Near-Infrared Spectroscopy Combined with Explainable Machine Learning for Storage Time Prediction of Frozen Antarctic Krill
Abstract
1. Introduction
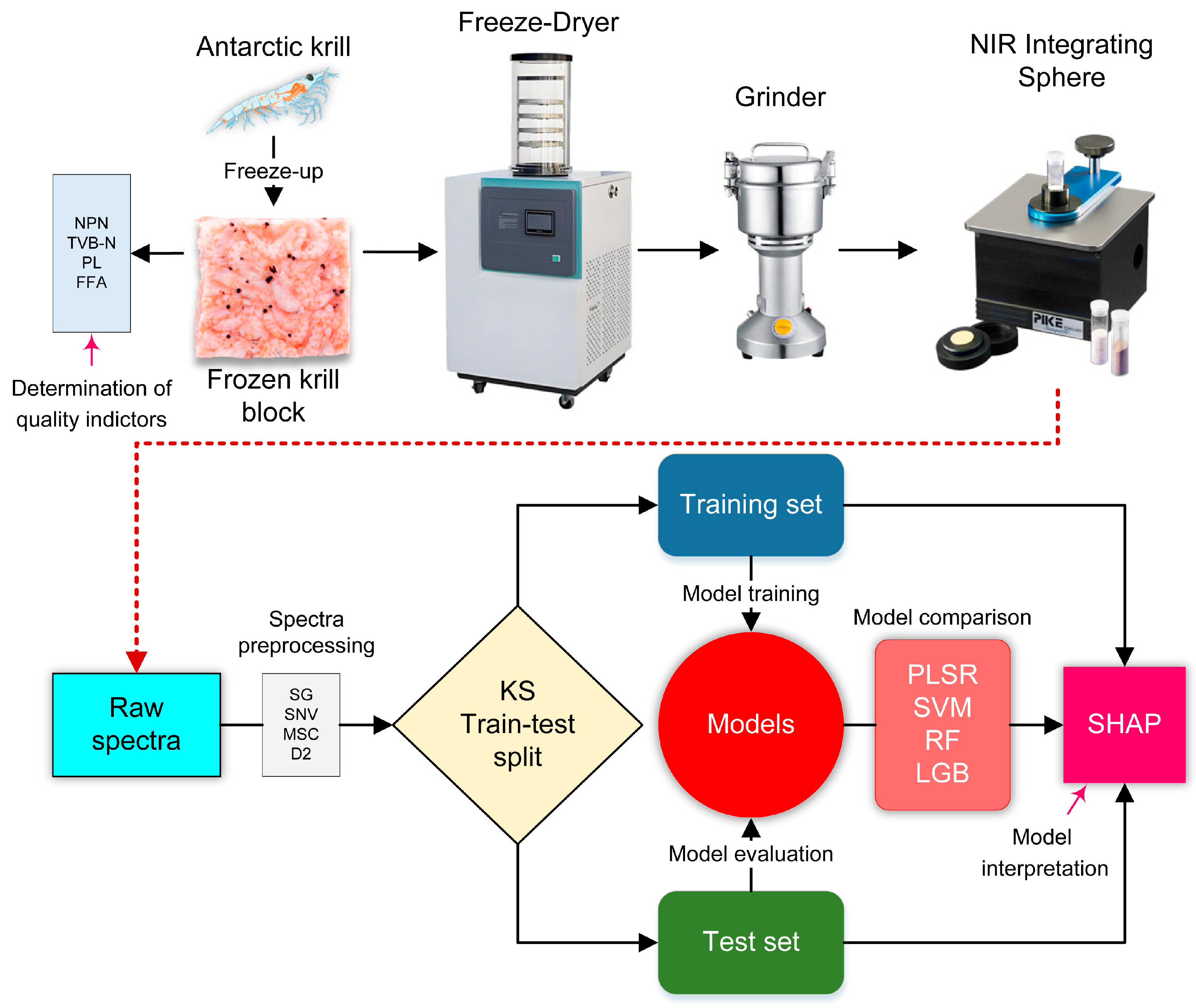
2. Materials and Methods
2.1. Materials
2.2. Measurement of Quality Indicators
2.3. NIR Spectra Acquisition
2.4. Spectra Preprocessing
2.5. Machine Learning Models
2.5.1. Partial Least Squares Regression (PLSR)
2.5.2. Support Vector Regression (SVR)
2.5.3. Random Forest (RF)
2.5.4. Light Gradient Boosting Machine (LightGBM)
2.6. Model Construction and Evaluation
2.7. Hyperparameters Optimization
2.8. Model Interpretation
2.9. Computing Implementation
3. Results and Discussion
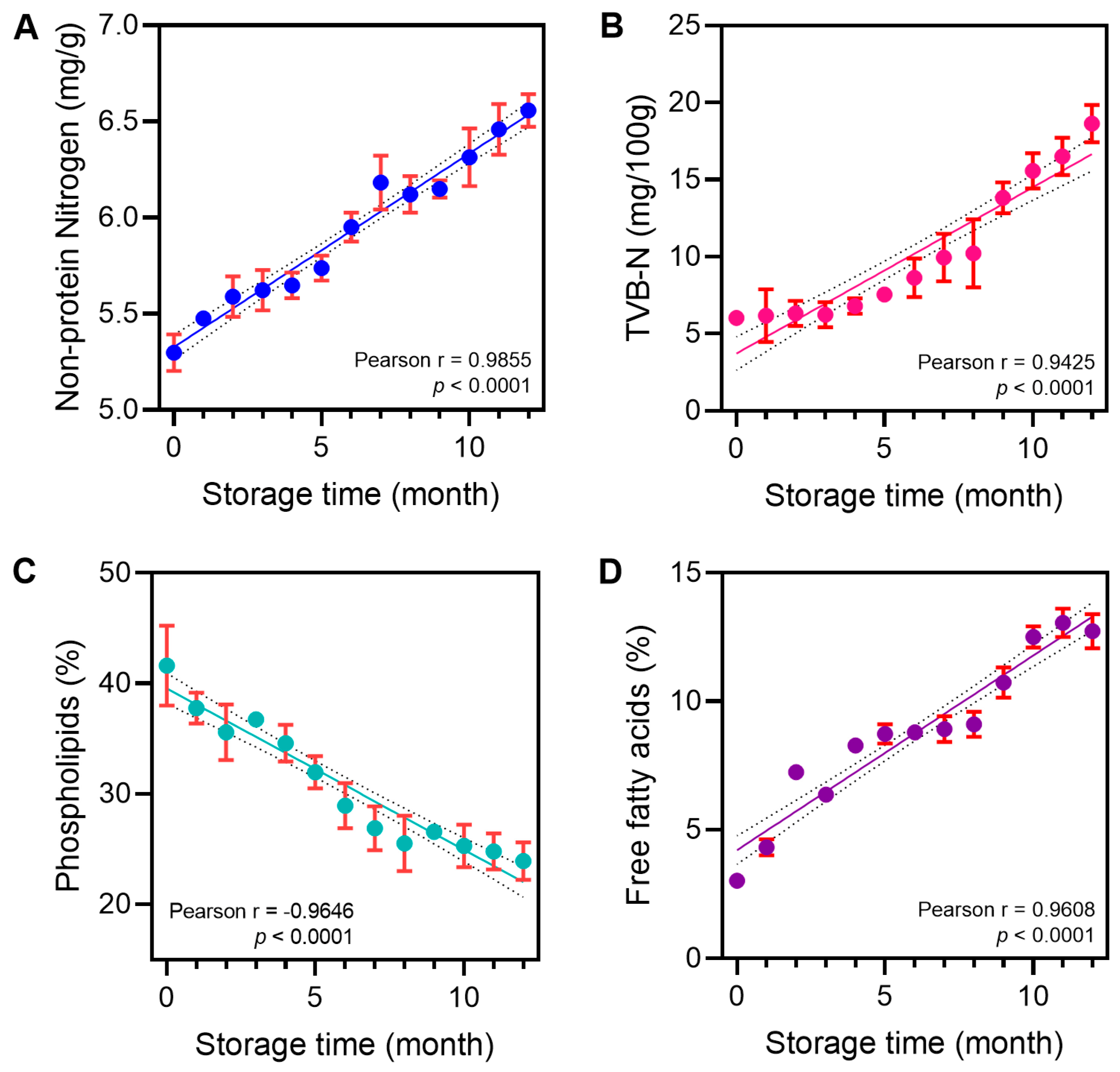
3.1. Correlation Between Krill Quality Indictors and Storage Time
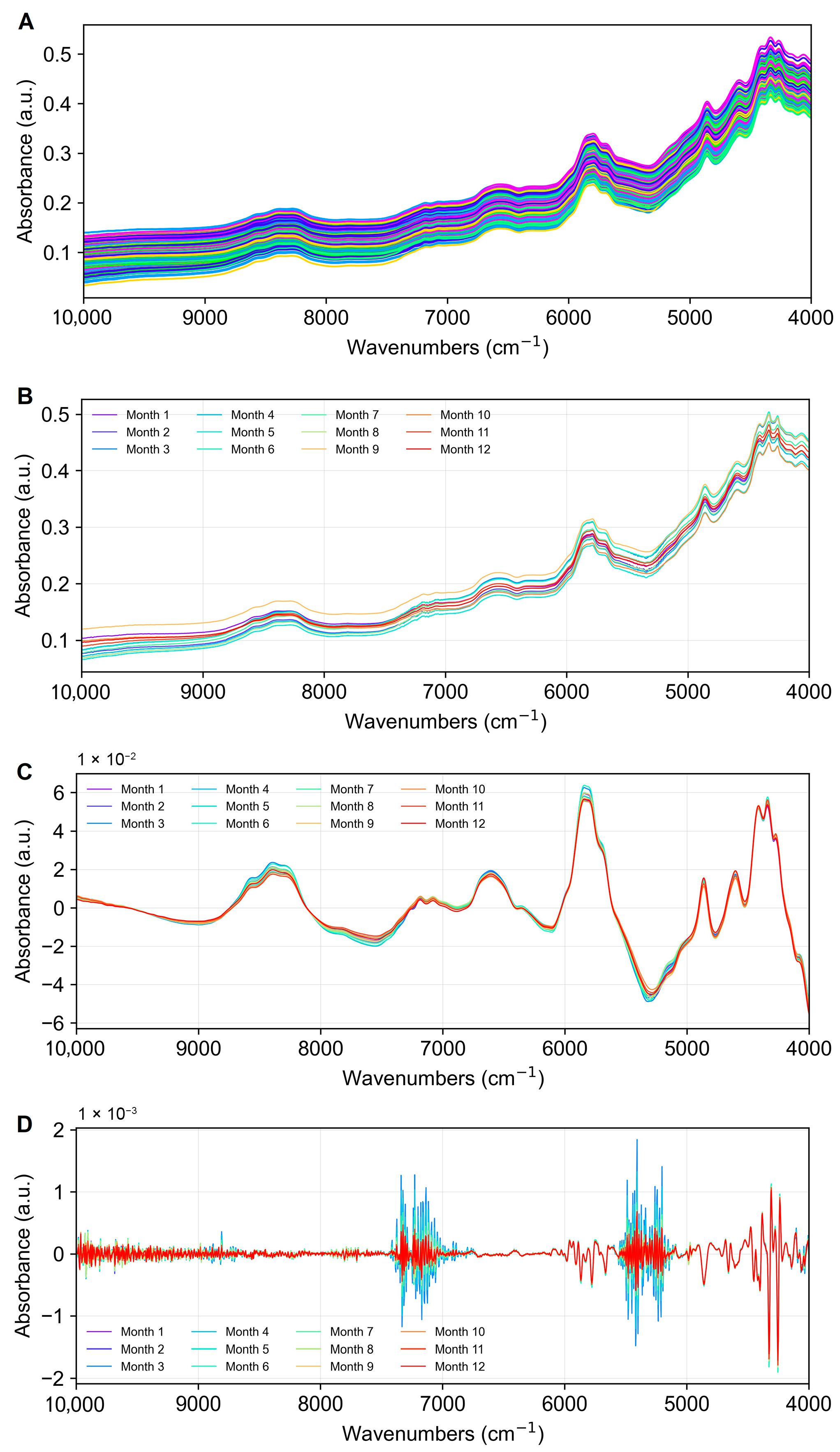
3.2. Preliminary Analysis of Krill Spectra
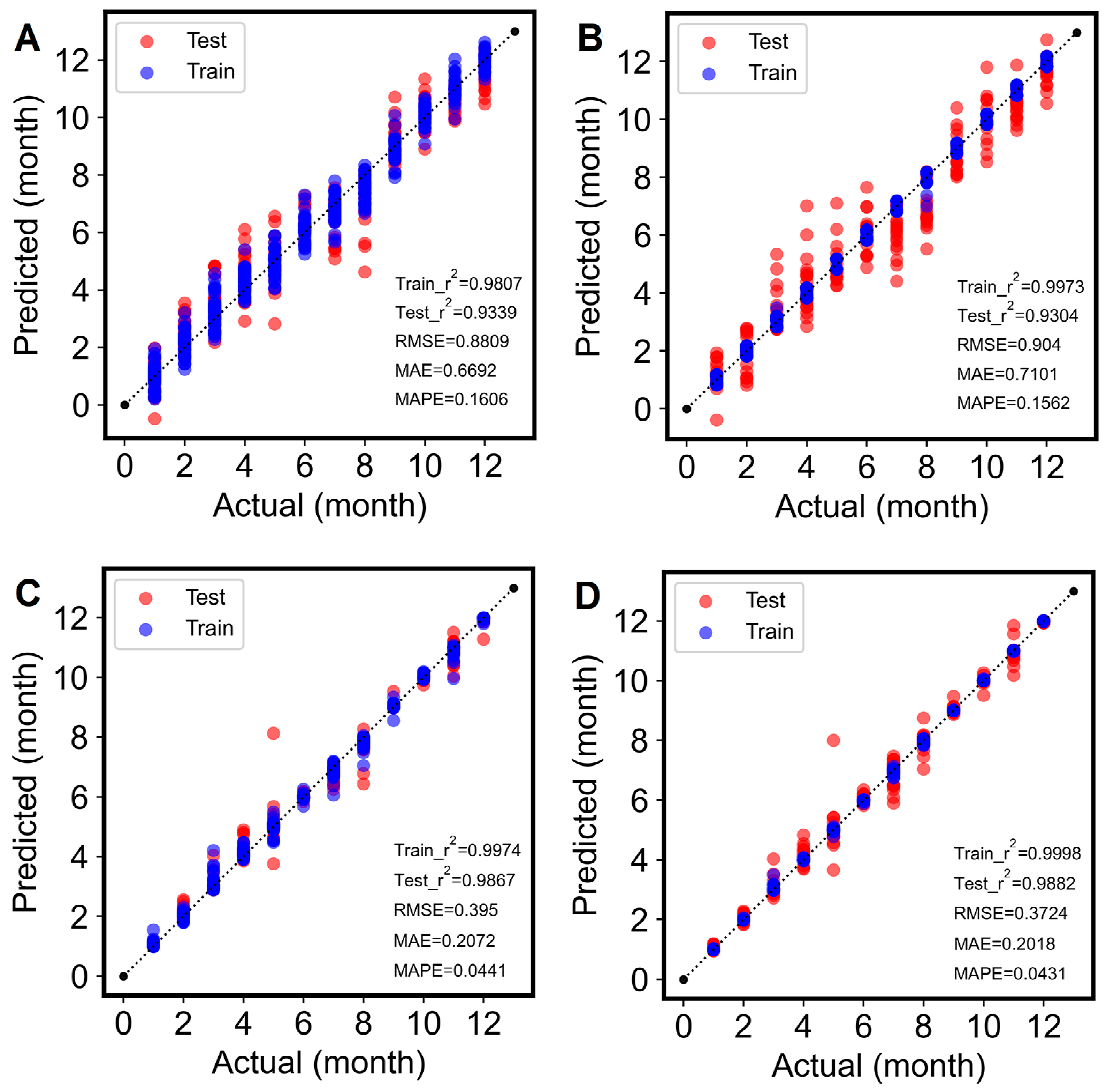
3.3. Prediction Modeling
3.3.1. Optimal Spectral Preprocessing Selection
3.3.2. Model Hyperparameters Optimization
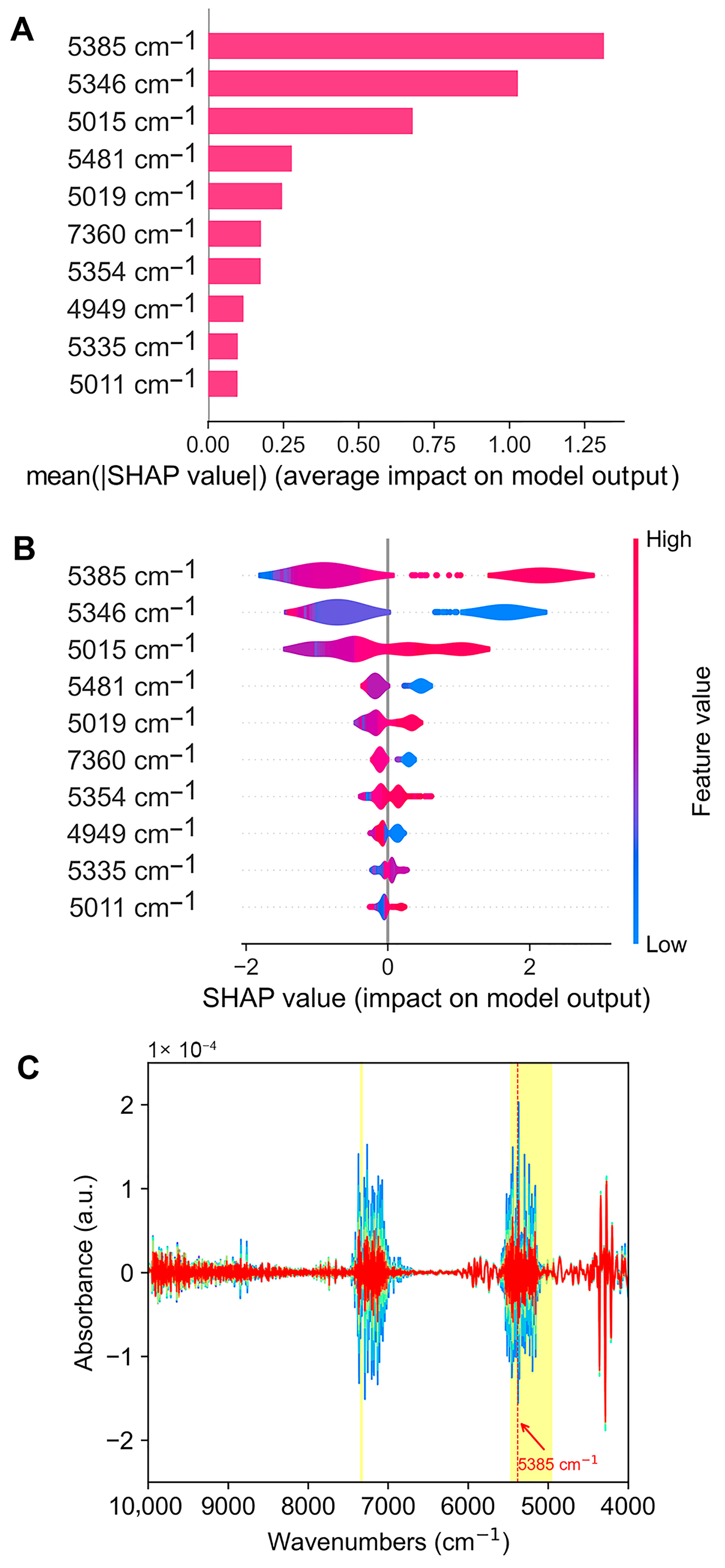
3.4. Interpretation of the Optimal Model
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| NIRS | near-infrared spectroscopy |
| NIR | near-infrared |
| LightGBM | light gradient boosting machine |
| TVB-N | total volatile base nitrogen |
| NPN | non-protein nitrogen |
| PL | phospholipids |
| FFA | free fatty acid |
| EPA | eicosapentaenoic acid |
| DHA | docosahexaenoic acid |
| ML | machine learning |
| PLSR | partial least squares regression |
| SVR | support vector regression |
| RF | random forest |
| XAI | explainable artificial intelligence |
| TCA | trichloroacetic acid |
| SD | standard deviation |
| MSC | multivariate scattering correction |
| SNV | standard normal variable |
| SG | Savitzky–Golay smoothing |
| D2 | second-order derivative |
| PCR | principal component regression |
| SVM | support vector machines |
| R2 | coefficient of determination |
| RMSE | root mean square error |
| MAE | mean absolute error |
| MAPE | mean absolute percentage error |
References
- Bar-On, Y.M.; Phillips, R.; Milo, R. The Biomass Distribution on Earth. Proc. Natl. Acad. Sci. USA 2018, 115, 6506–6511. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-C.; Tou, J.C.; Jaczynski, J. Amino Acid and Mineral Composition of Protein and Other Components and Their Recovery Yields from Whole Antarctic Krill (Euphausia superba) Using Isoelectric Solubilization/Precipitation. J. Food Sci. 2009, 74, H31–H39. [Google Scholar] [CrossRef] [PubMed]
- Tou, J.C.; Jaczynski, J.; Chen, Y.-C. Krill for Human Consumption: Nutritional Value and Potential Health Benefits. Nutr. Rev. 2007, 65, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Wang, J.J.; Li, Y.; Malakar, P.K.; Zhao, Y. Recent Advances in the Use of Antarctic Krill (Euphausia superba) as a Sustainable Source of High-Quality Protein: A Comprehensive Review. Trends Food Sci. Technol. 2024, 152, 104684. [Google Scholar] [CrossRef]
- Jiang, P.; Liu, Y.; Yang, J.; Cai, W.; Jiang, C.; Hang, J.; Miao, X.; Sun, N. Feasibility of Circular Fermentation as a New Strategy to Accelerate Fermentation and Enhance Flavor of Antarctic Krill Paste. Curr. Res. Food Sci. 2024, 9, 100838. [Google Scholar] [CrossRef]
- Sun, P.; Zhang, X.; Ren, X.; Cao, Z.; Zhao, Y.; Man, H.; Li, D. Effect of Basic Amino Acid Pretreatment on the Quality of Canned Antarctic Krill. Food Bioprocess Technol. 2023, 16, 1690–1702. [Google Scholar] [CrossRef]
- Teng, X.-N.; Wang, S.-C.; Zeb, L.; Xiu, Z.-L. Effects of Carboxymethyl Chitosan Adsorption on Bioactive Components of Antarctic Krill Oil. Food Chem. 2022, 388, 132995. [Google Scholar] [CrossRef]
- Chang, H.-C.; Chen, S.-D. Study on Microwave Freeze-Drying of Krill. Processes 2024, 12, 1366. [Google Scholar] [CrossRef]
- Ma, X.; Su, D.; Zhu, J.; Miao, J.; Liu, X.; Leng, K.; Wang, X.; Xie, W. Comparative Metabolomics Study on the Quality of Antarctic Krill (Euphausia superba) Stored at Different Temperatures. Int. J. Food Sci. Technol. 2024, 59, 4489–4499. [Google Scholar] [CrossRef]
- Kolakowski, E. Changes of Non-Protein Nitrogens Fractions in Antarctic Krill (Euphausia superba Dana) during Storage at 3 and 20 °C. Z. Lebensm. Unters. Forch. 1986, 183, 421–425. [Google Scholar] [CrossRef]
- Nielsen, N.S.; Lu, H.F.S.; Bruheim, I.; Jacobsen, C. Quality Changes of Antarctic Krill Powder during Long Term Storage. Eur. J. Lipid Sci. Technol. 2017, 119, 1600085. [Google Scholar] [CrossRef]
- Xue, M.; Zhu, G. Variation in Fatty Acids of Antarctic Krill (Euphausia superba) Preserved under Constant Dry Conditions: Does Storage Time and Ontogeny Matter? J. Food Process. Preserv. 2021, 45, e15357. [Google Scholar] [CrossRef]
- Wang, L.; Liu, Z.; Yang, H.; Huang, L.; Zeng, M. Optimal Modified Atmosphere for Packaging and Its Effects on Quality and Shelf-Life of Pacific White Shrimp (Litopenaeus vannamei) under Controlled Freezing-Point Storage at −0.8 °C. Food Sci. Technol. Res. 2016, 22, 173–183. [Google Scholar] [CrossRef]
- Xie, D.; Gong, M.; Wei, W.; Jin, J.; Wang, X.; Wang, X.; Jin, Q. Antarctic Krill (Euphausia superba) Oil: A Comprehensive Review of Chemical Composition, Extraction Technologies, Health Benefits, and Current Applications. Compr. Rev. Food Sci. Food Saf. 2019, 18, 514–534. [Google Scholar] [CrossRef]
- Govari, M.; Tryfinopoulou, P.; Parlapani, F.F.; Boziaris, I.S.; Panagou, E.Z.; Nychas, G.-J.E. Quest of Intelligent Research Tools for Rapid Evaluation of Fish Quality: FTIR Spectroscopy and Multispectral Imaging versus Microbiological Analysis. Foods 2021, 10, 264. [Google Scholar] [CrossRef]
- Liu, D.; Zeng, X.-A.; Sun, D.-W. NIR Spectroscopy and Imaging Techniques for Evaluation of Fish Quality—A Review. Appl. Spectrosc. Rev. 2013, 48, 609–628. [Google Scholar] [CrossRef]
- Ozaki, Y.; Genkawa, T.; Futami, Y. Near-Infrared Spectroscopy. In Encyclopedia of Spectroscopy and Spectrometry, 3rd ed.; Lindon, J.C., Tranter, G.E., Koppenaal, D.W., Eds.; Academic Press: Hoboken, NJ, USA, 2017; pp. 40–49. [Google Scholar] [CrossRef]
- Basile, T.; Marsico, A.D.; Perniola, R. NIR Analysis of Intact Grape Berries: Chemical and Physical Properties Prediction Using Multivariate Analysis. Foods 2021, 10, 113. [Google Scholar] [CrossRef]
- Beć, K.B.; Grabska, J.; Huck, C.W. Near-Infrared Spectroscopy in Bio-Applications. Molecules 2020, 25, 2948. [Google Scholar] [CrossRef]
- Jiao, X.; Guo, D.; Zhang, X.; Su, Y.; Ma, R.; Chen, L.; Tian, K.; Su, J.; Sahati, T.; Aierkenjiang, X.; et al. The Application of Near-Infrared Spectroscopy Combined with Chemometrics in the Determination of the Nutrient Composition in Chinese Cyperus esculentus L. Foods 2025, 14, 366. [Google Scholar] [CrossRef]
- Jordan, M.I.; Mitchell, T.M. Machine Learning: Trends, Perspectives, and Prospects. Science 2015, 349, 255–260. [Google Scholar] [CrossRef]
- Janiesch, C.; Zschech, P.; Heinrich, K. Machine Learning and Deep Learning. Electron. Mark. 2021, 31, 685–695. [Google Scholar] [CrossRef]
- Ninh, D.K.; Phan, K.D.; Vo, C.T.; Dang, M.N.; Le Thanh, N. Classification of Histamine Content in Fish Using Near-Infrared Spectroscopy and Machine Learning Techniques. Information 2024, 15, 528. [Google Scholar] [CrossRef]
- Shim, K.; Jeong, Y. Freshness Evaluation in Chub Mackerel (Scomber japonicus) Using near-Infrared Spectroscopy Determination of the Cadaverine Content. J. Food Prot. 2019, 82, 768–774. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Wang, L.; Wu, H.; Li, Q.; Li, M.; Zhang, Z.; Zhao, Y.; Lu, Z.; Zou, Z. Machine Learning Modeling and Prediction of Peanut Protein Content Based on Spectral Images and Stoichiometry. LWT 2022, 169, 114015. [Google Scholar] [CrossRef]
- Zaukuu, J.-L.Z.; Zimmermann, E.; Acquah, B.B.; Kwofie, E.D. Novel Detection Techniques for Shrimp Powder Adulteration Using Near Infrared Spectroscopy in Tandem Chemometric Tools and Multiple Spectral Preprocessing. Food Anal. Methods 2023, 16, 819–831. [Google Scholar] [CrossRef]
- Zou, Z.; Yuan, D.; Wu, Q.; Wang, Q.; Li, M.; Zhen, J.; Xu, C.; Yin, S.; Cui, Q.; Zhou, M.; et al. Salmon Origin Traceability Based on Hyperspectral Imaging Data Fusion Strategy and Improved Deep Learning Method. Food Control 2024, 166, 110740. [Google Scholar] [CrossRef]
- Cavallini, N.; Pennisi, F.; Giraudo, A.; Pezzolato, M.; Esposito, G.; Gavoci, G.; Magnani, L.; Pianezzola, A.; Geobaldo, F.; Savorani, F.; et al. Chemometric Differentiation of Sole and Plaice Fish Fillets Using Three Near-Infrared Instruments. Foods 2022, 11, 1643. [Google Scholar] [CrossRef]
- Cui, Y.; Pan, T.; Chen, S.; Zou, X. A Gender Classification Method for Chinese Mitten Crab Using Deep Convolutional Neural Network. Multimed. Tools Appl. 2020, 79, 7669–7684. [Google Scholar] [CrossRef]
- Huang, F.; Peng, Y.; Li, L.; Ye, S.; Hong, S. Near-Infrared Spectroscopy Combined with Machine Learning Methods for Distinguishment of the Storage Years of Rice. Infrared Phys. Technol. 2023, 133, 104835. [Google Scholar] [CrossRef]
- Shi, S.; Feng, J.; Yang, L.; Xing, J.; Pan, G.; Tang, J.; Wang, J.; Liu, J.; Cao, C.; Jiang, Y. Combination of NIR Spectroscopy and Algorithms for Rapid Differentiation between One-Year and Two-Year Stored Rice. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 291, 122343. [Google Scholar] [CrossRef]
- Guo, H.; Yan, F.; Li, P.; Li, M. Determination of Storage Period of Harvested Plums by Near-Infrared Spectroscopy and Quality Attributes. J. Food Process. Preserv. 2022, 46, e16504. [Google Scholar] [CrossRef]
- Shen, F.; Zhang, B.; Cao, C.; Jiang, X. On-Line Discrimination of Storage Shelf-Life and Prediction of Post-Harvest Quality for Strawberry Fruit by Visible and near Infrared Spectroscopy. J. Food Process Eng. 2018, 41, e12866. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, D.; Ren, X.; Shen, Y.; Cao, X.; Liu, H.; Li, J. Quality Changes and Shelf-Life Prediction Model of Postharvest Apples Using Partial Least Squares and Artificial Neural Network Analysis. Food Chem. 2022, 394, 133526. [Google Scholar] [CrossRef]
- Guan, S.; Shang, Y.; Zhao, C. Storage Time Detection of Torreya Grandis Kernels Using Near Infrared Spectroscopy. Sustainability 2023, 15, 7757. [Google Scholar] [CrossRef]
- Yan, Z.; Liu, H.; Li, J.; Wang, Y. Qualitative and Quantitative Analysis of Lanmaoa asiatica in Different Storage Years Based on FT-NIR Combined with Chemometrics. Microchem. J. 2023, 189, 108580. [Google Scholar] [CrossRef]
- Nori, H.; Jenkins, S.; Koch, P.; Caruana, R. InterpretML: A Unified Framework for Machine Learning Interpretability. Available online: https://arxiv.org/abs/1909.09223v1 (accessed on 10 March 2025).
- Lundberg, S.M.; Erion, G.; Chen, H.; DeGrave, A.; Prutkin, J.M.; Nair, B.; Katz, R.; Himmelfarb, J.; Bansal, N.; Lee, S.-I. From Local Explanations to Global Understanding with Explainable AI for Trees. Nat. Mach. Intell. 2020, 2, 56–67. [Google Scholar] [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why Should I Trust You?”: Explaining the Predictions of Any Classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016. [Google Scholar]
- Jeong, S.; Kim, Y.; Hur, S.H.; Bang, H.; Kim, H.; Chung, H. Explainable Extreme Gradient Boosting as a Machine Learning Tool for Discrimination of the Geographical Origin of Chili Peppers Using Laser Ablation-Inductively Coupled Plasma Mass Spectrometry, X-Ray Fluorescence, and near-Infrared Spectroscopy. J. Agric. Food Res. 2024, 18, 101446. [Google Scholar] [CrossRef]
- Kalopesa, E.; Karyotis, K.; Tziolas, N.; Tsakiridis, N.; Samarinas, N.; Zalidis, G. Estimation of Sugar Content in Wine Grapes via in Situ VNIR–SWIR Point Spectroscopy Using Explainable Artificial Intelligence Techniques. Sensors 2023, 23, 1065. [Google Scholar] [CrossRef]
- Ren, Z.-R.; Luo, L.; Na, B. Predicting the Air-Dry Density of Black Walnut Based on NIR Analysis. Holzforschung 2023, 77, 784–792. [Google Scholar] [CrossRef]
- Sun, Y.; Li, L.; Meng, Z.; Sun, H.; Cao, R.; Liu, R.; Wang, S.; Liu, N. Rapid On-Site Differentiation of Turbot from Different Culture Modes Using Miniaturized near Infrared Spectroscopy Coupled with Interpretable Machine Learning. Microchem. J. 2024, 207, 111981. [Google Scholar] [CrossRef]
- Standard No. GB 5009.228-2016; National Food Safety Standard: Determination of Volatile Salt Nitrogen in Foods. Standardization Administration of China (SAC): Beijing, China, 2016.
- Standard No. GB/T 5537-2008; Inspection of Grain and Oils-Determination of Phosphatide Content. Standardization Administration of China (SAC): Beijing, China, 2008.
- Lowry, R.R.; Tinsley, I.J. Rapid Colorimetric Determination of Free Fatty Acids. J. Am. Oil Chem. Soc. 1976, 53, 470–472. [Google Scholar] [CrossRef] [PubMed]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-Regression: A Basic Tool of Chemometrics. Chemom. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Tan, B.; You, W.; Tian, S.; Xiao, T.; Wang, M.; Zheng, B.; Luo, L. Soil Nitrogen Content Detection Based on Near-Infrared Spectroscopy. Sensors 2022, 22, 8013. [Google Scholar] [CrossRef]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.-Y. LightGBM: A Highly Efficient Gradient Boosting Decision Tree. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; Curran Associates Inc.: Red Hook, NY, USA, 2017; pp. 3149–3157. [Google Scholar]
- Kennard, R.W.; Stone, L.A. Computer Aided Design of Experiments. Technometrics 1969, 11, 137–148. [Google Scholar] [CrossRef]
- Akiba, T.; Sano, S.; Yanase, T.; Ohta, T.; Koyama, M. Optuna: A Next-Generation Hyperparameter Optimization Framework. Available online: https://arxiv.org/abs/1907.10902v1 (accessed on 30 January 2024).
- Srinivas, P.; Katarya, R. hyOPTXg: OPTUNA Hyper-Parameter Optimization Framework for Predicting Cardiovascular Disease Using XGBoost. Biomed. Signal Process. Control 2022, 73, 103456. [Google Scholar] [CrossRef]
- Nishimura, K.; Kawamura, Y.; Matoba, T.; Yonezawa, D. Deterioration of Antarctic Krill Muscle during Freeze Storage. Agric. Biol. Chem. 1983, 47, 2881–2888. [Google Scholar] [CrossRef]
- Huang, X.; Cao, R.; Li, Z. Changes in Protein Components of Antarctic Krill during Autolysis Process. Hunan Agric. Sci. 2015, 9, 76–78. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, M.; Bian, H.; Xu, W.; Xu, X.; Zhu, Y.; Liu, F.; Geng, Z.; Zhou, G. Changes of Phospholipase A2 and C Activities during Dry-Cured Duck Processing and Their Relationship with Intramuscular Phospholipid Degradation. Food Chem. 2014, 145, 997–1001. [Google Scholar] [CrossRef]
- Bao, J.; Chen, L.; Liu, T. Dandelion Polysaccharide Suppresses Lipid Oxidation in Antarctic Krill (Euphausia superba). Int. J. Biol. Macromol. 2019, 133, 1164–1167. [Google Scholar] [CrossRef] [PubMed]
- Nicolaï, B.M.; Beullens, K.; Bobelyn, E.; Peirs, A.; Saeys, W.; Theron, K.I.; Lammertyn, J. Nondestructive Measurement of Fruit and Vegetable Quality by Means of NIR Spectroscopy: A Review. Postharvest Biol. Technol. 2007, 46, 99–118. [Google Scholar] [CrossRef]
- Li, C.; Guo, H.; Zong, B.; He, P.; Fan, F.; Gong, S. Rapid and Non-Destructive Discrimination of Special-Grade Flat Green Tea Using near-Infrared Spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 206, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Zeng, Z.; Hou, Y.; Chen, A.; Xu, J.; Zhao, L.; Liu, X. Rapid Authentication of Variants of Gastrodia Elata Blume Using near-Infrared Spectroscopy Combined with Chemometric Methods. J. Pharm. Biomed. Anal. 2023, 235, 115592. [Google Scholar] [CrossRef]
- Wang, H.-P.; Chen, P.; Dai, J.-W.; Liu, D.; Li, J.-Y.; Xu, Y.-P.; Chu, X.-L. Recent Advances of Chemometric Calibration Methods in Modern Spectroscopy: Algorithms, Strategy, and Related Issues. TrAC Trends Anal. Chem. 2022, 153, 116648. [Google Scholar] [CrossRef]
- Mienye, I.D.; Sun, Y. A Survey of Ensemble Learning: Concepts, Algorithms, Applications, and Prospects. IEEE Access 2022, 10, 99129–99149. [Google Scholar] [CrossRef]
- Johnson, J.B.; Walsh, K.B.; Naiker, M.; Ameer, K. The Use of Infrared Spectroscopy for the Quantification of Bioactive Compounds in Food: A Review. Molecules 2023, 28, 3215. [Google Scholar] [CrossRef]
- Ma, L.; Peng, Y.; Pei, Y.; Zeng, J.; Shen, H.; Cao, J.; Qiao, Y.; Wu, Z. Systematic Discovery about NIR Spectral Assignment from Chemical Structural Property to Natural Chemical Compounds. Sci. Rep. 2019, 9, 9503. [Google Scholar] [CrossRef]
- Teppola, P. Near-Infrared Spectroscopy. Principles, Instruments, Applications. H. W. Siesler, Y. Ozaki, S. Kawata and H. M. Heise (Eds): Book Review. J. Chemom. 2002, 16, 636–638. [Google Scholar] [CrossRef]
| Model | Preprocess | R2 | RMSE | MAE | MAPE |
|---|---|---|---|---|---|
| PLSR | Raw | 0.1917 | 3.0800 | 2.6571 | 0.8505 |
| MSC | 0.5895 | 2.1949 | 1.7490 | 0.5652 | |
| SNV | 0.5869 | 2.2018 | 1.7579 | 0.5647 | |
| D2 | 0.8209 | 1.4497 | 1.2179 | 0.3065 | |
| D2+SG | 0.8211 | 1.4489 | 1.2060 | 0.2889 | |
| D2+MSC | 0.8016 | 1.5260 | 1.2391 | 0.3649 | |
| D2+SNV | 0.8016 | 1.5258 | 1.2929 | 0.3468 | |
| SVR | Raw | 0.1550 | 3.1492 | 2.5274 | 0.9071 |
| MSC | 0.0530 | 3.3339 | 2.8820 | 0.8304 | |
| SNV | 0.0529 | 3.3339 | 2.8821 | 0.8304 | |
| D2 | 0.8189 | 1.4580 | 1.1446 | 0.3752 | |
| D2+SG | 0.6521 | 2.0208 | 1.5529 | 0.5267 | |
| D2+MSC | 0.8239 | 1.4374 | 1.1276 | 0.3611 | |
| D2+SNV | 0.8240 | 1.4372 | 1.1275 | 0.3610 | |
| RF | Raw | 0.5843 | 2.2089 | 1.6499 | 0.5742 |
| MSC | 0.8096 | 1.4949 | 0.7728 | 0.2659 | |
| SNV | 0.8115 | 1.4874 | 0.7431 | 0.2567 | |
| D2 | 0.9732 | 0.5612 | 0.2563 | 0.0547 | |
| D2+SG | 0.9862 | 0.4027 | 0.1938 | 0.0415 | |
| D2+MSC | 0.9419 | 0.8255 | 0.3325 | 0.0667 | |
| D2+SNV | 0.9438 | 0.8122 | 0.3273 | 0.0670 | |
| LightGBM | Raw | 0.6134 | 2.1301 | 1.6006 | 0.5285 |
| MSC | 0.8352 | 1.3907 | 0.7733 | 0.2723 | |
| SNV | 0.8549 | 1.3049 | 0.7459 | 0.2602 | |
| D2 | 0.9823 | 0.4561 | 0.2513 | 0.0586 | |
| D2+SG | 0.9864 | 0.3999 | 0.2095 | 0.0436 | |
| D2+MSC | 0.9782 | 0.5063 | 0.3016 | 0.0705 | |
| D2+SNV | 0.9774 | 0.5154 | 0.3115 | 0.0763 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Cao, R.; Zhao, L.; Liu, N.; Sun, H.; Zhang, Z.; Sun, Y. Near-Infrared Spectroscopy Combined with Explainable Machine Learning for Storage Time Prediction of Frozen Antarctic Krill. Foods 2025, 14, 1293. https://doi.org/10.3390/foods14081293
Li L, Cao R, Zhao L, Liu N, Sun H, Zhang Z, Sun Y. Near-Infrared Spectroscopy Combined with Explainable Machine Learning for Storage Time Prediction of Frozen Antarctic Krill. Foods. 2025; 14(8):1293. https://doi.org/10.3390/foods14081293
Chicago/Turabian StyleLi, Lin, Rong Cao, Ling Zhao, Nan Liu, Huihui Sun, Zhaohui Zhang, and Yong Sun. 2025. "Near-Infrared Spectroscopy Combined with Explainable Machine Learning for Storage Time Prediction of Frozen Antarctic Krill" Foods 14, no. 8: 1293. https://doi.org/10.3390/foods14081293
APA StyleLi, L., Cao, R., Zhao, L., Liu, N., Sun, H., Zhang, Z., & Sun, Y. (2025). Near-Infrared Spectroscopy Combined with Explainable Machine Learning for Storage Time Prediction of Frozen Antarctic Krill. Foods, 14(8), 1293. https://doi.org/10.3390/foods14081293






