Diversity Among Clinical and Fresh Produce Isolates of Stenotrophomonas: Insights Through a One Health Perspective
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Detection of Stenotrophomonas Isolates
2.2. Multilocus Sequence Typing (MLST) and Analysis
2.3. Bacterial Growth Curve Determination at 37 °C
2.4. Assessment of Biofilm Production Through Crystal Violet Assay
2.5. Antimicrobial Susceptibility Testing
2.6. Genetic Characterization by Whole Genome Sequencing
2.7. Data Processing and Statistical Analysis
3. Results
3.1. Isolation and Taxonomic Classification of Stenotrophomonas spp. Isolates
3.2. MLST Analysis
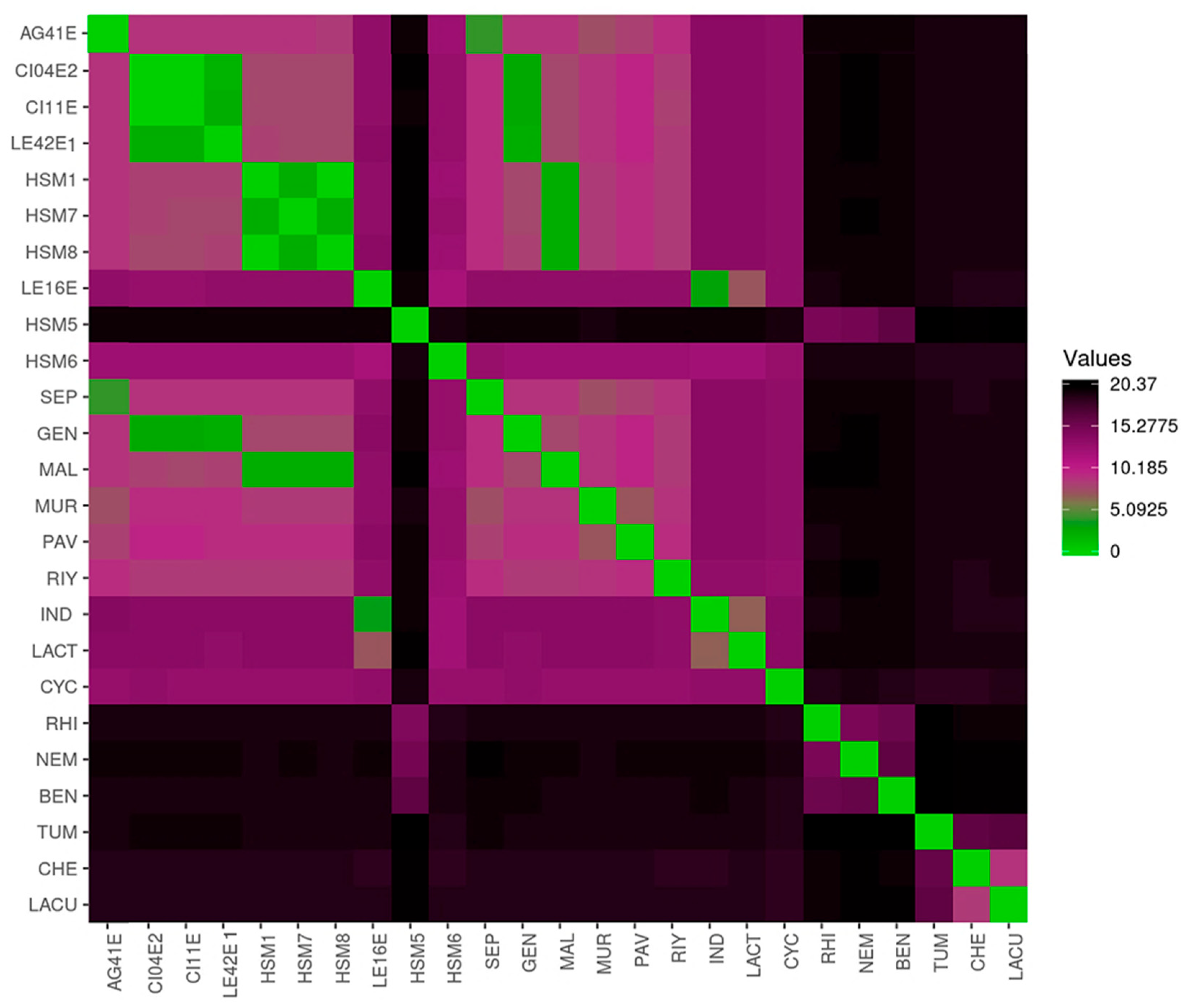
3.3. Average Nucleotide Identity (ANIb) Analysis
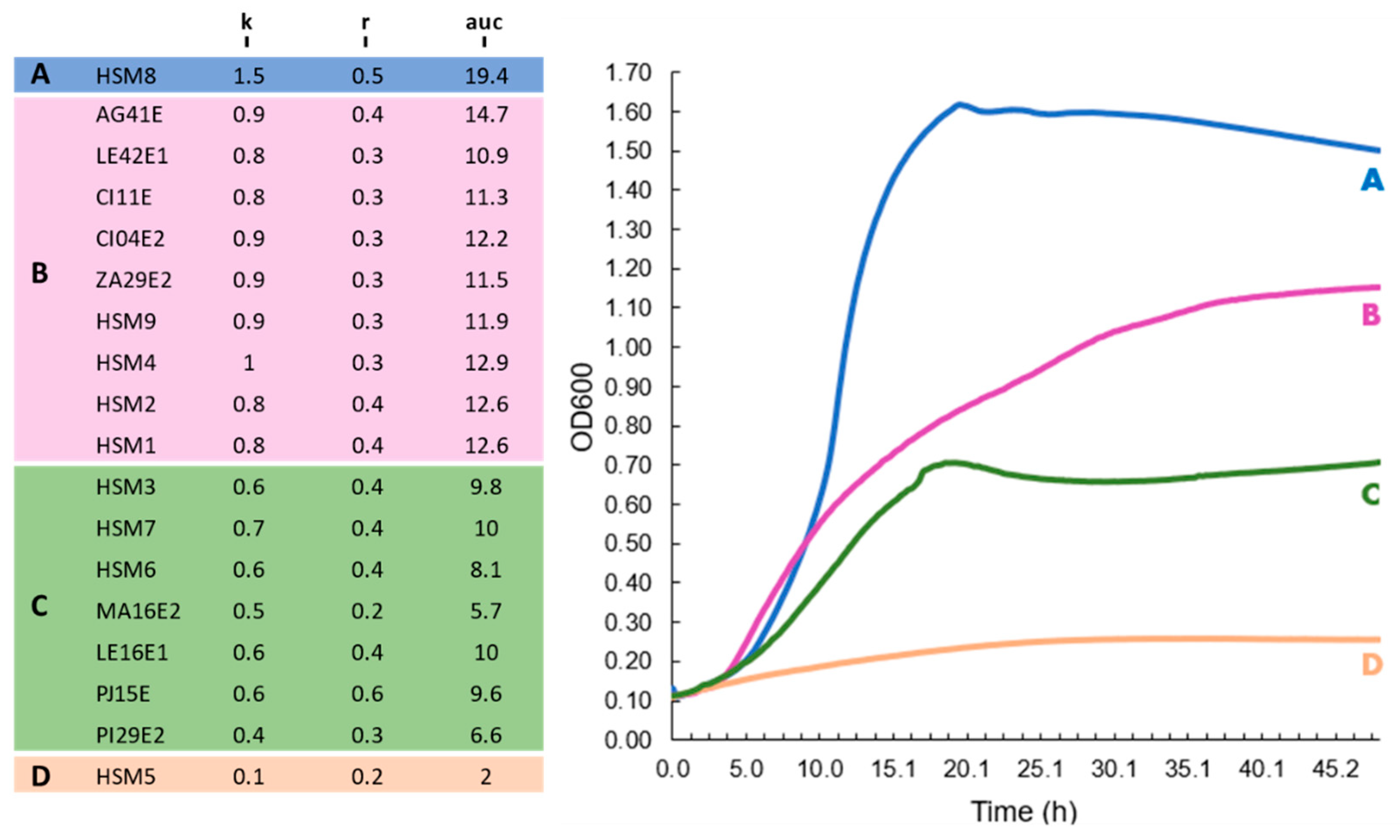
3.4. Growth Kinetics at 37 °C
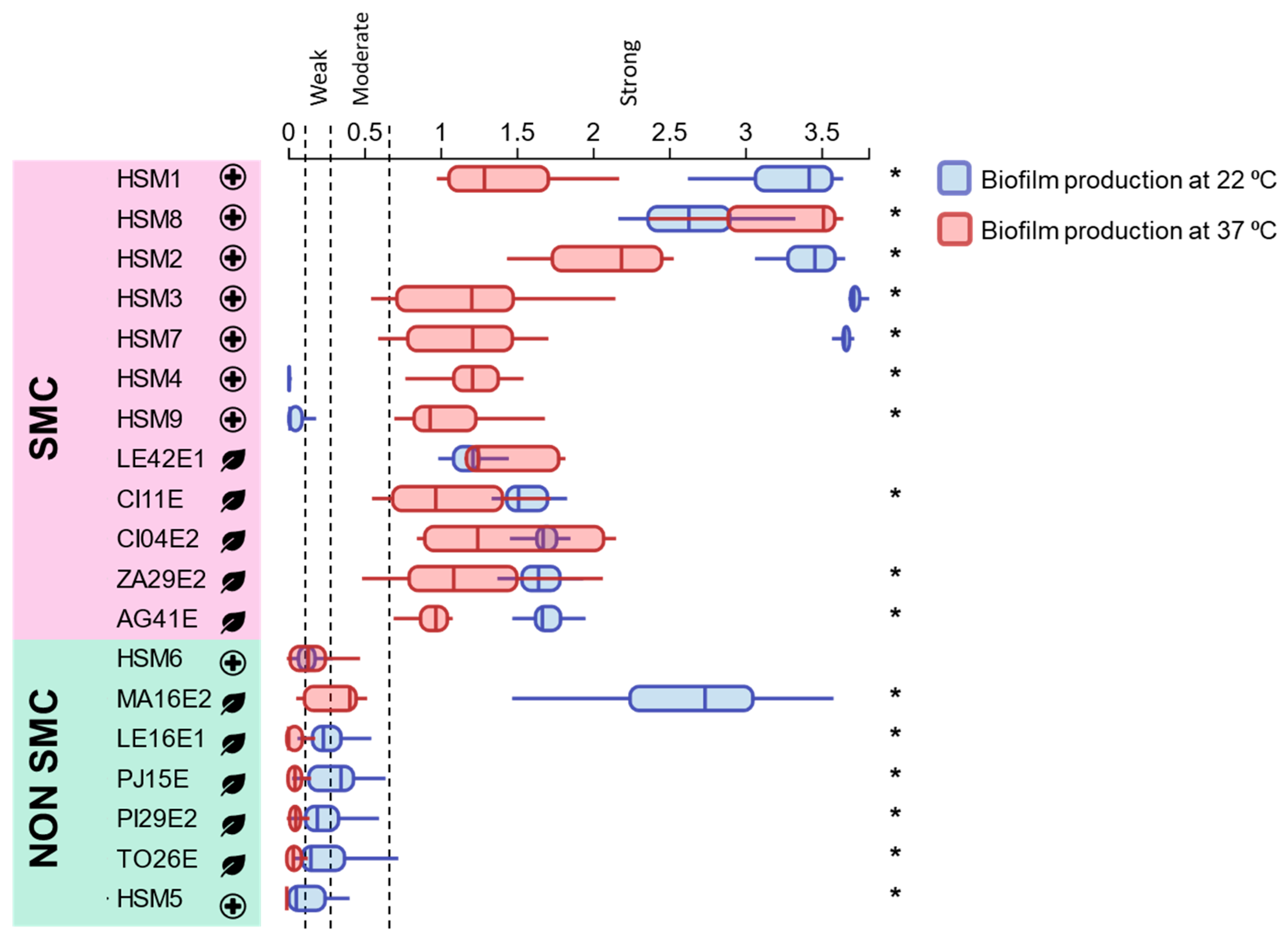
3.5. Biofilm-Forming Ability
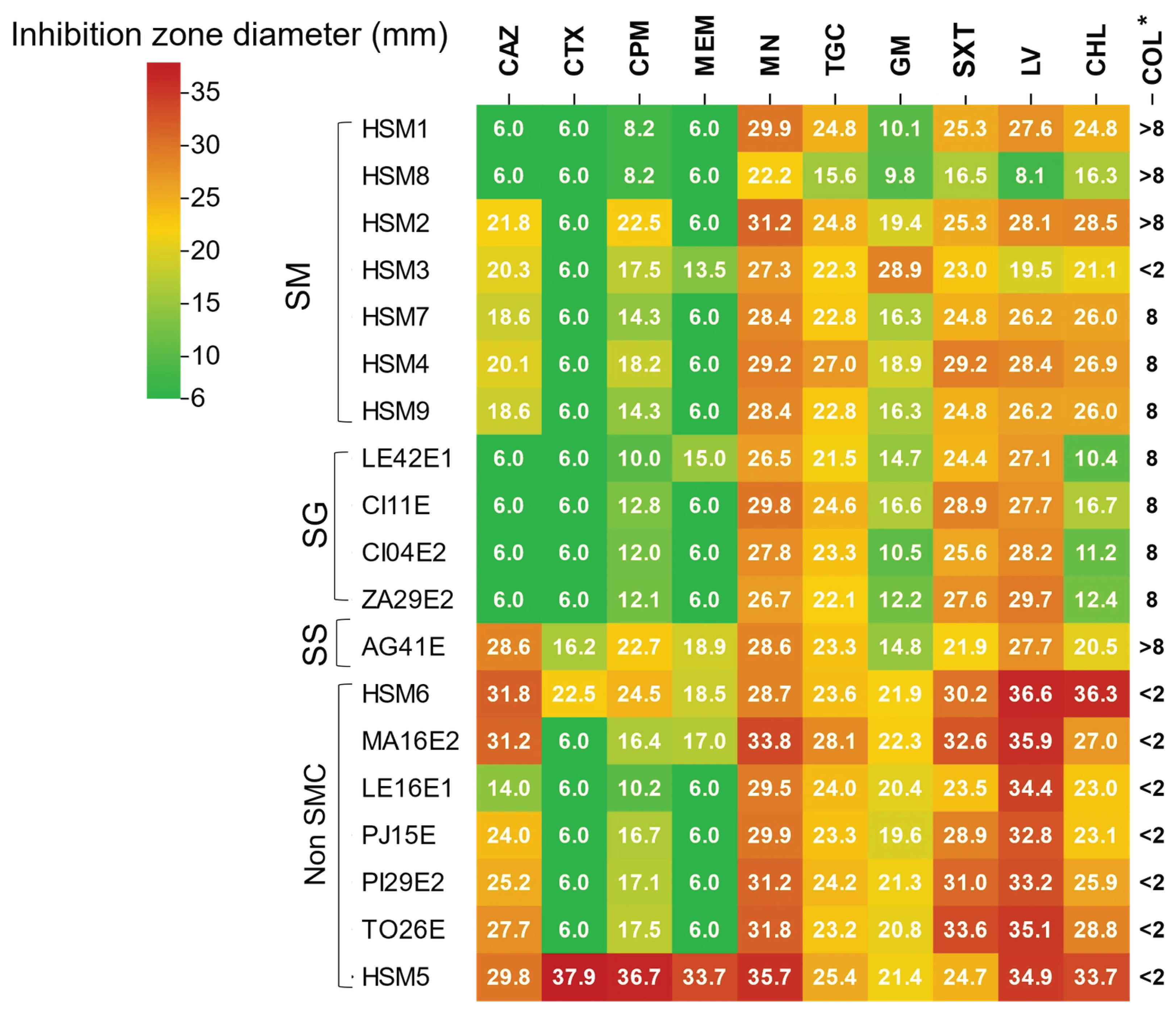
3.6. Antimicrobial Susceptibility Profiles of Stenotrophomonas Isolates
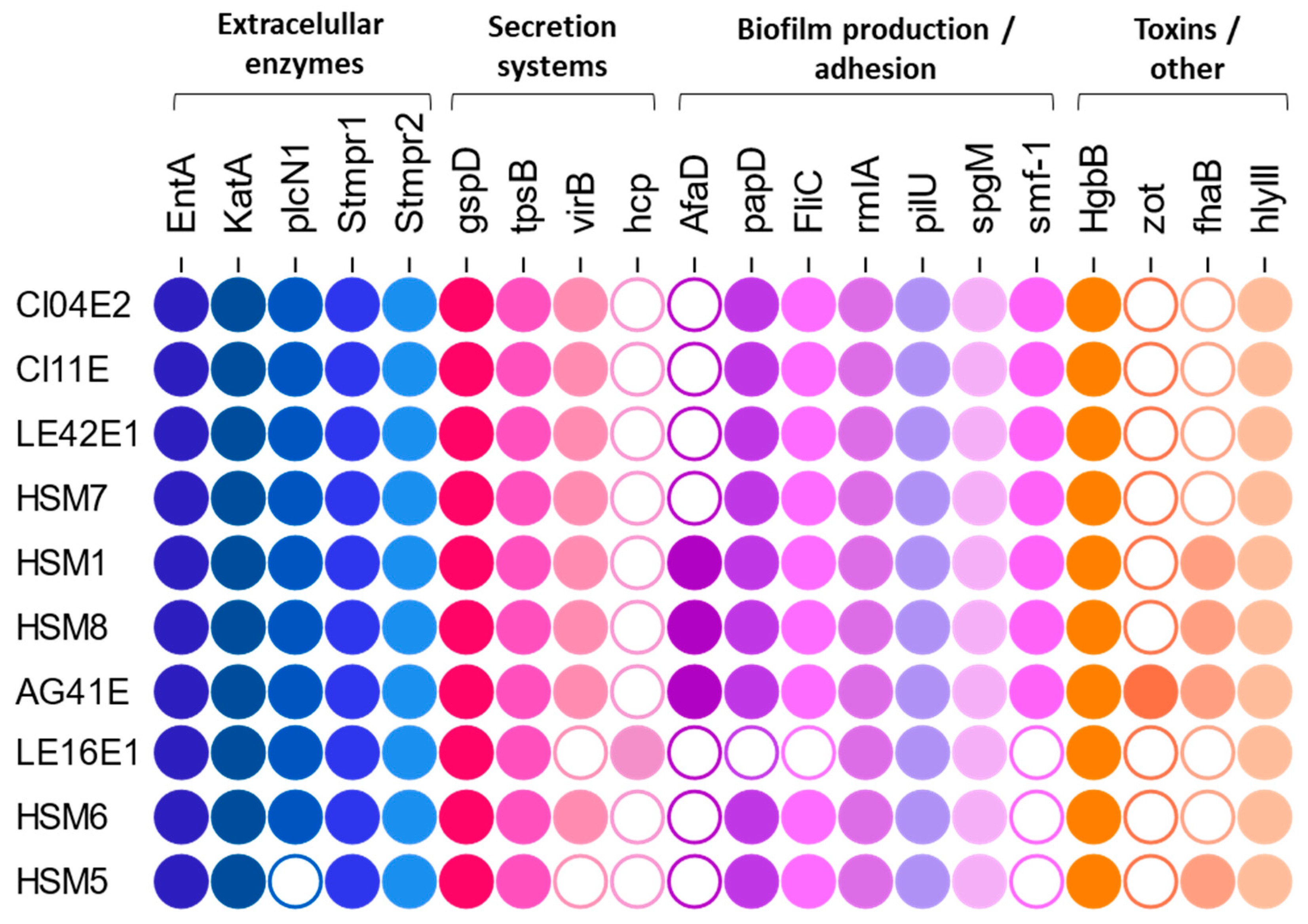
3.7. Detection of Virulence and Antibiotic Resistance Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| SMC | Stenotrophomonas maltophilia complex |
| ANI | Average nucleotide identity |
| MLST | Multilocus sequence typing |
| ST | Sequence type |
| AUC | Area under the curve |
| T2SS | Type II Secretion System |
| T4SS | Type IV Secretion System |
References
- Gales, A.C.; Seifert, H.; Gur, D.; Castanheira, M.; Jones, R.N.; Sader, H.S. Antimicrobial Susceptibility of Acinetobacter calcoaceticus–Acinetobacter baumannii Complex and Stenotrophomonas maltophilia Clinical Isolates: Results From the SENTRY Antimicrobial Surveillance Program (1997–2016). Open Forum Infect. Dis. 2019, 6, S34–S46. [Google Scholar] [CrossRef] [PubMed]
- Trifonova, A.; Strateva, T. Stenotrophomonas maltophilia—A Low-Grade Pathogen with Numerous Virulence Factors. Infect. Dis. 2019, 51, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Gautam, V.; Patil, P.P.; Bansal, K.; Kumar, S.; Kaur, A.; Singh, A.; Korpole, S.; Singhal, L.; Patil, P.B. Description of Stenotrophomonas sepilia sp. nov., Isolated from Blood Culture of a Hospitalized Patient as a New Member of Stenotrophomonas maltophilia Complex. New Microbes New Infect. 2021, 43, 100920. [Google Scholar] [CrossRef]
- Mikhailovich, V.; Heydarov, R.; Zimenkov, D.; Chebotar, I. Stenotrophomonas maltophilia Virulence: A Current View. Front. Microbiol. 2024, 15, 1385631. [Google Scholar] [CrossRef]
- Flores-Alvarez, L.J.; Martínez-Flores, I.; Bustos, P.; Gómez-García, A.; Gutiérrez-Castellanos, S.; Poot-Hernández, A.C.; Arredondo-Santoyo, M. Sequencing and Description of the Genome of a Strain of Stenotrophomonas geniculata Isolated from a Patient Infected with COVID-19 at Hospital Regional No.1 de Charo, Michoacán, México. MicroPubl. Biol. 2024, 2024, 10.17912/micropub.biology.001051. [Google Scholar] [CrossRef]
- Paez, J.I.G.; Costa, S.F. Risk Factors Associated with Mortality of Infections Caused by Stenotrophomonas maltophilia: A Systematic Review. J. Hosp. Infect. 2008, 70, 101–108. [Google Scholar] [CrossRef]
- Huang, C.; Lin, L.; Kuo, S. Risk Factors for Mortality in Stenotrophomonas maltophilia Bacteremia—A Meta-Analysis. Infect. Dis. 2024, 56, 335–347. [Google Scholar] [CrossRef]
- Gajdács, M.; Urbán, E. Prevalence and Antibiotic Resistance of Stenotrophomonas maltophilia in Respiratory Tract Samples: A 10-Year Epidemiological Snapshot. Health Serv. Res. Manag. Epidemiol. 2019, 6, 233339281987077. [Google Scholar] [CrossRef]
- Nicoletti, M.; Iacobino, A.; Prosseda, G.; Fiscarelli, E.; Zarrilli, R.; De Carolis, E.; Petrucca, A.; Nencioni, L.; Colonna, B.; Casalino, M. Stenotrophomonas maltophilia Strains from Cystic Fibrosis Patients: Genomic Variability and Molecular Characterization of Some Virulence Determinants. Int. J. Med. Microbiol. 2011, 301, 34–43. [Google Scholar] [CrossRef]
- Ryan, R.P.; Monchy, S.; Cardinale, M.; Taghavi, S.; Crossman, L.; Avison, M.B.; Berg, G.; van der Lelie, D.; Dow, J.M. The Versatility and Adaptation of Bacteria from the Genus Stenotrophomonas. Nat. Rev. Microbiol. 2009, 7, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Deredjian, A.; Alliot, N.; Blanchard, L.; Brothier, E.; Anane, M.; Cambier, P.; Jolivet, C.; Khelil, M.N.; Nazaret, S.; Saby, N.; et al. Occurrence of Stenotrophomonas maltophilia in Agricultural Soils and Antibiotic Resistance Properties. Res. Microbiol. 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, S.V.; Edwards, D.; Vaughn, E.L.; Escobar, V.; Ali, S.; Doss, J.H.; Steyer, J.T.; Scott, S.; Bchara, W.; Bruns, N.; et al. Expanding the Stenotrophomonas maltophilia Complex: Phylogenomic Insights, Proposal of Stenotrophomonas forensis sp. nov. and Reclassification of Two Pseudomonas Species. Int. J. Syst. Evol. Microbiol. 2024, 74, 006602. [Google Scholar] [CrossRef]
- Li, K.; Yu, K.; Huang, Z.; Liu, X.; Mei, L.; Ren, X.; Bai, X.; Gao, H.; Sun, Z.; Liu, X.; et al. Stenotrophomonas maltophilia Complex: Insights into Evolutionary Relationships, Global Distribution and Pathogenicity. Front. Cell. Infect. Microbiol. 2023, 13, 1325379. [Google Scholar] [CrossRef]
- Yu, Z.L.; Wang, R.B. Revised Taxonomic Classification of the Stenotrophomonas Genomes, Providing New Insights into the Genus Stenotrophomonas. Front. Microbiol. 2024, 15, 1488674. [Google Scholar] [CrossRef]
- Monardo, R.; Mojica, M.F.; Ripa, M.; Aitken, S.L.; Bonomo, R.A.; van Duin, D. How Do I Manage a Patient with Stenotrophomonas maltophilia Infection? Clin. Microbiol. Infect. 2025, 31, 1291–1297. [Google Scholar] [CrossRef] [PubMed]
- Bostanghadiri, N.; Sholeh, M.; Navidifar, T.; Dadgar-Zankbar, L.; Elahi, Z.; van Belkum, A.; Darban-Sarokhalil, D. Global Mapping of Antibiotic Resistance Rates among Clinical Isolates of Stenotrophomonas maltophilia: A Systematic Review and Meta-Analysis. Ann. Clin. Microbiol. Antimicrob. 2024, 23, 26. [Google Scholar] [CrossRef]
- Adamek, M.; Linke, B.; Schwartz, T. Virulence Genes in Clinical and Environmental Stenotrophonas maltophilia Isolates: A Genome Sequencing and Gene Expression Approach. Microb. Pathog. 2014, 67–68, 20–30. [Google Scholar] [CrossRef]
- Azimi, A.; Aslanimehr, M.; Yaseri, M.; Shadkam, M.; Douraghi, M. Distribution of smf-1, rmlA, spgM and rpfF Genes among Stenotrophomonas maltophilia Isolates in Relation to Biofilm-Forming Capacity. J. Glob. Antimicrob. Resist. 2020, 23, 321–326. [Google Scholar] [CrossRef]
- García, G.; Girón, J.A.; Yañez, J.A.; Cedillo, M.L. Stenotrophomonas maltophilia and Its Ability to Form Biofilms. Microbiol. Res. 2022, 14, 1–20. [Google Scholar] [CrossRef]
- Singhal, L.; Kaur, P.; Gautam, V. Stenotrophomonas maltophilia: From Trivial to Grievous. Indian. J. Med. Microbiol. 2017, 35, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Furlan, J.P.R.; Sanchez, D.G.; Gallo, I.F.L.; Stehling, E.G. Characterization of Acquired Antimicrobial Resistance Genes in Environmental Stenotrophomonas maltophilia Isolates from Brazil. Microb. Drug Resist. 2019, 25, 475–479. [Google Scholar] [CrossRef]
- Klimkaitė, L.; Armalytė, J.; Skerniškytė, J.; Sužiedėlienė, E. The Toxin-Antitoxin Systems of the Opportunistic Pathogen Stenotrophomonas maltophilia of Environmental and Clinical Origin. Toxins 2020, 12, 635. [Google Scholar] [CrossRef]
- Berg, G.; Martinez, J.L. Friends or Foes: Can We Make a Distinction between Beneficial and Harmful Strains of the Stenotrophomonas maltophilia Complex? Front. Microbiol. 2015, 6, 241. [Google Scholar] [CrossRef]
- Kaiser, S.; Biehler, K.; Jonas, D. A Stenotrophomonas maltophilia Multilocus Sequence Typing Scheme for Inferring Population Structure. J. Bacteriol. 2009, 191, 2934–2943. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-Access Bacterial Population Genomics: BIGSdb Software, the PubMLST.Org Website and Their Applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Stepanović, S.; Vuković, D.; Hola, V.; Di Bonaventura, G.; Djukić, S.; Ćirković, I.; Ruzicka, F. Quantification of Biofilm in Microtiter Plates: Overview of Testing Conditions and Practical Recommendations for Assessment of Biofilm Production by Staphylococci. APMIS 2007, 115, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R.; Oliver Glöckner, F.; Peplies, J. JSpeciesWS: A Web Server for Prokaryotic Species Circumscription Based on Pairwise Genome Comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, Y.; Ye, K.; Qiu, X.; Zhao, Q.; Ye, L.Y.; Yang, J. Molecular Epidemiology, Genetic Diversity, Antibiotic Resistance and Pathogenicity of Stenotrophomonas maltophilia Complex from Bacteremia Patients in a Tertiary Hospital in China for Nine Years. Front. Microbiol. 2024, 15, 1424241. [Google Scholar] [CrossRef]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-Enabled Heat Mapping for All. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef]
- Xie, J.; Chen, Y.; Cai, G.; Cai, R.; Hu, Z.; Wang, H. Tree Visualization by One Table (TvBOT): A Web Application for Visualizing, Modifying and Annotating Phylogenetic Trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef] [PubMed]
- Hagemann, M.; Hasse, D.; Berg, G. Detection of a Phage Genome Carrying a Zonula Occludens like Toxin Gene (Zot) in Clinical Isolates of Stenotrophomonas maltophilia. Arch. Microbiol. 2006, 185, 449–458. [Google Scholar] [CrossRef]
- Tamma, P.D.; Aitken, S.L.; Bonomo, R.A.; Mathers, A.J.; Van Duin, D.; Clancy, C.J. Infectious Diseases Society of America Guidance on the Treatment of AmpC β-Lactamase–Producing Enterobacterales, Carbapenem-Resistant Acinetobacter baumannii, and Stenotrophomonas maltophilia Infections. Clin. Infect. Dis. 2022, 74, 2089–2114. [Google Scholar] [CrossRef] [PubMed]
- Patil, P.P.; Kumar, S.; Midha, S.; Gautam, V.; Patil, P.B. Taxonogenomics Reveal Multiple Novel Genomospecies Associated with Clinical Isolates of Stenotrophomonas maltophilia. Microb. Genom. 2018, 4, e000207. [Google Scholar] [CrossRef] [PubMed]
- Chauviat, A.; Abrouk, D.; Brothier, E.; Muller, D.; Meyer, T.; Favre-Bonté, S. Genomic and Phylogenetic Re-Assessment of the Genus Stenotrophomonas: Description of Stenotrophomonas thermophila sp. nov., and the Proposal of Parastenotrophomonas gen. nov., Pseudostenotrophomonas gen. nov., Pedostenotrophomonas gen. nov., and Allostenotrophomonas gen. nov. Syst. Appl. Microbiol. 2025, 48, 126630. [Google Scholar] [CrossRef]
- Ochoa-Sánchez, L.E.; Vinuesa, P. Evolutionary Genetic Analysis Uncovers Multiple Species with Distinct Habitat Preferences and Antibiotic Resistance Phenotypes in the Stenotrophomonas maltophilia Complex. Front. Microbiol. 2017, 8, 245563. [Google Scholar] [CrossRef]
- Adamek, M.; Overhage, J.; Bathe, S.; Winter, J.; Fischer, R.; Schwartz, T. Genotyping of Environmental and Clinical Stenotrophomonas maltophilia Isolates and Their Pathogenic Potential. PLoS ONE 2011, 6, e27615. [Google Scholar] [CrossRef]
- Gröschel, M.I.; Meehan, C.J.; Barilar, I.; Diricks, M.; Gonzaga, A.; Steglich, M.; Conchillo-Solé, O.; Scherer, I.C.; Mamat, U.; Luz, C.F.; et al. The Phylogenetic Landscape and Nosocomial Spread of the Multidrug-Resistant Opportunist Stenotrophomonas maltophilia. Nat. Commun. 2020, 11, 2044. [Google Scholar] [CrossRef]
- Gautam, V.; Sharma, M.; Singhal, L.; Kumar, S.; Kaur, P.; Tiwari, R.; Ray, P. MALDI-TOF Mass Spectrometry: An Emerging Tool for Unequivocal Identification of Non-Fermenting Gram-Negative Bacilli. Indian J. Med. Res. 2017, 145, 665. [Google Scholar] [CrossRef]
- Mahnoor; Noor-Ul-Ain; Arshad, F.; Ahsan, T.; Alharbi, S.A.; Ansari, M.J.; Khan, I.; Alshiekheid, M.; Sabour, A.A.A. Whole Genome Analysis of Stenotrophomonas geniculata MK2 and Antagonism against Botrytis cinerea in Strawberry. Int. Microbiol. 2025, 28, 1255–1268. [Google Scholar] [CrossRef]
- Ercole, T.G.; Kava, V.M.; Petters-Vandresen, D.A.L.; Ribeiro, R.A.; Hungria, M.; Galli, L.V. Unveiling Agricultural Biotechnological Prospects: The Draft Genome Sequence of Stenotrophomonas geniculata LGMB417. Curr. Microbiol. 2024, 81, 247. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Ding, W.J.; Xu, L.; Sun, J.Q. A Comprehensive Comparative Genomic Analysis Revealed That Plant Growth Promoting Traits Are Ubiquitous in Strains of Stenotrophomonas. Front. Microbiol. 2024, 15, 1395477. [Google Scholar] [CrossRef]
- Omomowo, O.I.; Babalola, O.O. Genomic Insights into Two Endophytic Strains: Stenotrophomonas geniculata NWUBe21 and Pseudomonas carnis NWUBe30 from Cowpea with Plant Growth-Stimulating Attributes. Appl. Sci. 2022, 12, 12953. [Google Scholar] [CrossRef]
- Quach, N.T.; Nguyen, T.T.A.; Vu, T.H.N.; Le, T.T.X.; Nguyen, T.T.L.; Chu, H.H.; Phi, Q.T. Phenotypic and Genomic Analysis Deciphering Plant Growth Promotion and Oxidative Stress Alleviation of Stenotrophomonas sepilia ZH16 Isolated from Rice. Microbiology 2025, 94, 38–49. [Google Scholar] [CrossRef]
- Chakraborty, S.; Shekhar, N.; Singhal, L.; Rawat, R.S.; Duseja, A.; Verma, R.K.; Bansal, K.; Kour, I.; Biswas, S.; Rajni, E.; et al. Susceptibility of Clinical Isolates of Novel Pathogen Stenotrophomonas sepilia to Novel Benzoquinolizine Fluoroquinolone Levonadifloxacin. JAC Antimicrob. Resist. 2024, 6, dlae130. [Google Scholar] [CrossRef] [PubMed]
- Babalola, O.O.; Adeleke, B.S.; Ayangbenro, A.S.; Baltrus, D.A. Draft Genome Sequencing of Stenotrophomonas indicatrix BOVIS40 and Stenotrophomonas maltophilia JVB5, Two Strains with Identifiable Genes Involved in Plant Growth Promotion. Microbiol. Resour. Announc. 2021, 10, e00482-21. [Google Scholar] [CrossRef]
- Adeleke, B.S.; Ayangbenro, A.S.; Babalola, O.O. Genomic Assessment of Stenotrophomonas indicatrix for Improved Sunflower Plant. Curr. Genet. 2021, 67, 891–907. [Google Scholar] [CrossRef]
- Weber, M.; Schünemann, W.; Fuß, J.; Kämpfer, P.; Lipski, A. Stenotrophomonas lactitubi sp. nov. and Stenotrophomonas indicatrix sp. nov., Isolated from Surfaces with Food Contact. Int. J. Syst. Evol. Microbiol. 2018, 68, 1830–1838. [Google Scholar] [CrossRef]
- Steinmann, J.; Mamat, U.; Abda, E.M.; Kirchhoff, L.; Streit, W.R.; Schaible, U.E.; Niemann, S.; Kohl, T.A. Analysis of Phylogenetic Variation of Stenotrophomonas maltophilia Reveals Human-Specific Branches. Front. Microbiol. 2018, 9, 806. [Google Scholar] [CrossRef]
- Pinski, A.; Zur, J.; Hasterok, R.; Hupert-Kocurek, K. Comparative Genomics of Stenotrophomonas maltophilia and Stenotrophomonas rhizophila Revealed Characteristic Features of Both Species. Int. J. Mol. Sci. 2020, 21, 4922. [Google Scholar] [CrossRef]
- Zhang, T.; Wu, H.; Ma, C.; Yang, Y.; Li, H.; Yang, Z.; Zhou, S.; Shi, D.; Chen, T.; Yang, D.; et al. Emergence of Colistin-Resistant Stenotrophomonas maltophilia with High Virulence in Natural Aquatic Environments. Sci. Total Environ. 2024, 933, 173221. [Google Scholar] [CrossRef]
- Duan, Z.; Qin, J.; Liu, Y.; Li, C.; Ying, C. Molecular Epidemiology and Risk Factors of Stenotrophomonas maltophilia Infections in a Chinese Teaching Hospital. BMC Microbiol. 2020, 20, 294. [Google Scholar] [CrossRef]
- Li, Y.; Liu, X.; Chen, L.; Shen, X.; Wang, H.; Guo, R.; Li, X.; Yu, Z.; Zhang, X.; Zhou, Y.; et al. Comparative Genomics Analysis of Stenotrophomonas maltophilia Strains from a Community. Front. Cell. Infect. Microbiol. 2023, 13, 1266295. [Google Scholar] [CrossRef]
- Mojica, M.F.; Rutter, J.D.; Taracila, M.; Abriata, L.A.; Fouts, D.E.; Papp-Wallace, K.M.; Walsh, T.J.; Lipuma, J.J.; Vila, A.J.; Bonomo, R.A. Population Structure, Molecular Epidemiology, and β-Lactamase Diversity among Stenotrophomonas maltophilia Isolates in the United States. mBio 2019, 10, e00405-19. [Google Scholar] [CrossRef] [PubMed]
- Corlouer, C.; Lamy, B.; Desroches, M.; Ramos-Vivas, J.; Mehiri-Zghal, E.; Lemenand, O.; Delarbre, J.M.; Decousser, J.W.; Aberanne, S.; Belmonte, O.; et al. Stenotrophomonas maltophilia Healthcare-Associated Infections: Identification of Two Main Pathogenic Genetic Backgrounds. J. Hosp. Infect. 2017, 96, 183–188. [Google Scholar] [CrossRef]
- Cho, H.H.; Sung, J.Y.; Kwon, K.C.; Koo, S.H. Expression of Sme Efflux Pumps and Multilocus Sequence Typing in Clinical Isolates of Stenotrophomonas maltophilia. Ann. Lab. Med. 2011, 32, 38. [Google Scholar] [CrossRef]
- Alavi, P.; Starcher, M.R.; Thallinger, G.G.; Zachow, C.; Müller, H.; Berg, G. Stenotrophomonas Comparative Genomics Reveals Genes and Functions That Differentiate Beneficial and Pathogenic Bacteria. BMC Genom. 2014, 15, 482. [Google Scholar] [CrossRef] [PubMed]
- Pompilio, A.; Ranalli, M.; Piccirilli, A.; Perilli, M.; Vukovic, D.; Savic, B.; Krutova, M.; Drevinek, P.; Jonas, D.; Fiscarelli, E.V.; et al. Biofilm Formation among Stenotrophomonas maltophilia Isolates Has Clinical Relevance: The ANSELM Prospective Multicenter Study. Microorganisms 2021, 9, 49. [Google Scholar] [CrossRef]
- Klimkaitė, L.; Drevinskaitė, R.; Krinickis, K.; Sužiedėlienė, E.; Armalytė, J. Stenotrophomonas maltophilia of Clinical Origin Display Higher Temperature Tolerance Comparing with Environmental Isolates. Virulence 2025, 16, 2498669. [Google Scholar] [CrossRef] [PubMed]
- Jahid, I.K.; Ha, S. Do A Review of Microbial Biofilms of Produce: Future Challenge to Food Safety. Food Sci. Biotechnol. 2012, 21, 299–316. [Google Scholar] [CrossRef]
- Alcaraz, E.; García, C.; Friedman, L.; De Rossi, B.P. The Rpf/DSF Signalling System of Stenotrophomonas maltophilia Positively Regulates Biofilm Formation, Production of Virulence-Associated Factors and β-Lactamase Induction. FEMS Microbiol. Lett. 2019, 366, fnz069. [Google Scholar] [CrossRef]
- Cruz-Córdova, A.; Mancilla-Rojano, J.; Luna-Pineda, V.M.; Escalona-Venegas, G.; Cázares-Domínguez, V.; Ormsby, C.; Franco-Hernández, I.; Zavala-Vega, S.; Hernández, M.A.; Medina-Pelcastre, M.; et al. Molecular Epidemiology, Antibiotic Resistance, and Virulence Traits of Stenotrophomonas maltophilia Strains Associated With an Outbreak in a Mexican Tertiary Care Hospital. Front. Cell. Infect. Microbiol. 2020, 10, 50. [Google Scholar] [CrossRef]
- Rocco, F.; De Gregorio, E.; Colonna, B.; Di Nocera, P.P. Stenotrophomonas maltophilia Genomes: A Start-up Comparison. Int. J. Med. Microbiol. 2009, 299, 535–546. [Google Scholar] [CrossRef]
- Bhaumik, R.; Beard, A.; Harrigan, O.; Ramos-Hegazy, L.; Mattoo, S.; Anderson, G.G. Role of SMF-1 and Cbl Pili in Stenotrophomonas maltophilia Biofilm Formation. Biofilm 2025, 9, 100253. [Google Scholar] [CrossRef]
- Gallo, S.W.; Figueiredo, T.P.; Bessa, M.C.; Pagnussatti, V.E.; Ferreira, C.A.S.; Oliveira, S.D. Isolation and Characterization of Stenotrophomonas maltophilia Isolates from a Brazilian Hospital. Microb. Drug Resist. 2016, 22, 688–695. [Google Scholar] [CrossRef]
- Goodman, A.L.; Kulasekara, B.; Rietsch, A.; Boyd, D.; Smith, R.S.; Lory, S. A Signaling Network Reciprocally Regulates Genes Associated with Acute Infection and Chronic Persistence in Pseudomonas aeruginosa. Dev. Cell 2004, 7, 745–754. [Google Scholar] [CrossRef]
- Furukawa, S.; Kuchma, S.L.; O’Toole, G.A. Keeping Their Options Open: Acute versus Persistent Infections. J. Bacteriol. 2006, 188, 1211–1217. [Google Scholar] [CrossRef] [PubMed]
- Di Bonaventura, G.; Prosseda, G.; Del Chierico, F.; Cannavacciuolo, S.; Cipriani, P.; Petrucca, A.; Superti, F.; Ammendolia, M.G.; Concato, C.; Fiscarelli, E.; et al. Molecular Characterization of Virulence Determinants of Stenotrophomonas maltophilia Strains Isolated from Patients Affected by Cystic Fibrosis. Int. J. Immunopathol. Pharmacol. 2007, 20, 529–537. [Google Scholar] [CrossRef] [PubMed]
- Bhaumik, R.; Aungkur, N.Z.; Anderson, G.G. A Guide to Stenotrophomonas maltophilia Virulence Capabilities, as We Currently Understand Them. Front. Cell. Infect. Microbiol. 2023, 13, 1322853. [Google Scholar] [CrossRef] [PubMed]
- Uzzau, S.; Cappuccinelli, P.; Fasano, A. Expression of Vibrio Cholerae Zonula Occludens Toxin and Analysis of Its Subcellular Localization. Microb. Pathog. 1999, 27, 377–385. [Google Scholar] [CrossRef]
- Petrova, M.; Shcherbatova, N.; Kurakov, A.; Mindlin, S. Genomic Characterization and Integrative Properties of PhiSMA6 and PhiSMA7, Two Novel Filamentous Bacteriophages of Stenotrophomonas maltophilia. Arch. Virol. 2014, 159, 1293–1303. [Google Scholar] [CrossRef]
- Gillespie, J.J.; Phan, I.Q.H.; Scheib, H.; Subramanian, S.; Edwards, T.E.; Lehman, S.S.; Piitulainen, H.; Rahman, M.S.; Rennoll-Bankert, K.E.; Staker, B.L.; et al. Structural Insight into How Bacteria Prevent Interference between Multiple Divergent Type IV Secretion Systems. mBio 2015, 6, e01867-15. [Google Scholar] [CrossRef]
- Nas, M.Y.; White, R.C.; DuMont, A.L.; Lopez, A.E.; Cianciottoa, N.P. Stenotrophomonas maltophilia Encodes a VirB/VirD4 Type IV Secretion System That Modulates Apoptosis in Human Cells and Promotes Competition against Heterologous Bacteria, Including Pseudomonas aeruginosa. Infect. Immun. 2019, 87, e00457-19. [Google Scholar] [CrossRef] [PubMed]
- Nas, M.Y.; Gabell, J.; Cianciotto, N.P. Effectors of the Stenotrophomonas maltophilia Type IV Secretion System Mediate Killing of Clinical Isolates of Pseudomonas aeruginosa. mBio 2021, 12, e01502-21. [Google Scholar] [CrossRef]
- Phillips, M.C.; Lee, B.; Miller, S.; Yan, J.; Maeusli, M.; She, R.; Luna, B.M.; Spellberg, B. Ceftazidime Retains in Vivo Efficacy against Strains of Stenotrophomonas maltophilia for Which Traditional Testing Predicts Resistance. Open Forum Infect Dis 2023, 10, ofad500.086. [Google Scholar] [CrossRef]
- Flores-Treviño, S.; Bocanegra-Ibarias, P.; Camacho-Ortiz, A.; Morfín-Otero, R.; Salazar-Sesatty, H.A.; Garza-González, E. Stenotrophomonas maltophilia Biofilm: Its Role in Infectious Diseases. Expert Rev. Anti-Infect. Ther. 2019, 17, 877–893. [Google Scholar] [CrossRef] [PubMed]
- Gibb, J.; Wong, D.W. Antimicrobial Treatment Strategies for Stenotrophomonas maltophilia: A Focus on Novel Therapies. Antibiotics 2021, 10, 1226. [Google Scholar] [CrossRef] [PubMed]
- Falagas, M.E.; Valkimadi, P.E.; Huang, Y.T.; Matthaiou, D.K.; Hsueh, P.R. Therapeutic Options for Stenotrophomonas maltophilia Infections beyond Co-Trimoxazole: A Systematic Review. J. Antimicrob. Chemother. 2008, 62, 889–894. [Google Scholar] [CrossRef]
- Dadashi, M.; Hajikhani, B.; Nazarinejad, N.; Noorisepehr, N.; Yazdani, S.; Hashemi, A.; Hashemizadeh, Z.; Goudarzi, M.; Fatemeh, S. Global Prevalence and Distribution of Antibiotic Resistance among Clinical Isolates of Stenotrophomonas maltophilia: A Systematic Review and Meta-Analysis. J. Glob. Antimicrob. Resist. 2023, 34, 253–267. [Google Scholar] [CrossRef]
- Delgarm Shams-Abadi, A.; Mohammadian-Hafshejani, A.; Paterson, D.L.; Arash, R.; Asadi Farsani, E.; Taji, A.; Heidari, H.; Shahini Shams Abadi, M. The Prevalence of Colistin Resistance in Clinical Stenotrophomonas maltophilia Isolates Worldwide: A Systematic Review and Meta-Analysis. BMC Microbiol. 2023, 23, 200. [Google Scholar] [CrossRef]
- Zhao, J.; Liu, Y.; Liu, Y.; Wang, D.; Ni, W.; Wang, R.; Liu, Y.; Zhang, B. Frequency and Genetic Determinants of Tigecycline Resistance in Clinically Isolated Stenotrophomonas maltophilia in Beijing, China. Front. Microbiol. 2018, 9, 337267. [Google Scholar] [CrossRef] [PubMed]
- Gil-Gil, T.; Martínez, J.L.; Blanco, P. Mechanisms of Antimicrobial Resistance in Stenotrophomonas maltophilia: A Review of Current Knowledge. Expert Rev. Anti-Infect. Ther. 2020, 18, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.F.; Chang, X.; Ye, Y.; Wang, Z.X.; Shao, Y.B.; Shi, W.; Li, X.; Li, J. Bin Stenotrophomonas maltophilia Resistance to Trimethoprim/Sulfamethoxazole Mediated by Acquisition of Sul and DfrA Genes in a Plasmid-Mediated Class 1 Integron. Int. J. Antimicrob. Agents 2011, 37, 230–234. [Google Scholar] [CrossRef]
- Lin, Y.T.; Huang, Y.W.; Chen, S.J.; Chang, C.W.; Yang, T.C. The SmeYZ Efflux Pump of Stenotrophomonas maltophilia Contributes to Drug Resistance, Virulence-Related Characteristics, and Virulence in Mice. Antimicrob. Agents Chemother. 2015, 59, 4067–4073. [Google Scholar] [CrossRef]
- Wu, R.X.; Yu, C.M.; Hsu, S.T.; Wang, C.H. Emergence of Concurrent Levofloxacin- and Trimethoprim/Sulfamethoxazole-Resistant Stenotrophomonas maltophilia: Risk Factors and Antimicrobial Sensitivity Pattern Analysis from a Single Medical Center in Taiwan. J. Microbiol. Immunol. Infect. 2022, 55, 107–113. [Google Scholar] [CrossRef]
- Chauviat, A.; Meyer, T.; Favre-Bonté, S. Versatility of Stenotrophomonas maltophilia: Ecological Roles of RND Efflux Pumps. Heliyon 2023, 9, e14639. [Google Scholar] [CrossRef]
- Cerezer, V.G.; Bando, S.Y.; Pasternak, J.; Franzolin, M.R.; Moreira-Filho, C.A. Phylogenetic Analysis of Stenotrophomonas spp. Isolates Contributes to the Identification of Nosocomial and Community-Acquired Infections. BioMed Res. Int. 2014, 2014, 151405. [Google Scholar] [CrossRef] [PubMed]
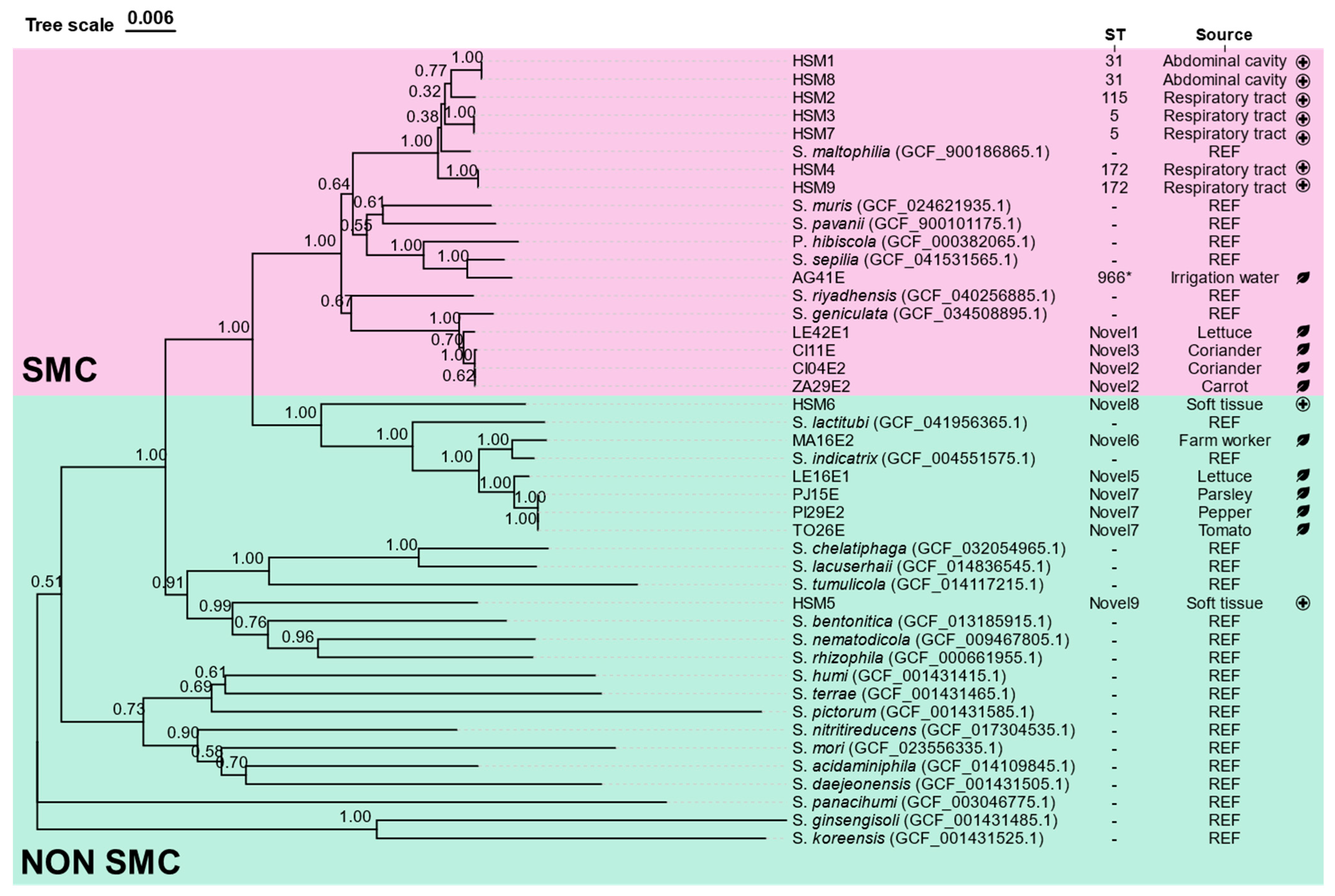
| Isolate | MALDI-TOF | MLSA/AniB | Source | |
| AG41E | Stenotrophomonas maltophilia | Stenotrophomonas sepilia | Irrigation Water | Environmental/Fresh produce |
| CI04E2 | Stenotrophomonas maltophilia | Stenotrophomonas geniculata | Coriander | |
| CI11E | Stenotrophomonas maltophilia | Stenotrophomonas geniculata | Coriander | |
| LE16E1 | Stenotrophomonas sp. | Stenotrophomonas indicatrix | Lettuce | |
| LE42E1 | Stenotrophomonas maltophilia | Stenotrophomonas geniculata | Lettuce | |
| MA16E2 | Stenotrophomonas sp. | Stenotrophomonas indicatrix | Farm Worker | |
| PI29E2 | Stenotrophomonas sp. | Stenotrophomonas indicatrix | Pepper | |
| PJ15E | Stenotrophomonas sp. | Stenotrophomonas indicatrix | Parsley | |
| TO26E | Stenotrophomonas sp. | Stenotrophomonas indicatrix | Tomato | |
| ZA29E2 | Stenotrophomonas maltophilia | Stenotrophomonas geniculata | Carrot | |
| HSM1 | Stenotrophomonas maltophilia | Stenotrophomonas maltophilia | Abdominal cavity | Clinical |
| HSM2 | Stenotrophomonas maltophilia | Stenotrophomonas maltophilia | Respiratory tract | |
| HSM3 | Stenotrophomonas maltophilia | Stenotrophomonas maltophilia | Respiratory tract | |
| HSM4 | Stenotrophomonas maltophilia | Stenotrophomonas maltophilia | Respiratory tract | |
| HSM5 | Stenotrophomonas rhizophila | Stenotrophomonas sp. | Soft tissue | |
| HSM6 | Stenotrophomonas maltophilia | Stenotrophomonas sp. | Soft tissue | |
| HSM7 | Stenotrophomonas maltophilia | Stenotrophomonas maltophilia | Respiratory tract | |
| HSM8 | Stenotrophomonas maltophilia | Stenotrophomonas maltophilia | Abdominal cavity | |
| HSM9 | Stenotrophomonas maltophilia | Stenotrophomonas maltophilia | Sputum | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Pintor-Cora, A.; Alegría, Á.; López-Medrano, R.; Rodríguez-Calleja, J.M.; Santos, J.A. Diversity Among Clinical and Fresh Produce Isolates of Stenotrophomonas: Insights Through a One Health Perspective. Foods 2026, 15, 23. https://doi.org/10.3390/foods15010023
Pintor-Cora A, Alegría Á, López-Medrano R, Rodríguez-Calleja JM, Santos JA. Diversity Among Clinical and Fresh Produce Isolates of Stenotrophomonas: Insights Through a One Health Perspective. Foods. 2026; 15(1):23. https://doi.org/10.3390/foods15010023
Chicago/Turabian StylePintor-Cora, Alberto, Ángel Alegría, Ramiro López-Medrano, Jose M. Rodríguez-Calleja, and Jesús A. Santos. 2026. "Diversity Among Clinical and Fresh Produce Isolates of Stenotrophomonas: Insights Through a One Health Perspective" Foods 15, no. 1: 23. https://doi.org/10.3390/foods15010023
APA StylePintor-Cora, A., Alegría, Á., López-Medrano, R., Rodríguez-Calleja, J. M., & Santos, J. A. (2026). Diversity Among Clinical and Fresh Produce Isolates of Stenotrophomonas: Insights Through a One Health Perspective. Foods, 15(1), 23. https://doi.org/10.3390/foods15010023









