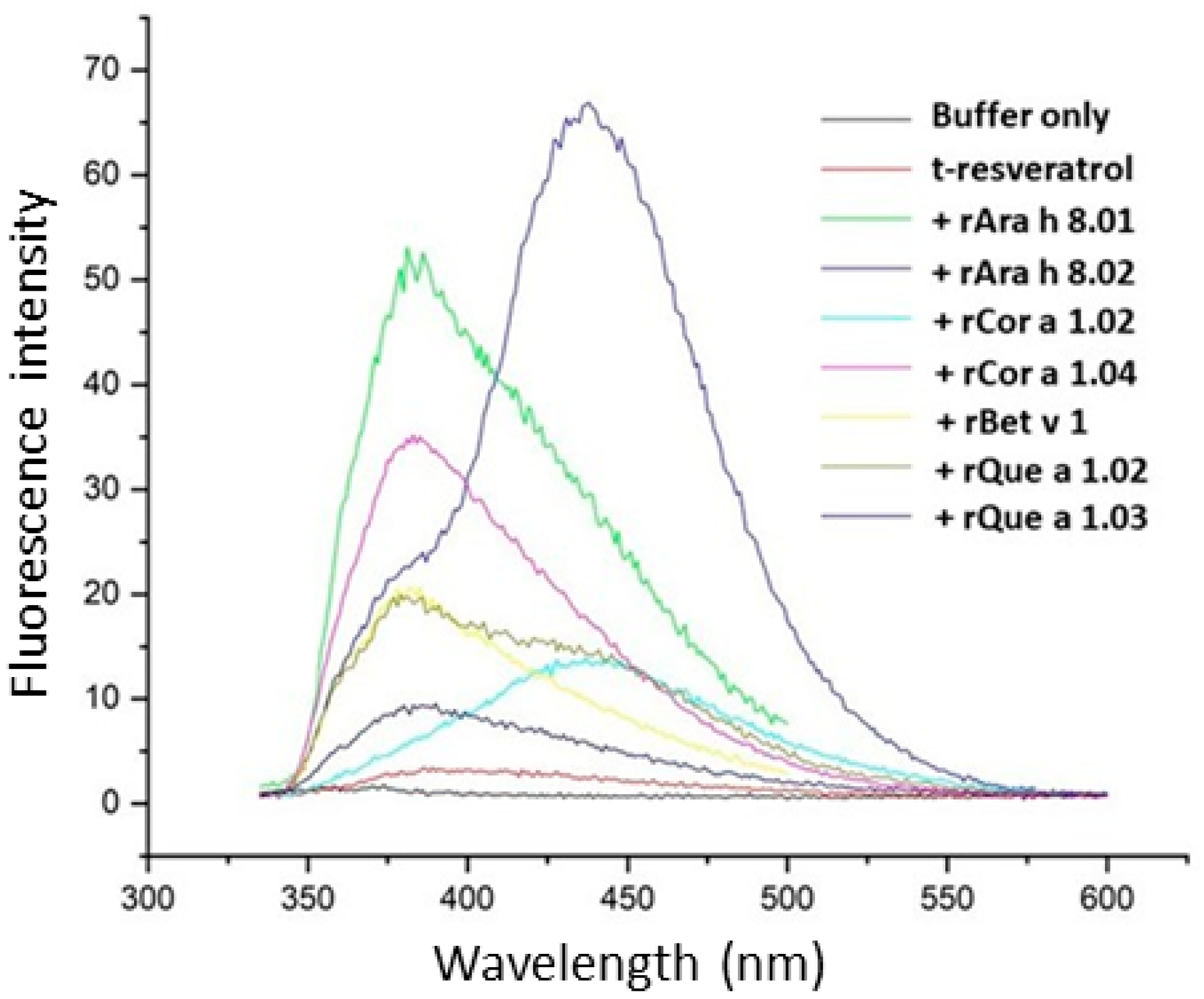
3.3. Ligand Binding of PR-10 Proteins
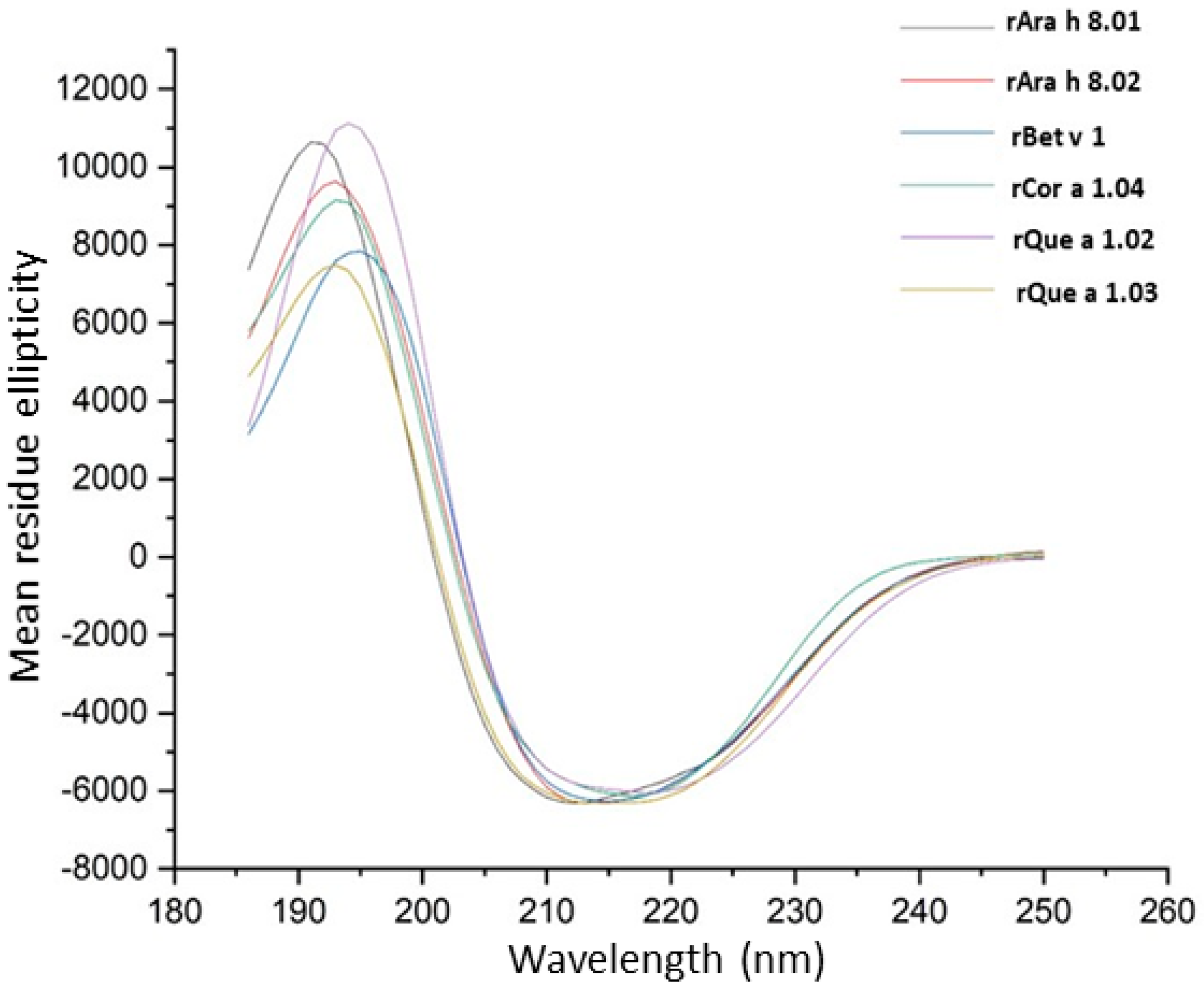
3.4. Far UV Circular Dichroism Spectroscopy
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kondo, Y.; Urisu, A. Oral allergy syndrome. Allergol. Int. 2009, 58, 485–491. [Google Scholar] [CrossRef]
- Fritsch, R.; Wiedermann, U.; Jahn-Schmid, B.; Krebitz, M.; Breiteneder, H.; Kraft, D.; Ebner, C. Bet v 1, the major birch pollen allergen, and Mal d 1, the major apple allergen, cross-react at the level of allergen-specific T helper cells. J. Allergy Clin. Immunol. 1998, 102, 679–686. [Google Scholar] [CrossRef]
- Scheurer, S.; Son, D.Y.; Boehm, M.; Karamloo, F.; Franke, S.; Hoffmann, A.; Haustein, D.; Vieths, S. Cross-reactivity and epitope analysis of Pru a 1, the major cherry allergen. Mol. Immunol. 1999, 36, 155–167. [Google Scholar] [CrossRef]
- Mittag, D.; Akkerdaas, J.; Ballmer-Weber, B.K.; Vogel, L.; Wensing, M.; Becker, W.M.; Koppelman, S.J.; Knulst, A.C.; Helbling, A.; Hefle, S.L.; et al. Ara h 8, a Bet v 1-homologous allergen from peanut, is a major allergen in patients with combined birch pollen allergy. J. Allergy Clin. Immunol. 2004, 114, 1410–1417. [Google Scholar] [CrossRef] [PubMed]
- Bollen, M.A.; Garcia, A.; Cordewener, J.H.; Wichers, H.J.; Helsper, J.P.; Savelkoul, H.F.; van Boekel, M.A. Purification and characterization of natural Bet v 1 from birch pollen and related allergens from carrot and celery. Mol. Nutr. Food Res. 2007, 51, 1527–1536. [Google Scholar] [CrossRef]
- Karamloo, F.; Scheurer, S.; Wangorsch, A.; May, S.; Haustein, D.; Vieths, S. Pyr c 1, the major allergen from pear (Pyrus communis), is a new member of the Bet v 1 allergen family. J. Chromatogr. B 2001, 756, 281–293. [Google Scholar] [CrossRef]
- Berkner, H.; Neudecker, P.; Mittag, D.; Ballmer-Weber, B.K.; Schweimer, K.; Vieths, S.; Rösch, P. Cross-reactivity of pollen and food allergens: Soybean Gly m 4 is a member of the Bet v 1 superfamily and closely resembles yellow lupine proteins. Biosci. Rep. 2009, 29, 183–192. [Google Scholar] [CrossRef]
- Ipsen, H.; Lowenstein, H. Isolation and immunochemical characterization of the major allergen of birch pollen (Betula verrucosa). J. Allergy Clin. Immunol. 1983, 72, 150–159. [Google Scholar] [CrossRef]
- Breiteneder, H.; Ebner, C. Molecular and biochemical classification of plant-derived food allergens. J. Allergy Clin. Immunol. 2000, 106, 27–36. [Google Scholar] [CrossRef]
- Gajhede, M.; Osmark, P.; Poulsen, F.M.; Ipsen, H.; Larsen, J.N.; Joost van Neerven, R.J.; Schou, C.; Løwenstein, H.; Spangfort, M.D. X-ray and NMR structure of Bet v 1, the origin of birch pollen allergy. Nat. Struct. Biol. 1996, 3, 1040–1045. [Google Scholar] [CrossRef]
- Vieths, S.; Reindl, J.; Müller, U.; Hoffmann, A.; Haustein, D. Digestibility of peanut and hazelnut allergens investigated by a simple in vitro procedure. Eur. Food Res. Technol. 1999, 209, 379–388. [Google Scholar] [CrossRef]
- Liu, J.; Ekramoddoullah, A.K.M.; Yu, X. Differential expression of multiple PR10 proteins in western white pine following wounding, fungal infection and cold-hardening. Physiol. Plant. 2003, 119, 544–553. [Google Scholar] [CrossRef]
- Bufe, A.; Spangfort, M.D.; Kahlert, H.; Schlaak, M.; Becker, W.M. The major birch pollen allergen, Bet v 1, shows ribonuclease activity. Planta 1996, 199, 413–415. [Google Scholar] [CrossRef] [PubMed]
- Zubini, P.; Zambelli, B.; Musiani, F.; Ciurli, S.; Bertolini, P.; Baraldi, E. The RNA hydrolysis and the cytokinin binding activities of PR-10 proteins are differently performed by two isoforms of the Pru p 1 peach major allergen and are possibly functionally related. Plant Physiol. 2009, 150, 1235–1247. [Google Scholar] [CrossRef]
- Fernandes, H.; Michalska, K.; Sikorski, M.; Jaskolski, M. Structural and functional aspects of PR-10 proteins. FEBS J. 2013, 280, 1169–1199. [Google Scholar] [CrossRef]
- Andrade, L.B.S.; Oliveira, A.S.; Ribeiro, J.K.C.; Kiyota, S.; Vasconcelos, I.M.; de Oliveira, J.T.A.; de Sales, M.P. Effects of a novel Pathogenesis-Related Class 10 (PR-10) protein from Crotalaria pallida roots with papain inhibitory activity against root-knot nematode Meloidogyne incognita. J. Agric. Food Chem. 2010, 58, 4145–4152. [Google Scholar] [CrossRef]
- Mogensen, J.E.; Wimmer, R.; Larsen, J.N.; Spangfort, M.D.; Otzen, D.E. The major birch allergen, Bet v 1, shows affinity for a broad spectrum of physiological ligands. J. Biol. Chem. 2002, 277, 23684–23692. [Google Scholar] [CrossRef]
- Hurlburt, B.K.; Offermann, L.R.; McBride, J.K.; Majorek, K.A.; Maleki, S.J.; Chruszcz, M. Structure and function of the peanut panallergen Ara h 8. J. Biol. Chem. 2013, 288, 36890–36901. [Google Scholar] [CrossRef]
- Mogensen, J.E.; Ferreras, M.; Wimmer, R.; Petersen, S.V.; Enghild, J.J.; Otzen, D.E. The major allergen from birch tree pollen, Bet v 1, binds and permeabilizes membranes. Biochemistry 2007, 46, 3356–3365. [Google Scholar] [CrossRef]
- Fernandes, H.; Pasternak, O.; Bujacz, G.; Bujacz, A.; Sikorski, M.M.; Jaskolski, M. Lupinus luteus pathogenesis-related protein as a reservoir for cytokinin. J. Mol. Biol. 2008, 378, 1040–1051. [Google Scholar] [CrossRef]
- Fernandes, H.; Bujacz, A.; Bujacz, G.; Jelen, F.; Jasinski, M.; Kachlicki, P.; Otlewski, J.; Sikorski, M.M.; Jaskolski, M. Cytokinin-induced structural adaptability of a Lupinus luteus PR-10 protein. FEBS J. 2009, 276, 1596–1609. [Google Scholar] [CrossRef] [PubMed]
- Sliwiak, J.; Dauter, Z.; Jaskolski, M. Crystal structure of Hyp-1, a Hypericum perforatum PR-10 protein, in complex with melatonin. Front. Plant Sci. 2016, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kofler, S.; Asam, C.; Eckhard, U.; Wallner, M.; Ferreira, F.; Brandstetter, H. Crystallographically mapped ligand binding differs in high and low IgE binding isoforms of birch pollen allergen Bet v 1. J. Mol. Biol. 2012, 422, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Casañal, A.; Zander, U.; Muñoz, C.; Dupeux, F.; Luque, I.; Botella, M.A.; Schwab, W.; Valpuesta, V.; Marquez, J.A. The strawberry Pathogenesis-related 10 (PR-10) Fra a proteins control flavonoid biosynthesis by binding to metabolic intermediates. J. Biol. Chem. 2013, 288, 35322–35332. [Google Scholar] [CrossRef] [PubMed]
- Ferreyra, M.L.F.; Rius, S.P.; Casati, P. Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 2012, 3, 1–15. [Google Scholar]
- Rijavec, T.; Dermastia, M. Cytokinins and their function in developing seeds. Acta Chim. Slov. 2010, 57, 617–629. [Google Scholar]
- Tsujishita, Y.; Hurley, J.H. Structure and lipid transport mechanism of a StAR-related domain. Nat. Struct. Biol. 2000, 7, 408–414. [Google Scholar]
- Scala, E.; Abeni, D.; Cecchi, L.; Guerra, E.C.; Locanto, M.; Pirrotta, L.; Giani, M.; Asero, R. Molecular recognition profiles and clinical patterns of PR-10 sensitization in a birch-free Mediterranean area. Int. Arch. Allergy Immunol. 2017, 173, 138–146. [Google Scholar] [CrossRef]
- Blankestijn, M.A.; Knulst, A.C.; Knol, E.F.; Thuy-My, L.; Rockmann, H.; Otten, H.G.; Kleman, R.J.B. Sensitization to PR-10 proteins is indicative of distinctive sensitization patterns in adults with a suspected food allergy. Clin. Transl. Allergy 2017, 7, 1–8. [Google Scholar] [CrossRef]
- Jeong, K.Y.; Son, M.; Park, J.H.; Park, K.H.; Park, H.J.; Lee, J.H.; Hong, C.S.; Park, J.W. Cross-reactivity between oak and birch pollens in Korean tree pollinosis. J. Korean Med. Sci. 2016, 31, 1202–1207. [Google Scholar] [CrossRef]
- Petersen, A.; Rennert, S.; Kull, S.; Becker, W.M.; Notbohm, H.; Goldmann, T.; Jappe, U. Roasting and lipid binding provide allergenic and proteolytic stability to the peanut allergen Ara h 8. J. Biol. Chem. 2014, 395, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Hillis, W.E.; Ishikura, N. The chromatographic and spectral properties of stilbene derivatives. J. Chromatogr. 1968, 32, 323–336. [Google Scholar] [CrossRef]
- Micsonai, A.; Wien, F.; Kernya, L.; Lee, Y.H.; Goto, Y.; Réfrégiers, M.; Kardos, J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. USA 2015, 112, 3095–3103. [Google Scholar] [CrossRef] [PubMed]
- José Arnau, J.; Lauritzen, C.; Petersen, G.E.; Pedersen, J. Current strategies for the use of affinity tags and tag removal for the purification of recombinant proteins. Protein Expr. Purif. 2006, 48, 1–13. [Google Scholar] [CrossRef]
- Hurlburt, B.K.; McBride, J.K.; Nesbit, J.B.; Ruan, S.; Maleki, S.J. Purification of recombinant peanut allergen Ara h 1 and comparison of IgE binding to the natural protein. Foods 2014, 3, 642–657. [Google Scholar] [CrossRef]
- Ferreira, F.; Hirtenlehner, K.; Jilek, A.; Godnik-Var, J.; Breiteneder, H.; Grimm, R.; Hoffmann-Sommergruber, K.; Scheiner, O.; Kraft, D.; Breitenbach, M.; et al. Dissection of immunoglobin E and T lymphocyte reactivity of isoforms of the major birch pollen allergen Bet v 1: Potential use of hypoallergenic isoforms for immunotherapy. J. Exp. Med. 1996, 183, 599–609. [Google Scholar] [CrossRef]
- Tscheppe, A.; Breiteneder, H. Recombinant allergens in structural biology, diagnosis, and immunotherapy. Int. Arch. Allergy Immunol. 2017, 172, 187–202. [Google Scholar] [CrossRef]
- Walsh, G. Post-translational modifications of protein biopharmaceuticals. Drug Discov. Today 2010, 15, 773–780. [Google Scholar] [CrossRef]
- Johnson, P.E.; Sayers, R.L.; Gethings, L.A.; Balasundaram, A.; Marsh, J.T.; Langridge, J.I.; Mills, E.N. Quantitative proteomic profiling of peanut allergens in food ingredients used for oral food challenges. Anal. Chem. 2016, 88, 5689–5695. [Google Scholar] [CrossRef]
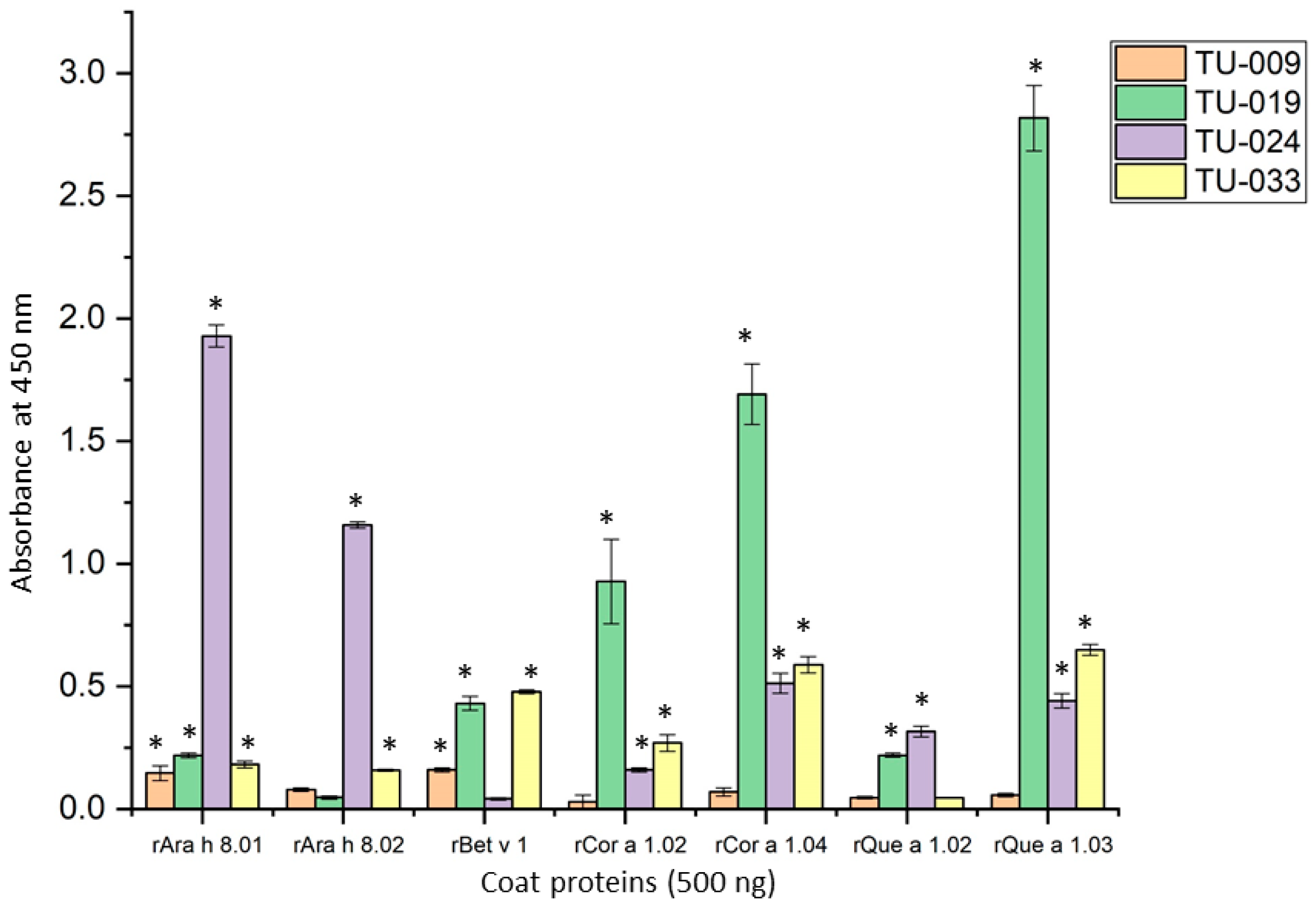
| Patient ID | Age | Sex | Ara h 8.01 | Bet v 1 | Cor a 1.04 | Known Food Allergies |
|---|---|---|---|---|---|---|
| TU-009 | 56 | M | 1.04 | 2.76 | 0.63 | Walnut, pecan, almond, brazil nut, pine nut, hazelnut, macadamia nut, apricot, apple, pear, plum, similar fruits |
| TU-019 | 36 | F | 1.61 | 84.7 | 35.0 | Soy, peach, pear, apple, cherry, pine nut |
| TU-024 | 24 | F | 8.04 | 7.5 | 7.47 | Cashew, peanut, walnut, pecan, other tree nuts |
| TU-033 | 26 | F | 1.28 | 14.7 | 5.14 | Peanut, brazil nut, other tree nuts, mango, peach, plum, melons |
| Protein Name | Calculated MW (Da) | Calculated PI | [Ammonium Sulfate] |
|---|---|---|---|
| rAra h 8.01 | 16,952 | 4.955 | 60% of saturation |
| rAra h 8.02 | 16,412 | 5.003 | 50% of saturation |
| rBet v 1.01 | 17,571 | 5.411 | 60% of saturation |
| rCor a 1.02 | 17,417 | 5.931 | 60% of saturation |
| rCor a 1.04 | 17,581 | 6.448 | 50% of saturation |
| rQue a 1.02 | 17,303 | 5.741 | 45% of saturation |
| rQue a 1.03 | 17,478 | 5.402 | 40% of saturation |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
McBride, J.K.; Cheng, H.; Maleki, S.J.; Hurlburt, B.K. Purification and Characterization of Pathogenesis Related Class 10 Panallergens. Foods 2019, 8, 609. https://doi.org/10.3390/foods8120609
McBride JK, Cheng H, Maleki SJ, Hurlburt BK. Purification and Characterization of Pathogenesis Related Class 10 Panallergens. Foods. 2019; 8(12):609. https://doi.org/10.3390/foods8120609
Chicago/Turabian StyleMcBride, Jane K., Hsiaopo Cheng, Soheila J. Maleki, and Barry K. Hurlburt. 2019. "Purification and Characterization of Pathogenesis Related Class 10 Panallergens" Foods 8, no. 12: 609. https://doi.org/10.3390/foods8120609
APA StyleMcBride, J. K., Cheng, H., Maleki, S. J., & Hurlburt, B. K. (2019). Purification and Characterization of Pathogenesis Related Class 10 Panallergens. Foods, 8(12), 609. https://doi.org/10.3390/foods8120609





