Identification and Classification of Snack-Type Watermelon (Citrullus lanatus) Genotypes Using Seed Morphology and Machine Learning Techniques
Abstract
1. Introduction
- •
- The genotyping of nine watermelon genotypes of the seed stock based on red, white, and black seed coats was performed using the morphological, physical, and colorimetric (L, a, b) characteristics of the seeds.
- •
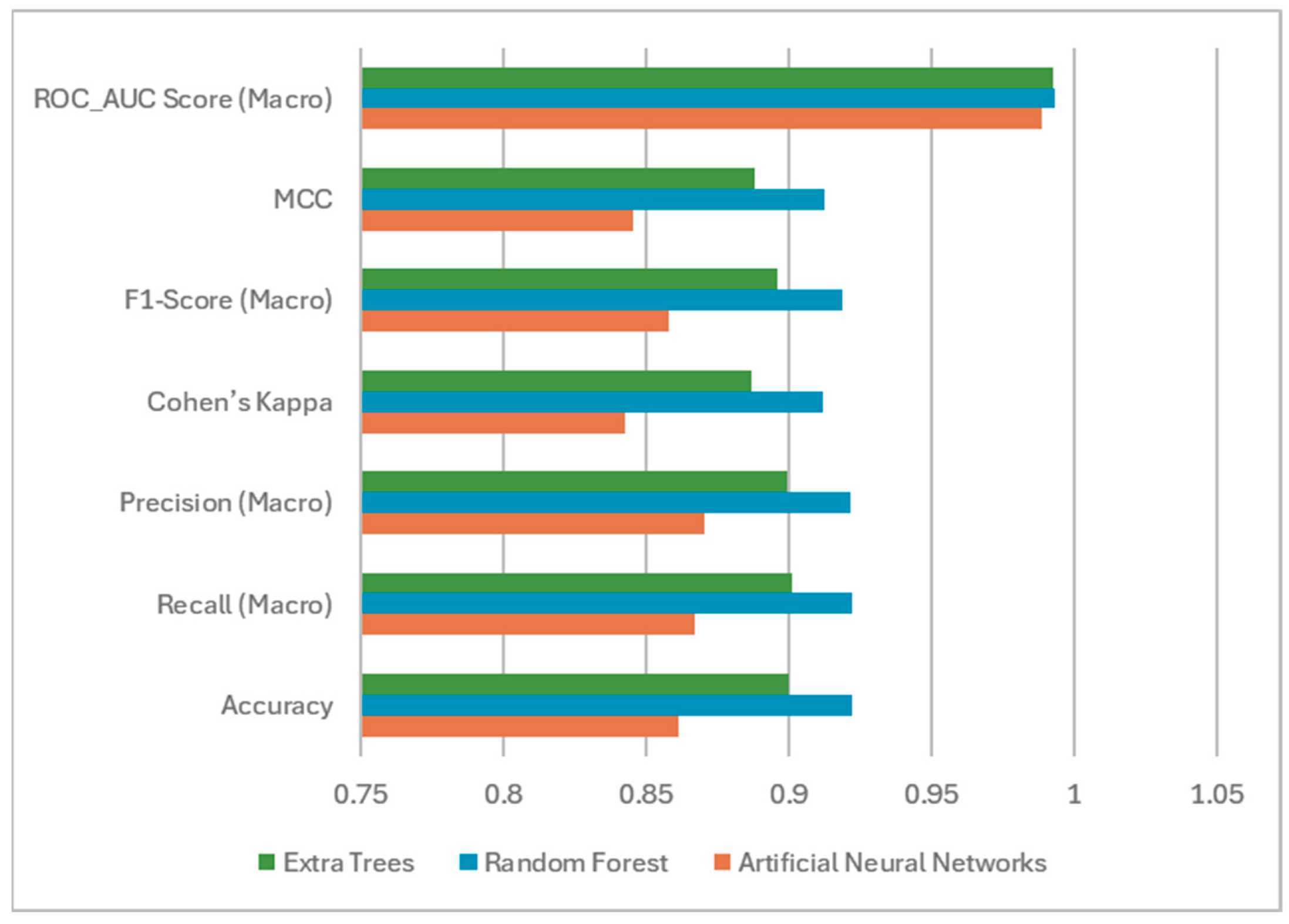
- ANN, Random Forest (RF), and Extra Trees (ET) algorithms were relatively compared, and it was shown that the best performance was achieved by the RF model with an accuracy of 92.22 and a Cohen Kappa value of 0.9118.
- •
- The Wilcoxon signed-rank test statistically proved that the RF and ET models were significantly superior to the ANN model, which demonstrates the high stability of the ensemble-based approaches.
- •
- The suggested ML-based framework can be used to offer a fast, accurate, and non-destructive classification framework able to fulfill the high-throughput needs of the seed processing industry, the breeding programs enhancing varietal traceability, and the quality regulation in commercial seed production.
- •
- The research paper helps to fill the gap between traditional methods and more modern machine learning applications providing concrete evidence of the relevance of computer vision and artificial intelligence, especially in the domain of horticultural seed science.
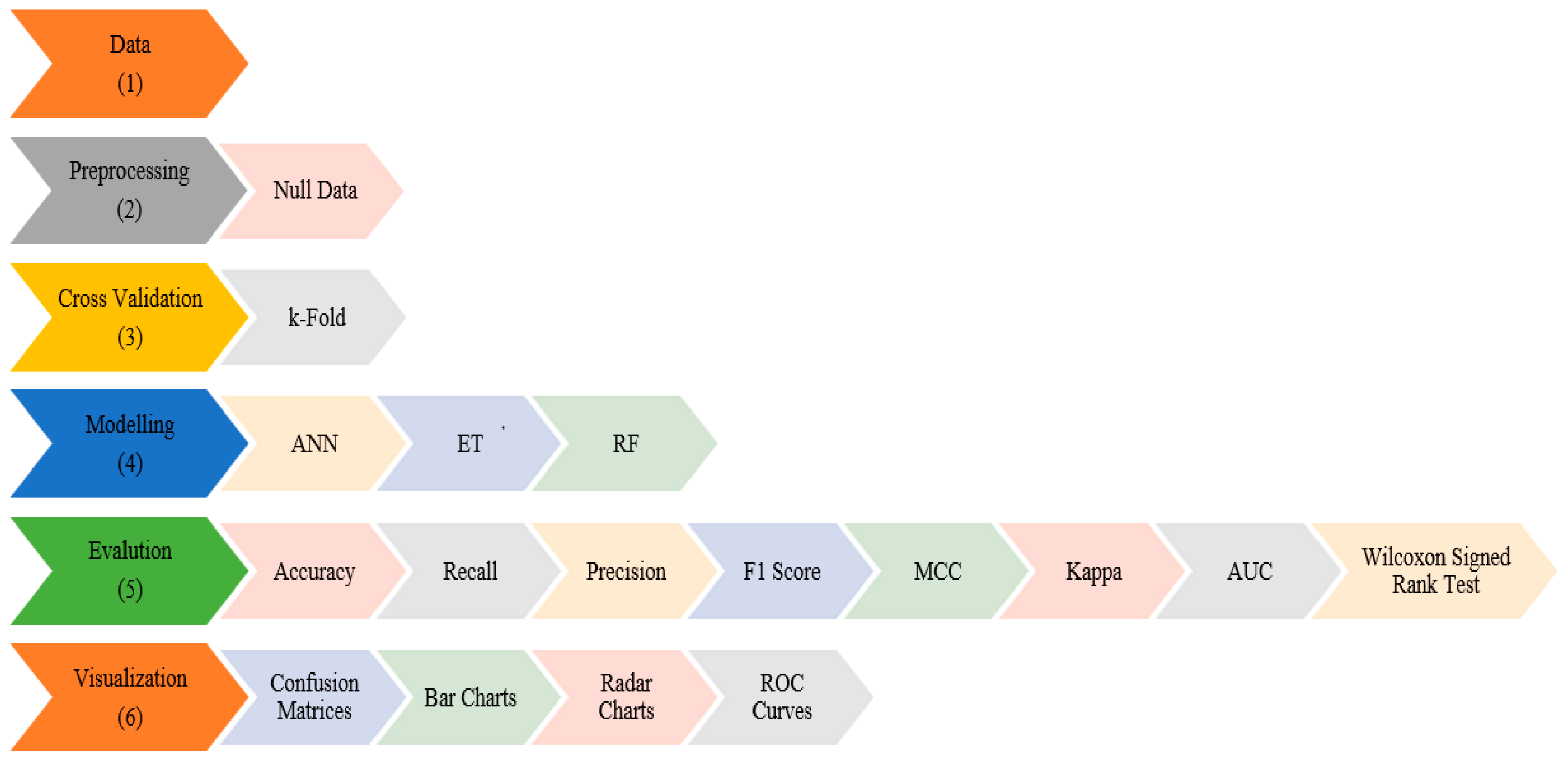
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Data Collection Site
2.3. Machine Learning
2.4. Random Forests
2.5. Artificial Neural Networks
2.6. Extra Trees
3. Results and Discussion
3.1. Statistical Analysis
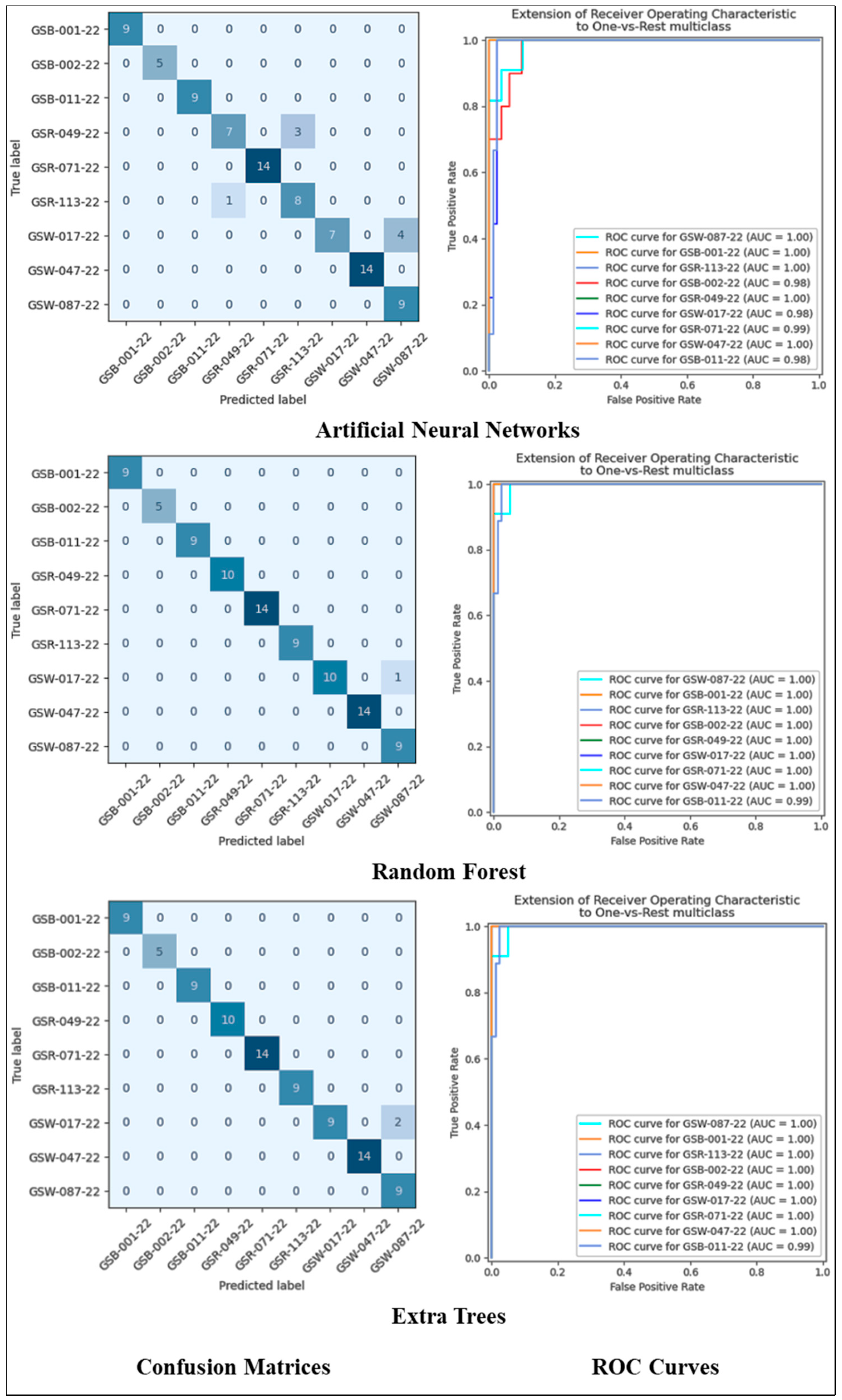
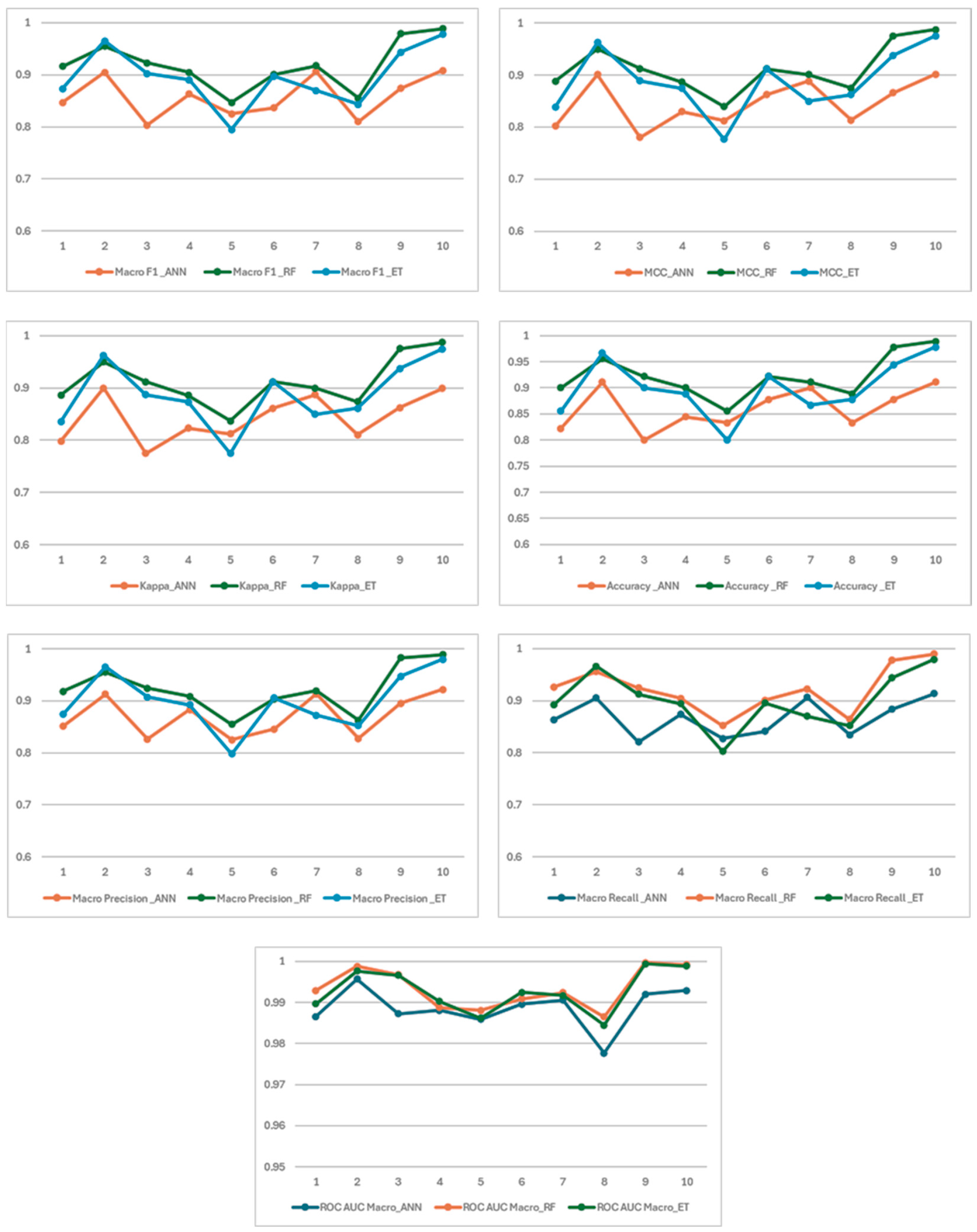
3.2. Fold Basis Results and Interpretation
3.3. Based on Machine Learning Method and Evaluation Metrics
3.4. Statistical Analysis: Significance Between Model Results
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ANN | Artificial Neural Networks |
| RF | Random Forest |
| ET | Extra Trees |
| ML | Machine Learning |
| CNNs | Convolutional Neural Networks |
| SVM | Support Vector Machines |
| LR | Logistic Regression |
| kNN | k-Nearest Neighbors |
| NIR-HSI | Near-Infrared Hyperspectral Imaging |
References
- Maoto, M.M.; Beswa, D.; Jideani, A.I. Watermelon as a potential fruit snack. Int. J. Food Prop. 2019, 22, 355–370. [Google Scholar] [CrossRef]
- Wani, A.A.; Sogi, D.S.; Singh, P.; Wani, I.A.; Shivhare, U.S. Characterisation and functional properties of watermelon (Citrullus lanatus) seed proteins. J. Sci. Food Agric. 2011, 91, 113–121. [Google Scholar] [CrossRef]
- Siol, M.; Witkowska, B.; Mańko-Jurkowska, D.; Makouie, S.; Bryś, J. Comprehensive evaluation of the nutritional quality of stored watermelon seed oils. Appl. Sci. 2025, 15, 830. [Google Scholar] [CrossRef]
- Benmeziane, F.; Derradji, F. Composition, bioactive potential and food applications of watermelon (Citrullus lanatus) seeds–a review. J. Food Meas. Charact. 2023, 17, 5045–5061. [Google Scholar] [CrossRef]
- Sathish, S.; Vikram, S.; Suraj, R.J.N.E. Effectiveness of turbidity removal from synthetic and tannery wastewater by using seeds of a natural coagulant Citrullus lanatus. Nat. Environ. Pollut. Technol. 2018, 17, 551–553. [Google Scholar]
- Köçeroğlu, D. Kavurma İşleminin Karpuz Çekirdeği Yağının Oksidasyonu Üzerine Etkisi. Master’s Thesis, Van Yüzüncü Yıl University, Institute of Science, Department of Food Engineering, Van, Türkiye, 2018; 56p. [Google Scholar]
- El-Adawy, T.A.; Taha, K.M. Characteristics and composition of watermelon, pumpkin, and paprika seed oils and flours. J. Agric. Food Chem. 2001, 49, 1253–1259. [Google Scholar] [CrossRef] [PubMed]
- Tabiri, B.; Agbenorhevi, J.K.; Wireko-Manu, F.D.; Ompouma, E.I. Watermelon seeds as food: Nutrient composition, phytochemicals and antioxidant activity. Int. J. Nutr. Food Sci. 2016, 5, 18. [Google Scholar] [CrossRef]
- Koklu, M.; Sarigil, S.; Ozbek, O. The use of machine learning methods in classification of pumpkin seeds (Cucurbita pepo L.). Genet. Resour. Crop Evol. 2021, 68, 2713–2726. [Google Scholar] [CrossRef]
- Mukasa, P.; Wakholi, C.; Faqeerzada, M.A.; Amanah, H.Z.; Kim, H.; Joshi, R.; Suh, H.K.; Kim, G.; Lee, H.; Kim, M.S.; et al. Nondestructive discrimination of seedless from seeded watermelon seeds by using multivariate and deep learning image analysis. Comput. Electron. Agric. 2022, 194, 106799. [Google Scholar] [CrossRef]
- Taspinar, Y.S.; Dogan, M.; Cinar, I.; Kursun, R.; Ozkan, I.A.; Koklu, M. Computer vision classification of dry beans (Phaseolus vulgaris L.) based on deep transfer learning techniques. Eur. Food Res. Technol. 2022, 248, 2707–2725. [Google Scholar] [CrossRef]
- de Medeiros, A.D.; Capobiango, N.P.; da Silva, J.M.; da Silva, L.J.; da Silva, C.B.; dos Santos Dias, D.C.F. Interactive machine learning for soybean seed and seedling quality classification. Sci. Rep. 2020, 10, 11267. [Google Scholar] [CrossRef]
- Xu, P.; Yang, R.; Zeng, T.; Zhang, J.; Zhang, Y.; Tan, Q. Varietal classification of maize seeds using computer vision and machine learning techniques. J. Food Process Eng. 2021, 44, e13846. [Google Scholar] [CrossRef]
- Ermiş, S.; Ercan, U.; Kabaş, A.; Kabaş, Ö.; Moiceanu, G. Machine learning-based morphological classification and diversity analysis of ornamental pumpkin seeds. Foods 2025, 14, 1498. [Google Scholar] [CrossRef]
- Yasmin, J.; Ahmed, M.R.; Wakholi, C.; Lohumi, S.; Mukasa, P.; Kim, G.; Kim, J.; Lee, H.; Cho, B.K. Near-infrared hyperspectral imaging for online measurement of the viability detection of naturally aged watermelon seeds. Front. Plant Sci. 2022, 13, 986754. [Google Scholar] [CrossRef]
- Qi, H.; He, M.; Huang, Z.; Yan, J.; Zhang, C. Application of hyperspectral imaging for watermelon seed classification using deep learning and scoring mechanism. J. Food Qual. 2024, 2024, 7313214. [Google Scholar] [CrossRef]
- Jin, C.; Zhou, L.; Pu, Y.; Zhang, C.; Qi, H.; Zhao, Y. Application of deep learning for high-throughput phenotyping of seed: A review. Artif. Intell. Rev. 2025, 58, 76. [Google Scholar] [CrossRef]
- Mohialden, Y.M.; Hussien, N.M.; Salman, S.A.; Alwahhab, A.B.A.; Ali, M. Enhancing agriculture crop classification with deep learning. Babylon. J. Artif. Intell. 2024, 2024, 20–26. [Google Scholar] [CrossRef]
- Sable, A.; Singh, P.; Kaur, A.; Driss, M.; Boulila, W. Quantifying soybean defects: A computational approach to seed classification using deep learning techniques. Agronomy 2024, 14, 1098. [Google Scholar] [CrossRef]
- Kumar, V.; Aydav, P.S.S.; Minz, S. Crop seeds classification using traditional machine learning and deep learning techniques: A comprehensive survey. SN Comput. Sci. 2024, 5, 1031. [Google Scholar] [CrossRef]
- Rajalakshmi, R.; Faizal, S.; Sivasankaran, S.; Geetha, R. RiceSeedNet: Rice seed variety identification using deep neural network. J. Agric. Food Res. 2024, 16, 101062. [Google Scholar] [CrossRef]
- Çiftci, B.; Çetin, N.; Günaydın, S.; Kaplan, M. Machine learning approaches for binary classification of sorghum (Sorghum bicolor L.) seeds from image color features. J. Food Compos. Anal. 2025, 140, 107208. [Google Scholar] [CrossRef]
- Sanmiguel, J.; Andrade, V.; Vargas-Tierras, Y.; Samaniego, I.; Paredes-Arcos, F.; Vásquez-Castillo, W.; Viera-Arroyo, W. Physical–chemical characterization of fruit harvested at different maturity stages of grafted yellow pitahaya (Selenicereus megalanthus Haw.). Plants 2025, 14, 178. [Google Scholar] [CrossRef] [PubMed]
- Pivatto, M.S.; Funes, G.; Ferreras, A.E.; Gurvich, D.E. Seed mass, germination and seedling traits for some central Argentinian cacti. Seed Sci. Res. 2014, 24, 71–77. [Google Scholar] [CrossRef]
- Shahin, M.A.; Symons, S.J.; Poysa, V.W. Determining soya bean seed size uniformity with image analysis. Biosyst. Eng. 2006, 94, 191–198. [Google Scholar] [CrossRef]
- Felix, F.C.; Chagas, K.P.T.D.; Araújo, F.D.S.; Medeiros, J.A.D.D.; Vieira, F.D.A.; Torres, S.B.; Pacheco, M.V. Image analysis of seeds and machine learning as a tool for distinguishing populations: Applied to an invasive tree species. Acta Sci. Agron. 2024, 46, e62658. [Google Scholar] [CrossRef]
- Ma, F.; Wang, L.; Wang, C.; Wang, Q.; Lu, C. Study on impact soil movement experiments on wheat seeds based on EDEM. Agriculture 2025, 15, 400. [Google Scholar] [CrossRef]
- Berrar, D. Cross-validation. Encycl. Bioinform. Comput. Biol. 2019, 1, 542–545. [Google Scholar] [CrossRef]
- Kohavi, R. A study of cross-validation and bootstrap for accuracy estimation and model selection. In Proceedings of the 14th International Joint Conference on Artificial Intelligence (IJCAI’95), Montreal, QC, Canada, 20–25 August 1995; pp. 1137–1143. [Google Scholar]
- Ercan, U. İnternetten alışveriş yapan hanelerin rastgele orman yöntemiyle tahmin edilmesi. Kafkas Üniversitesi İktisadi İdari Bilim. Fakültesi Derg. 2021, 12, 728–752. [Google Scholar] [CrossRef]
- Rodriguez-Galiano, V.F.; Ghimire, B.; Rogan, J.; Chica-Olmo, M.; Rigol-Sanchez, J.P. An assessment of the effectiveness of a random forest classifier for land-cover classification. ISPRS J. Photogramm. Remote Sens. 2012, 67, 93–104. [Google Scholar] [CrossRef]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef]
- Ercan, U.; Kabas, O.; Kabaş, A.; Moiceanu, G. Classification of dragon fruit varieties based on morphological properties: Multi-class classification approach. Sustainability 2025, 17, 62629. [Google Scholar] [CrossRef]
- Patel, A.M.; Suthar, A. Adaboosted extra trees classifier for object-based multispectral image classification of urban fringe area. Int. J. Image Graph. 2022, 22, 2140006. [Google Scholar] [CrossRef]
- Speybroeck, N.; Berkvens, D.; Mfoukou-Ntsakala, A.; Aerts, M.; Hens, N.; Van Huylenbroeck, G.; Thys, E. Classification trees versus multinomial models in the analysis of urban farming systems in Central Africa. Agric. Syst. 2004, 80, 133–149. [Google Scholar] [CrossRef]
- Kumar, P.; Gupta, D.K.; Mishra, V.N.; Prasad, R. Comparison of support vector machine, artificial neural network, and spectral angle mapper algorithms for crop classification using LISS IV data. Int. J. Remote Sens. 2015, 36, 1604–1617. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Kocer, A. Prediction of the higher heating values of biomass using machine learning methods based on proximate and ultimate analysis. J. Mech. Sci. Technol. 2024, 38, 1569–1574. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R J. 2002, 2, 18–22. Available online: https://journal.r-project.org/articles/RN-2002-022/RN-2002-022.pdf (accessed on 9 October 2025).
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning representations by back-propagating errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the dimensionality of data with neural networks. Science 2006, 313, 504–507. [Google Scholar] [CrossRef]
- Zhang, G.; Patuwo, B.E.; Hu, M.Y. Forecasting with artificial neural networks: The state of the art. Int. J. Forecast. 1998, 14, 35–62. [Google Scholar] [CrossRef]
- Ahmad, M.W.; Reynolds, J.; Rezgui, Y. Predictive modelling for solar thermal energy systems: A comparison of support vector regression, random forest, extra trees and regression trees. J. Clean. Prod. 2018, 203, 810–821. [Google Scholar] [CrossRef]
- John, V.; Liu, Z.; Guo, C.; Mita, S.; Kidono, K. Real-time lane estimation using deep features and extra trees regression. In Image and Video Technology; Bräunl, T., McCane, B., Rivera, M., Yu, X., Eds.; Springer: Cham, Switzerland, 2016; Volume 9431, pp. 721–733. [Google Scholar] [CrossRef]
- Biau, G.; Scornet, E. A random forest guided tour. Test 2016, 25, 197–227. [Google Scholar] [CrossRef]
- Geurts, P.; Ernst, D.; Wehenkel, L. Extremely randomized trees. Mach. Learn. 2006, 63, 3–42. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar] [CrossRef]
- Refaeilzadeh, P.; Tang, L.; Liu, H. Cross-validation. In Encyclopedia of Database Systems; Springer: New York, NY, USA, 2009; pp. 532–538. [Google Scholar] [CrossRef]
- Ferri, C.; Hernández-Orallo, J.; Modroiu, R. An experimental comparison of performance measures for classification. Pattern Recognit. Lett. 2009, 30, 27–38. [Google Scholar] [CrossRef]
- Sokolova, M.; Lapalme, G. A systematic analysis of performance measures for classification tasks. Inf. Process. Manag. 2009, 45, 427–437. [Google Scholar] [CrossRef]
- Opitz, J.; Burst, S. Macro f1 and Macro f1. arXiv 2019, arXiv:1911.03347. [Google Scholar]
- Takahashi, K.; Yamamoto, K.; Kuchiba, A.; Koyama, T. Confidence interval for micro-averaged F1 and macro-averaged F1 scores. Appl. Intell. 2022, 52, 4961–4972. [Google Scholar] [CrossRef]
- Grandini, M.; Bagli, E.; Visani, G. Metrics for multi-class classification: An overview. arXiv 2020, arXiv:2008.05756. [Google Scholar] [CrossRef]
- Chicco, D.; Jurman, G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genom. 2020, 21, 6. [Google Scholar] [CrossRef]
- Fawcett, T. An introduction to ROC analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Ahmed, M.R.; Yasmin, J.; Park, E.; Kim, G.; Kim, M.S.; Wakholi, C.; Mo, C.; Cho, B.K. Classification of watermelon seeds using morphological patterns of X-ray imaging: A comparison of conventional machine learning and deep learning. Sensors 2020, 20, 6753. [Google Scholar] [CrossRef]
- Gulzar, Y.; Hamid, Y.; Soomro, A.B.; Alwan, A.A.; Journaux, L. A convolution neural network-based seed classification system. Symmetry 2020, 12, 2018. [Google Scholar] [CrossRef]
- Ropelewska, E.; Rady, A.M.; Watson, N.J. Apricot stone classification using image analysis and machine learning. Sustainability 2023, 15, 9259. [Google Scholar] [CrossRef]
- Yurdakul, M.; Atabaş, İ.; Taşdemir, Ş. Almond (Prunus dulcis) varieties classification with genetic designed lightweight CNN architecture. Eur. Food Res. Technol. 2024, 250, 2625–2638. [Google Scholar] [CrossRef]
- Kraljevski, I.; Ju, Y.C.; Ivanov, D.; Tschöpe, C.; Wolff, M. How to Do Machine Learning with Small Data?—A Review from an Industrial Perspective. arXiv 2023, arXiv:2311.07126. [Google Scholar]
- Bailly, A.; Blanc, C.; Francis, É.; Guillotin, T.; Jamal, F.; Wakim, B.; Roy, P. Effects of dataset size and interactions on the prediction performance of logistic regression and deep learning models. Comput. Methods Programs Biomed. 2022, 213, 106504. [Google Scholar] [CrossRef]



| Color Group | Genotype Code | Genotype ID | Description |
|---|---|---|---|
| Red | GSR-49-22 | K1 | Red seed coat |
| GSR-71-22 | K2 | Red seed coat | |
| GSR-113-22 | K3 | Red seed coat | |
| White | GSW-87-22 | B1 | White seed coat |
| GSW-17-22 | B2 | White seed coat | |
| GSW-47-22 | B3 | White seed coat | |
| Black | GSB-1-22 | B1 | Black seed coat |
| GSB-2-22 | B2 | Black seed coat | |
| GSB-11-22 | B3 | Black seed coat |
| Model | Parameter Settings |
|---|---|
| ANN | hidden_layer_sizes = (64,128, 64), activation = ‘relu’, solver = ‘sgd’, alpha = 0.0001, batch_size = ‘auto’, learning_rate = ‘adaptive’, max_iter = 3000, learning_rate_init = 0.001, |
| ET | n_estimators = 250, criterion = ‘log_loss’, max_depth = 10, min_samples_split = 2, min_samples_leaf = 1, min_weight_fraction_leaf = 0.0, max_features = ‘log2’, bootstrap = False, |
| RF | n_estimators = 250, criterion = ‘log_loss’, max_depth = 10, min_samples_split = 2, min_samples_leaf = 1, min_weight_fraction_leaf = 0.0, max_features = ‘log2’, min_impurity_decrease = 0.0, verbose = 0, ccp_alpha = 0.0, bootstrap = True, |
| Average | SD | Min | Max | Skewness | Kurtosis | |
|---|---|---|---|---|---|---|
| Length (mm) | 13.62 | 1.04 | 10.32 | 16.88 | 0.65 | 0.77 |
| Width (mm) | 8.35 | 0.58 | 6.51 | 10.23 | −0.12 | −0.03 |
| Thickness (mm) | 2.42 | 0.23 | 1.87 | 3.70 | 0.56 | 1.14 |
| Mass (g) | 0.14 | 0.03 | 0.04 | 0.21 | 0.00 | 0.22 |
| L | 50.98 | 18.95 | 25.67 | 85.88 | 0.60 | −1.33 |
| a | 10.28 | 7.68 | 1.46 | 29.10 | 1.01 | −0.48 |
| b | 11.25 | 7.90 | −2.90 | 27.71 | −0.48 | −1.32 |
| Area (mm2) | 89.60 | 11.31 | 53.33 | 121.31 | 0.30 | 0.24 |
| Perimeter (mm) | 35.03 | 2.31 | 26.87 | 41.79 | 0.33 | 0.58 |
| Ratio of Height/Width | 1.64 | 0.12 | 1.25 | 2.01 | 0.17 | −0.02 |
| Compactness | 6.56 | 0.46 | 5.11 | 8.03 | −0.12 | −0.03 |
| Roundness | 0.91 | 0.02 | 0.84 | 0.98 | −0.29 | −0.07 |
| Equivalent diameter (mm) | 10.66 | 0.67 | 8.24 | 12.43 | 0.09 | 0.35 |
| ANN | RF | ET | ANN | RF | ET | |||
|---|---|---|---|---|---|---|---|---|
| Macro F1 | Min | 0.8035 | 0.8470 | 0.7951 | Macro Precision | 0.8251 | 0.8546 | 0.7981 |
| Max | 0.9082 | 0.9889 | 0.9778 | 0.9216 | 0.9889 | 0.9798 | ||
| Mean | 0.8578 | 0.9187 | 0.8958 | 0.8702 | 0.9217 | 0.8994 | ||
| SD | 0.0398 | 0.0467 | 0.0557 | 0.0393 | 0.0446 | 0.0549 | ||
| MCC | Min | 0.7802 | 0.8392 | 0.7764 | Macro Recall | 0.8206 | 0.8524 | 0.8025 |
| Max | 0.9018 | 0.9875 | 0.9753 | 0.9139 | 0.9899 | 0.9798 | ||
| Mean | 0.8457 | 0.9127 | 0.8879 | 0.8670 | 0.9220 | 0.9009 | ||
| SD | 0.0438 | 0.0461 | 0.0609 | 0.0349 | 0.0445 | 0.0534 | ||
| Kappa | Min | 0.7745 | 0.8372 | 0.7749 | Accuracy | 0.8000 | 0.8556 | 0.8000 |
| Max | 0.8997 | 0.9874 | 0.9748 | 0.9111 | 0.9889 | 0.9778 | ||
| Mean | 0.8428 | 0.9118 | 0.8868 | 0.8611 | 0.9222 | 0.9000 | ||
| SD | 0.0449 | 0.0467 | 0.0614 | 0.0396 | 0.0412 | 0.0544 |
| Metrics | Artificial Neural Networks | Random Forest | Extra Trees |
|---|---|---|---|
| Accuracy | 0.8611 | 0.9222 | 0.9000 |
| Recall (Macro) | 0.8670 | 0.9220 | 0.9009 |
| Precision (Macro) | 0.8702 | 0.9217 | 0.8994 |
| Cohen’s Kappa | 0.8428 | 0.9118 | 0.8868 |
| F1-Score (Macro) | 0.8578 | 0.9187 | 0.8958 |
| Matthews Correlation Coefficient | 0.8457 | 0.9127 | 0.8879 |
| ROC-AUC Score (Macro) | 0.9886 | 0.9934 | 0.9928 |
| Metrics | Comparison Models | Wilcoxon Signed-Rank Test with Benjamini–Hochberg Correction | Significant |
|---|---|---|---|
| Accuracy | RF vs. ANNs | 0.015 | + |
| ET vs. ANNs | 0.015 | + | |
| RF vs. ET | 0.161 | - | |
| Recall (Macro) | RF vs. ANNs | 0.015 | + |
| ET vs. ANNs | 0.015 | + | |
| RF vs. ET | 0.140 | - | |
| Precision (Macro) | RF vs. ANNs | 0.015 | + |
| ET vs. ANNs | 0.015 | + | |
| RF vs. ET | 0.285 | - | |
| Cohen’s Kappa | RF vs. ANNs | 0.015 | + |
| ET vs. ANNs | 0.015 | + | |
| RF vs. ET | 0.161 | - | |
| Matthews Correlation Coefficient | RF vs. ANNs | 0.015 | + |
| ET vs. ANNs | 0.015 | + | |
| RF vs. ET | 0.161 | - | |
| ROC-AUC Score (Macro) | RF vs. ANNs | 0.015 | + |
| ET vs. ANNs | 0.015 | + | |
| RF vs. ET | 0.441 | - | |
| F1-Score (Macro) | RF vs. ANNs | 0.015 | + |
| ET vs. ANNs | 0.015 | + | |
| RF vs. ET | 0.139 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ercan, U.; Ermiş, S.; Kabas, O.; Öktem, G.; Kabas, A.; Paraschiv, G. Identification and Classification of Snack-Type Watermelon (Citrullus lanatus) Genotypes Using Seed Morphology and Machine Learning Techniques. Foods 2025, 14, 4069. https://doi.org/10.3390/foods14234069
Ercan U, Ermiş S, Kabas O, Öktem G, Kabas A, Paraschiv G. Identification and Classification of Snack-Type Watermelon (Citrullus lanatus) Genotypes Using Seed Morphology and Machine Learning Techniques. Foods. 2025; 14(23):4069. https://doi.org/10.3390/foods14234069
Chicago/Turabian StyleErcan, Uğur, Sıtkı Ermiş, Onder Kabas, Güleda Öktem, Aylin Kabas, and Gigel Paraschiv. 2025. "Identification and Classification of Snack-Type Watermelon (Citrullus lanatus) Genotypes Using Seed Morphology and Machine Learning Techniques" Foods 14, no. 23: 4069. https://doi.org/10.3390/foods14234069
APA StyleErcan, U., Ermiş, S., Kabas, O., Öktem, G., Kabas, A., & Paraschiv, G. (2025). Identification and Classification of Snack-Type Watermelon (Citrullus lanatus) Genotypes Using Seed Morphology and Machine Learning Techniques. Foods, 14(23), 4069. https://doi.org/10.3390/foods14234069










