Antibiofilm Effect of DNase against Single and Mixed Species Biofilm
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture Maintenance
2.2. Biofilm Formation Assay
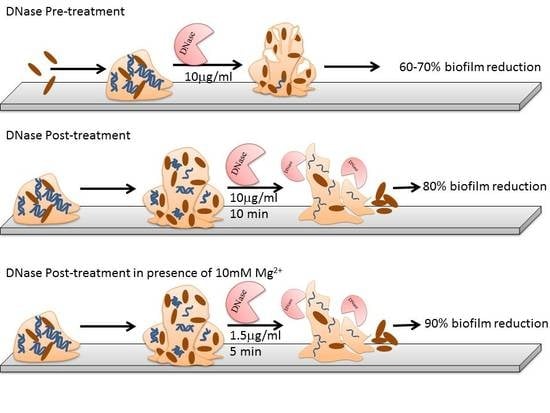
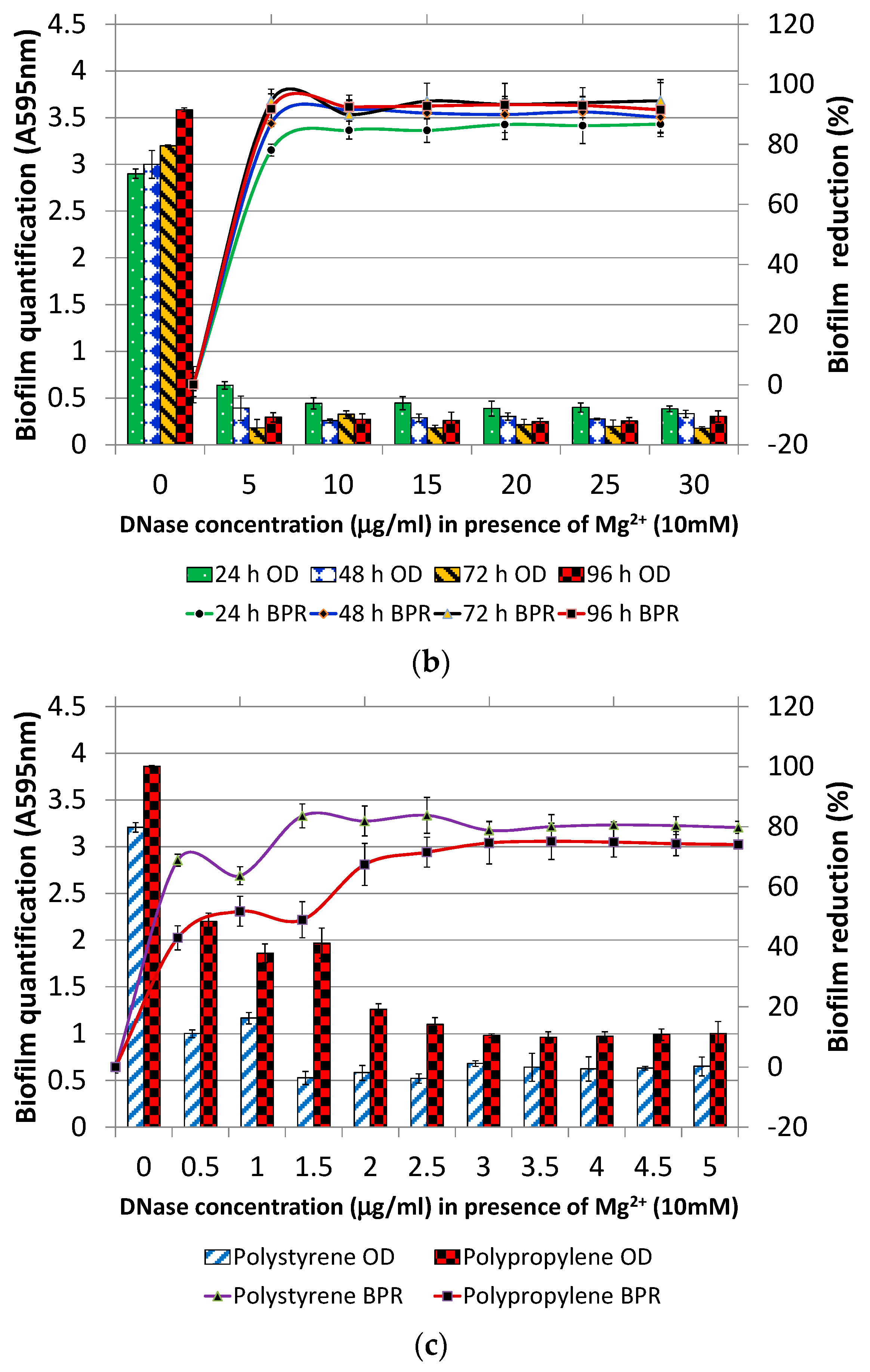
2.3. Optimization of DNase I Concentration for Pre-Treatment
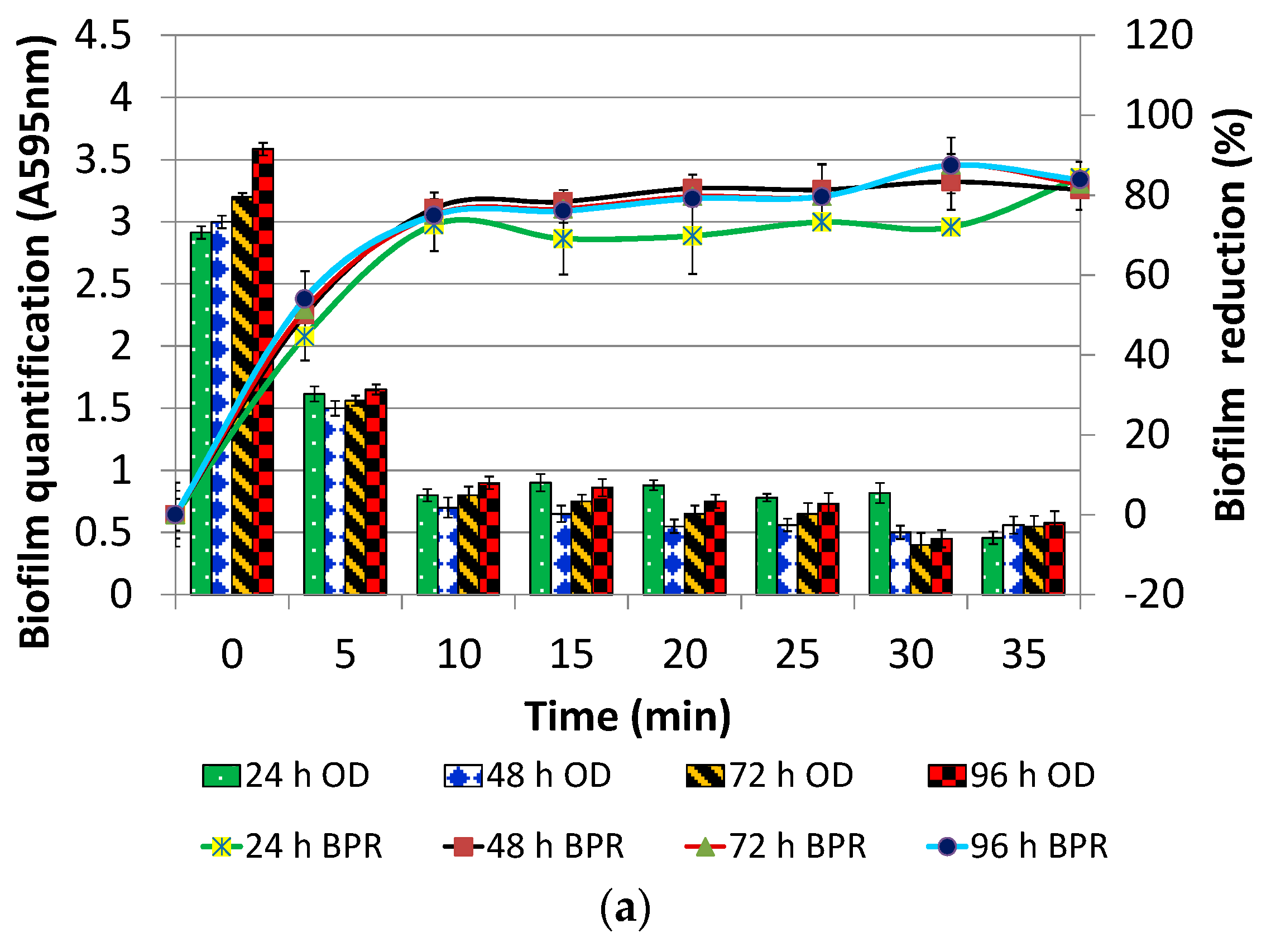
2.4. Optimization of Contact Time and Concentration of DNase I for Post Treatment
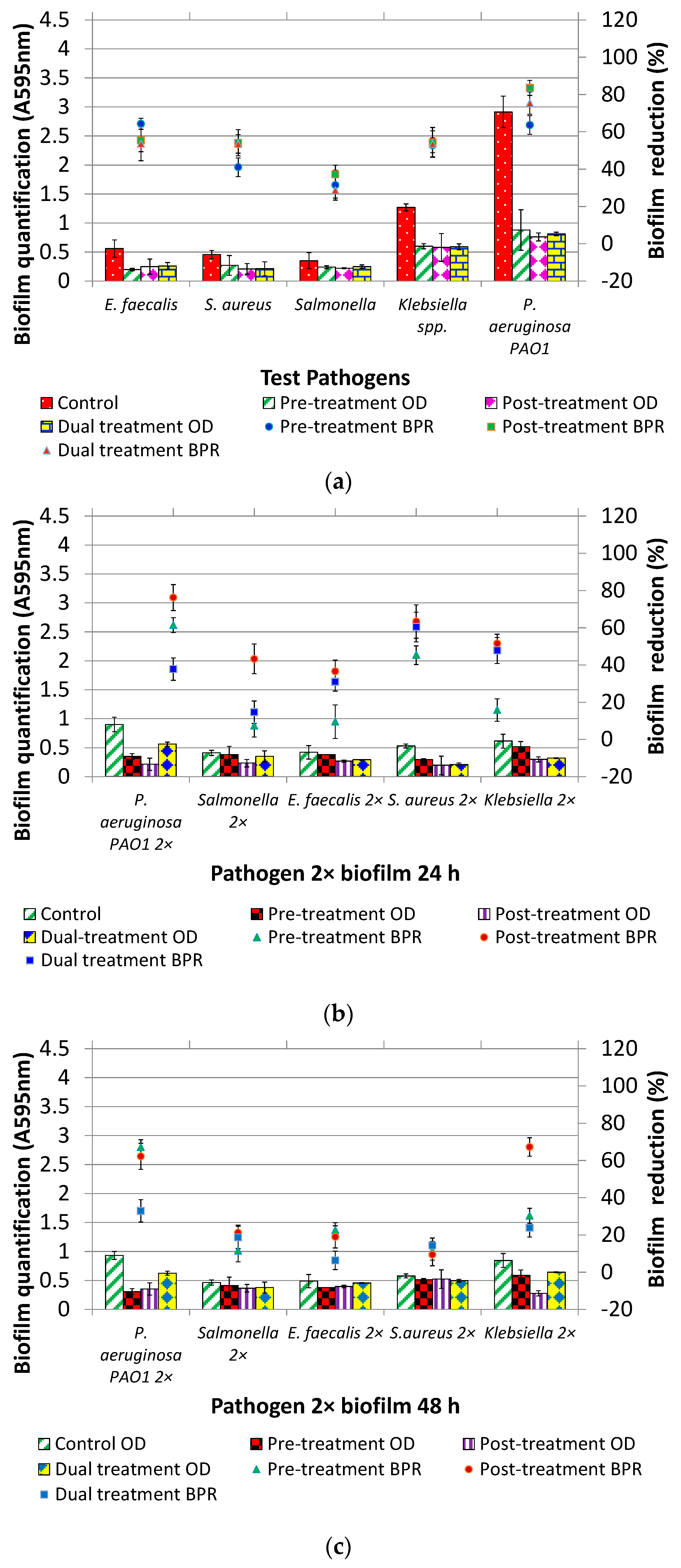
2.5. Pre-Treatment, Post-Treatment and Dual Treatment of Microbial Biofilms by DNase I
2.6. Pre-Treatment, Post-Treatment and Dual Treatment of Mixed Species Biofilm by DNase I
2.7. Statistical Analysis
3. Results
3.1. Optimization of Pre-Treatment and Post-Treatment
3.1.1. DNase I Concentration for Pre-Treatment
3.1.2. DNase I Contact Time for Post-Treatment
3.2. DNase I Treatment (Pre, Post and Dual) of Individual and Mixed Species Biofilms
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lebeaux, D.; Chauhan, A.; Rendueles, O.; Beloin, C. From in vitro to in vivo Models of Bacterial Biofilm-Related Infections. Pathogens 2013, 2, 288–356. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.P.; Singh, J. Antibiofilm strategies. In Biofilms in Bioengineering, 1st ed.; Simões, M., Mergulhão, F., Eds.; Nova Biomedical; Nova Science Publishers: Porto, Portugal, 2013; Volume 1, p. 363. ISBN 978-1-62948-161-6. [Google Scholar]
- Lasarre, B.; Federle, M.J. Exploiting Quorum Sensing to Confuse Bacterial. Pathogens 2013, 77, 73–111. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Schauder, S.; Potier, N.; Van Dorsselaer, A.; Pelczer, I.; Bassler, B.L.; Hughson, F.M. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 2002, 415, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Zhou, J.; Zhu, X.; Yu, S.; Chen, L.; Jin, H.; Cai, Z. Strain identification and quorum sensing inhibition characterization of marine-derived Rhizobium NAO1. R. Soc. Open Sci. 2017, 77, 73–111. [Google Scholar] [CrossRef]
- Pagedar, A.; Singh, J.; Batish, V.K. Efflux mediated adaptive and cross resistance to ciprofloxacin and benzalkonium chloride in Pseudomonas aeruginosa of dairy origin. J. Basic Microbiol. 2011, 51, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Pagedar, A.; Singh, J. Influence of physiological cell stages on biofilm formation by Bacillus cereus of dairy origin. Int. Dairy J. 2012, 23, 30–35. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J. Extracellular Polymeric Substances (EPS): Structural, Ecological and Technical aspects. In Encyclopedia of Environmental Microbiology; Bitton, G., Ed.; John Wiley & Sons: New York, NY, USA, 2002; pp. 1223–1231. [Google Scholar]
- Montanaro, L.; Poggi, A.; Visai, L.; Ravaioli, S.; Campoccia, D.; Speziale, P.; Arciola, C.R. Extracellular DNA in biofilms. Int. J. Artif. Organ. 2011, 34, 824–831. [Google Scholar] [CrossRef] [PubMed]
- Sahu, P.K.; Iyer, P.S.; Oak, A.M.; Pardesi, K.R.; Chopade, B. Characterization of eDNA from the Clinical Strain Acinetobacter baumannii AIIMS 7 and Its Role in Biofilm Formation. Sci. World J. 2012, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Whitchurch, C.B.; Tolker-Nielsen, T.; Ragas, P.C.; Mattick, J.S. Extracellular DNA required for bacterial biofilm formation. Science 2002, 295, 1487. [Google Scholar] [CrossRef] [PubMed]
- Fleming, D.; Rumbaugh, K. Approaches to Dispersing Medical Biofilms. Microorganisms 2017, 5, 15. [Google Scholar] [CrossRef] [PubMed]
- Beenken, K.E.; Spencer, H.; Griffin, L.M.; Smelter, S.M.; Camilli, A. Impact of extracellular nuclease production on the biofilm phenotype of Staphylococcus aureus under in vitro and in vivo conditions. Infect. Immun. 2012, 80, 1634–1638. [Google Scholar] [CrossRef] [PubMed]
- Gnanadhas, D.P.; Elango, M.; Datey, A. Chronic lung infection by Pseudomonas aeruginosa biofilm is cured by L-Methionine in combination with antibiotic therapy. Sci. Rep. 2015, 5, 16043. [Google Scholar] [CrossRef] [PubMed]
- Izano, E.; Amarante, M.; Kher, W.B.; Kaplan, J.B. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl. Environ. Microbiol. 2008, 74, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, J.B.; Lovetri, K.; Cardona, S.T.; Madhyastha, S.; Sadovskaya, I.; Jabbouri, S.; Izano, E.A. Recombinant human DNase I decreases biofilm and increases antimicrobial susceptibility in staphylococci. J. Antibiot. 2012, 65, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Klare, W.; Das, T.; Ibugo, A.; Buckle, E.; Manefield, M.; Manos, J. Glutathione-disrupted biofilms of clinical Pseudomonas aeruginosa Strains Exhibit an Enhanced Antibiotic Effect and a Novel Biofilm. Antimicrob. Agents Chemother. 2016, 60, 4539–4551. [Google Scholar] [CrossRef] [PubMed]
- Brown, H.L.; Reuter, M.; Hanman, K.; Betts, R.P.; Van Vliet, A.H.M. Prevention of biofilm formation and removal of existing biofilms by extracellular DNase of Campylobacter jejuni. PLoS ONE 2015, 10, e0121680. [Google Scholar] [CrossRef] [PubMed]
- Brown, H.L.; Hanman, K.; Reuter, M.; Betts, R.P.; van Vliet, A.H.M. Campylobacter jejuni biofilms contain extracellular DNA and are sensitive to DNase I treatment. Front. Microbiol. 2015, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Fuxman Bass, J.I.; Russo, D.M.; Gabelloni, M.L.; Geffner, J.R.; Giordano, M.; Catalano, M.; Zorreguieta, A.; Trevani, A.S. Extracellular DNA: A Major Proinflammatory Component of Pseudomonas aeruginosa Biofilms. J. Immunol. 2010, 184, 6386–6395. [Google Scholar] [CrossRef] [PubMed]
- Tetz, G.V.; Artemenko, N.K.; Tetz, V.V. Effect of DNase and antibiotics on biofilm characteristics. Antimicrob. Agents Chemother. 2009, 53, 1204–1209. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, U.T.; Burrows, L.L. DNase I and proteinase K impair Listeria monocytogenes biofilm formation and induce dispersal of pre-existing biofilms. Int. J. Food Microbiol. 2014, 187, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Fredheim, E.G.A.; Klingenberg, C.; Rohde, H.; Frankenberger, S.; Gaustad, P.; Flaegstad, T.; Sollid, J.E. Biofilm formation by Staphylococcus shaemolyticus. J. Clin. Microbiol. 2009, 47, 1172–1180. [Google Scholar] [CrossRef] [PubMed]
- Cavaliere, R.; Ball, J.L.; Turnbull, L.; Whitchurch, C.B. The biofilm matrix destabilizers, EDTA and DNaseI, enhance the susceptibility of nontypeable Hemophilus influenzae biofilms to treatment with ampicillin and ciprofloxacin. Microbiol. Open 2014, 3, 557–567. [Google Scholar] [CrossRef] [PubMed]
- Kawarai, T.; Narisawa, N.; Suzuki, Y.; Nagasawa, R.; Senpuku, H. Streptococcus mutans biofilm formation is dependent on extracellular DNA in primary low pH conditions. J. Oral Biosci. 2016, 58, 55–61. [Google Scholar] [CrossRef]
- Arslan, S.; Eyi, A.; Özdemir, F. Spoilage potentials and antimicrobial resistance of Pseudomonas spp. isolated from cheeses. J. Dairy Sci. 2011, 94, 5851–5856. [Google Scholar] [CrossRef] [PubMed]
- Guidelines for Drinking-water Quality. Available online: http://apps.who.int/iris/bitstream/10665/254637/1/9789241549950-eng.pdf?ua=1 (accessed on 16 March 2018).
- O’Toole, G. Microtiter Dish Biofilm Formation Assay. J. Vis. Exp. 2011, 10–11. [Google Scholar] [CrossRef] [PubMed]
- XLstatistics Add-in Tool for Excel. Available online: http://www.deakin.edu.au/~rodneyc/XLStatistics/ (accessed on 16 March 2018).
- Jahid, I.K.; Ha, S.D. The Paradox of Mixed-Species Biofilms in the Context of Food Safety. Comp. Rev. Food Sci. Food Saf. 2014, 13, 990–1011. [Google Scholar] [CrossRef]
- Sharma, K.; Singh, J.; Singh, A.P. Combating biofilm mediated antimicrobial resistance using efflux pump inhibitor and Deoxyribonuclease. Int. J. Mgmt. Appl. Sci. 2017, 5, 16–19. [Google Scholar]
- Tetz, V.V.; Tetz, G.V. Effect of extracellular DNA destruction by DNase I on characteristics of forming biofilms. DNA Cell Biol. 2010, 29, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Alhede, M.; Kragh, K.N.; Qvortrup, K.; Allesen-Holm, M.; van Gennip, M.; Christensen, L.D.; Jensen, P.Ø.; Nielsen, A.K.; Parsek, M.; Wozniak, D.; et al. Phenotypes of non-attached Pseudomonas aeruginosa aggregates resemble surface attached biofilm. PLoS ONE 2011, 6, e27943. [Google Scholar] [CrossRef] [PubMed]
- Schlafer, S.; Meyer, R.L.; Dige, I.; Regina, V.R. Extracellular DNA Contributes to Dental Biofilm Stability. Caries Res. 2017, 51, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Grande, R.; Giulio, M.; Bessa, L.J.; Di Campli, E.; Baffoni, M.; Guarnieri, S.; Cellini, L. Extracellular DNA in Helicobacter pylori biofilm: A backstairs rumour. J. Appl. Microbiol. 2011, 110, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, H.; Charron-Mazenod, L.; Lewenza, S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2008, 4, e1000213. [Google Scholar] [CrossRef] [PubMed]
- Zimelis, V.M.; Jackson, G.G. Activity of Aminoglycoside Antibiotics against Pseudomonas aeruginosa specificity and site of calcium and magnesium antagonism. J. Infect. Dis. 1973, 127, 663–669. [Google Scholar] [CrossRef] [PubMed]
- Beware Mixing Antibiotics with Magnesium. Available online: https://www.newsmax.com/Health/Health-Wire/magnesium-supplements-antibiotics-mixing/2015/05/27/id/646969/ (accessed on 16 March 2018).
- Rendueles, O.; Ghigo, J.M. Multi-species biofilms: How to avoid unfriendly neighbors. FEMS Microbiol. Rev. 2012, 36, 972–989. [Google Scholar] [CrossRef] [PubMed]
- Varposhti, M.; Entezari, F.; Feizabadi, M.M. Synergistic interactions in mixed-species biofilms of pathogenic bacteria from the respiratory tract. Rev. Soc. Bras. Med. Trop. 2014, 47, 649–652. [Google Scholar] [CrossRef] [PubMed]
- Pammi, M.; Liang, R.; Hicks, J.; Mistretta, T.A.; Versalovic, J. Biofilm extracellular DNA enhances mixed species biofilms of Staphylococcus epidermidis and Candida albicans. BMC Microbiol. 2013, 13, 257. [Google Scholar] [CrossRef] [PubMed]
- Jack, A.A.; Daniels, D.E.; Jepson, M.A.; Vickerman, M.M.; Lamont, R.J.; Jenkinson, H.F.; Nobbs, A.H. Streptococcus gordonii com CDE (competence) operon modulates biofilm formation with Candida albicans. Microbiology 2015, 161, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-López, P.; Carballo-Justo, A.; Draper, L.A.; Cabo, ML. Removal of Listeria monocytogenes dual-species biofilms using combined enzyme-benzalkonium chloride treatments. Biofouling 2017, 33, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Deoxyribonuclease I from Bovine Pancreas, Product Information. Available online: https://www.sigmaaldrich.com/content/dam/sigma-aldrich/docs/Sigma/Product_Information_Sheet/dneppis.pdf (accessed on 16 March 2018).
- Thermo Fisher Scientific. Available online: https://www.thermofisher.com/in/en/home/references/ambion-tech-support/nuclease-enzymes/general-articles/dnase-i-demystified.html (accessed on 28 October 2017).
- Torbic, H.; Hacobian, G. Evaluation of Inhaled Dornase Alfa Administration in Non-Cystic Fibrosis Patients at a Tertiary Academic Medical Center. J. Pharm. Pract. 2016, 29, 480–483. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, R.E.; Loomis, E.C. Fibrinolysin-Desoxyribonuclease for Enzymatic Debridement. Available online: https://www.google.co.in/patents/US3208908 (accessed on 28 October 2017).
- Hanson, D.P. Composition for Enzymatic Debridement. Available online: https://www.google.com/patents/US20130156745 (accessed on 28 October 2017).
- Swartjes, J.J.; Das, T.; Sharifi, S.; sharifi, S.; Subbiahdoss, G.; Sharma, P.K.; Krom, P.B.; Busscher, H.J.; Vander Mei, H.C. A functional DNase I coating to prevent adhesion of bacteria and the formation of biofilm. Adv. Funct. Mater. 2013, 23, 2843–2849. [Google Scholar] [CrossRef]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, K.; Pagedar Singh, A. Antibiofilm Effect of DNase against Single and Mixed Species Biofilm. Foods 2018, 7, 42. https://doi.org/10.3390/foods7030042
Sharma K, Pagedar Singh A. Antibiofilm Effect of DNase against Single and Mixed Species Biofilm. Foods. 2018; 7(3):42. https://doi.org/10.3390/foods7030042
Chicago/Turabian StyleSharma, Komal, and Ankita Pagedar Singh. 2018. "Antibiofilm Effect of DNase against Single and Mixed Species Biofilm" Foods 7, no. 3: 42. https://doi.org/10.3390/foods7030042
APA StyleSharma, K., & Pagedar Singh, A. (2018). Antibiofilm Effect of DNase against Single and Mixed Species Biofilm. Foods, 7(3), 42. https://doi.org/10.3390/foods7030042