Abstract
Sant’Agostino green table olives, traditionally processed in Apulia and flavoured with Foeniculum vulgare, represent a niche product whose microbial ecology remains largely unexplored. This study aimed to characterise the microbiota of the final product (both brine and fruit) after six months of storage with wild fennel. Four production batches were analysed using a combined culture-dependent and culture-independent approach. Microbiological counts revealed variable levels of aerobic mesophilic microorganisms, yeasts, lactic acid bacteria (LAB), and staphylococci, with yeasts and LAB being predominant. Ten LAB strains were identified, including Enterococcus faecium, Leuconostoc mesenteroides subsp. jonggajibkimchii, Leuconostoc mesenteroides subsp. cremoris, Leuconostoc pseudomesenteroides, Lactiplantibacillus plantarum, and Lactiplantibacillus pentosus. Yeast isolates belonged to Candida tropicalis, Torulaspora delbrueckii, and Saccharomyces cerevisiae. Amplicon sequencing (MiSeq Illumina) revealed distinct bacterial profiles between fruit and brine samples, with taxa from Actinobacteria, Bacteroidetes, Enterococcus, Lactobacillus, Leuconostoc, Alphaproteobacteria, Enterobacteriaceae, and other Gammaproteobacteria. Enterococcus and Leuconostoc were consistently detected, while Lactobacillus sensu lato appeared only in one fruit and one brine sample. These findings provide new insights into the microbial diversity of Sant’Agostino olives and contribute to the understanding of their fermentation ecology and potential for quality and safety enhancement.
1. Introduction
From 1990 to 2023, global production of table olives has markedly increased, rising from 950,000 t to over 3000 million t [1]. In 2023, the European Union (EU) led the world in production with 825,000 t, followed by Turkey with 605,000 t and Egypt with 600,000 t. Within the EU, Spain was the top producer at 414,000 t, followed by Greece (175,000 t), Italy (62,500 t), and Portugal (25,500 t) [2]. According to the International Olive Council [3], the EU, Turkey, and Egypt were the largest consumers of table olives in 2022/23. The production and consumption volumes highlight the widespread culinary use of table olives [4]. Different olive varieties worldwide are suitable for table olive production [5]. By employing various processing methods, it is possible to produce olives with diverse colours, tastes, smells, and textures [6]. However, freshly harvested olive drupes are unpalatable due to bitterness due to the presence of oleuropein, a polyphenol that affects their edibility [7]. The two main processing methods that degrade oleuropein, rendering olives edible are the Greek (natural) method and the Spanish (Seville) method. Both methods involve microorganisms that hydrolyse oleuropein during fermentation through beta-glucosidase activity [8]. The Spanish method also includes partially debittering the olives with soda before lactic acid fermentation [9]. Various processing styles may vary according to local customs and traditions [10].
Among the traditional methods with mainly local distribution, Sant’Agostino table olives from Apulia (Cerignola, Foggia, Italy) are processed similarly to the Seville style. After a generally “spontaneous” fermentation process, the olives are packed in glass jars and flavoured with wild fennel stems and inflorescences (Foeniculum vulgare). The addition of spices to table olives during packing is relatively rare. Campo Real table olives, produced near Madrid, are similar and are mainly flavoured with thyme, oregano, and fennel [11]. In some recipes, Cobrançosa table olives are immersed in brine enriched with herbs like thyme, oregano, and mint after fermentation [12]. Chalkidiki green table olives, produced using the Spanish low-salt method, are occasionally preserved in olive oil infused with essential oils of various spices to control undesired microorganisms [13]. While the antimicrobial properties of wild fennel against various microorganisms are well-known [14], in oil production, F. vulgare extracts are sometimes used to flavour and preserve oil from oxidative processes [15].
Recent studies have demonstrated that the microbial communities involved in the fermentation of vegetables, including table olives, are influenced by ecological and technological factors and can play an active role in product preservation through the production of antimicrobial metabolites. Lactic acid bacteria and specific yeasts are recognised for their capacity to inhibit the growth of both pathogenic and spoilage microorganisms, thereby ensuring microbiological stability and promoting food safety [16,17,18]. The addition of spices and aromatic plants, such as F. vulgare, has been demonstrated to significantly influence the microbial composition of the fermented product. Extracts and essential oils of F. vulgare have been shown to possess significant antimicrobial activity against Gram-positive and Gram-negative bacteria, attributable to the presence of phenolic and terpenic compounds [19,20]. These interactions have also been observed in other fermented vegetables, where the use of plant ingredients with antimicrobial properties has helped to improve the microbiological stability and shelf life of the product.
The specific microorganisms present in Sant’Agostino table olives flavoured with wild fennel are not documented in the literature. Since this product undergoes “spontaneous” fermentation, the microorganisms involved can vary by season and processing environment. To improve the quality of these olives, a deeper understanding of their microbiological aspects is essential. The spontaneous fermentation of Sant’Agostino table olives traditionally takes place under consistent environmental and processing conditions, which may result in the recurrence of particular microbial groups. Nevertheless, the final microbial composition can vary depending on the native microbiota of the olives and their surrounding environment.
This study aimed to evaluate the microbial diversity of Sant’Agostino green olives flavored with wild fennel using a combined, culture-dependent and culture-independent, multi-phase approach.
2. Materials and Methods
2.1. Table Olive Processing Method and Sampling
Drupes of the Sant’Agostino olive variety were harvested in the first ten days of October 2023 while still unripe. After an initial washing in tap water to remove dust and soil residues, they were placed in a 2% (w/v) NaOH solution (Farmalabor, Canosa di Puglia, Italy) for 6–7 h. The olives were then washed five times with potable water to remove excess sodium hydroxide. Fermentation occurred in fully openable 220 L high-density polyethylene (HDPE) drums with metal tie closures (Pack Service, Liscate, Italy) and lasted one month. Each drum contained 150 kg of olives immersed in approximately 70 L of a 10% (w/v) NaCl brine solution (Sali Alimentari e Industriali SRL, Margherita di Savoia, Italy). Fermentation was carried out at ambient temperature (21 °C ± 2 °C). During the wild fennel harvest in November, the olives were transferred from the drums into 5 L glass jars with screw caps (Vetrobari SRL, Bari, Italy) in the same brine (Figure S1). Twenty g of fresh wild fennel was added to each jar. The jars were kept in a cool, dark environment for about 5 months prior to consumption (with a maximum storage duration of six months). For microbiological analysis, one jar was taken from each of four independent production batches (n = 4), all of which were processed using the same spontaneous fermentation method. The olive fruit and brine from each batch were separated and labelled P1–P4 and S1–S4, respectively. All samples were collected at the same time after six months of storage.
2.2. pH Measurement
The pH of the brine samples was measured at the end of the storage period using a calibrated digital pH meter (model HI98165, Hanna Instruments, Ronchi di Villafranca Padovana, Italy) [21]. Measurements were performed in triplicate at room temperature (20 ± 1 °C) immediately after opening the jars. The average pH values were recorded and used to evaluate the acidification level of the final product.
2.3. Microbiological Analysis
Microbiological analyses were conducted on both the olive fruit (P1, P2, P3, and P4) and the brine (S1, S2, S3, and S4). For each sample, 10 g of olive fruit and 25 mL of brine [21] were diluted 1:10 in sterile Ringer solution (Thermo Scientific™, Segrate, Italy) and homogenised for 2 min using a BagMixer® 400 stomacher (Interscience, Saint Nom, France). The following microbial groups were enumerated by serial dilution and plating in Petri dishes: mesophilic aerobic microorganisms in plate count agar (PCA; Biotec, Grosseto, Italy) at 30 °C for 48 h [22]; mesophilic lactobacilli in de Man-Rogosa-Sharpe (MRS; Condalab, Torrejón de Ardoz, Spain) agar with cycloheximide (10 mg/mL) and incubated under microaerophilic conditions at 30 °C for 48 h [21]; mesophilic lactococci incubated at microaerophilic conditions on M17 agar (Condalab, Torrejón de Ardoz, Spain) supplemented with cycloheximide (10 mg/mL) at 30 °C for 48 h [23]; pseudomonads on Pseudomonas agar base (PAB; Microbiol, Cagliari, Italy) with CFC supplement (PCFC) incubated aerobically at 20 °C for 48 h [24]; staphylococci on mannitol salt agar (MSA; Microbiol, Cagliari, Italy) incubated aerobically at 32 °C for 78 h [25]; spore-forming bacteria were determined by subjecting the samples (fruit and brine) to a heat treatment in a thermostatic water bath (model 740, Asal SRL, Cernusco sul Naviglio, Italy) at 78 °C for 20 min to inactivate vegetative cells. After cooling, the samples were plated on tryptic soy agar (TSA, Condalab, Torrejón de Ardoz, Spain) and incubated at 30 °C for 24 h [26]; Enterobacteriaceae on violet red bile salt glucose agar (VRBGA; Microbiol, Cagliari, Italy) at 37 °C for 24 h [21]; coliforms in violet red bile salt agar (VRBA; Oxoid, Milan, Italy) at 37 °C for 24 h [27]; Salmonella spp. and Shigella spp. were detected using a multi-step enrichment protocol [28]. Ten g of olive pulp or 25 mL of brine were pre-enriched in 90 mL of buffered peptone water (BPW; Oxoid, Milan, Italy) and incubated at 37 °C for 18–24 h. For Salmonella spp., 1 mL of the pre-enrichment culture was transferred into 10 mL of Rappaport-Vassiliadis (RV) broth (Oxoid, Milan, Italy) and incubated at 42 °C for 24 h. For Shigella spp., a separate aliquot was inoculated into 10 mL of Gram-negative (GN) broth (Oxoid, Milan, Italy) and incubated at 37 °C for 24 h. After selective enrichment, cultures were streaked onto Hektoen Enteric Agar (HEA; Condalab, Torrejón de Ardoz, Spain) and incubated at 37 °C for 24 h; yeasts in dichloran-Rose-Bengal chloramphenicol (DRBC; Condalab, Torrejón de Ardoz, Spain) agar incubated at 25 °C for 96 h [29]. Microbial counts were expressed as log CFU/mL or g as the mean of three replicates for each sample.
2.4. Isolation, Phenotypic and Genotypic Characterisation of Lactic Acid Bacteria and Spore-Forming Bacteria
Bacterial colonies were collected on MRS, M17 and TSA from the highest dilution of the cell suspension of each sample from each batch. When the macroscopic morphology was found to be identical, at least three colonies were collected. These colonies were initially subjected to Gram staining [30] and the catalase test [31]. Presumptive lactic acid bacteria (LAB) were identified as Gram-positive and catalase-negative colonies isolated from MRS and M17 agar. These presumptive LAB and spore-forming isolates were purified by streaking on the same medium used for plate counting. The pure cultures (n = 80) were then transferred to the corresponding liquid medium. After incubating at 30 °C for 48 h, they were supplemented with 20% (v/v) glycerol and stored at −80 °C for further characterisation, for a maximum of 12 months. Cell morphology was observed using an Axiophot light microscope (Carl Zeiss, Oberkochen, Germany). All isolates were tested for their ability to grow at 15 °C and 45 °C. The ability of enterococci to grow at pH 9.2 and in 6.5% NaCl was also evaluated [32]. The production of CO2 from glucose was assessed using Durham tubes containing MRS broth, incubated at 30 °C for 48 h. Gas formation was recorded as an indicator of heterofermentative metabolism. The sporulation ability was determined using the methodology described by Manetsberger et al. [26]. Resistance to increasing NaCl concentrations (0 to 12%) was assessed in tryptone soy broth (TSB) tubes incubated at 30 °C for 48 h. The ability to grow in acidic or alkaline conditions (pH 0 to 12) was evaluated by growth tests in TSB tubes at 30 °C for 48 h. Additionally, the ability of spore-forming isolates to utilize and ferment glucose and hydrolyze starch was assessed following the method outlined by Vashist et al. [33].
Genomic DNA was extracted from all bacterial isolates using the Instagene Matrix kit (Bio-Rad, Hercules, CA, USA) after 24 h of growth in their respective optimal media. This process followed the instructions provided by the manufacturer and included cell lysis by heat treatment in the presence of a chelating resin and the subsequent removal of cell debris by centrifugation. The resulting DNA-containing supernatant could then be used directly for further investigations.
Strain typing was conducted using randomly amplified polymorphic DNA (RAPD)-PCR analysis with M13, AB106, and AB111 primers [34,35,36]. The PCR conditions and electrophoresis gel visualization followed the methods described by Alfonzo et al. [37]. RAPD profiles were analyzed using Gelcompare II software version 6.5 (Applied-Maths, Sint-Marten-Latem, Belgium). Isolates with unique patterns were identified as different strains. All bacterial strains were genetically identified by sequencing the 16S rRNA gene and comparing the sequences to public databases (GenBank and EZ-taxon) using a BLAST v2.16.0 search. PCR reactions were performed according to Allioui et al. [38] using the primer pair fD1 (5′-AGAGTTTGATCCTGGCTCAG-3′)/rD1 (5′-AAGGAGGTGATCCAGCC-3′). PCR products were purified with the QIAquick purification kit (Quiagen S.p.a., Milan, Italy) and sequenced at BMR Genomics (Padova, Italy) using the same primers as for PCR amplification, after confirming the molecular size of the amplicons (approximately 1600 bp) on agarose gels. All strains belonging to the Lactiplantibacillus plantarum group were subjected to the recA gene-based multiplex PCR technique described by Torriani et al. [39] with an objective to unequivocally distinguish between L. plantarum, Lactiplantibacillus paraplantarum and Lactiplantibacillus pentosus.
2.5. Isolation, Phenotypic and Molecular Characterisation of Yeasts
Yeast colonies were collected by DRBC from the highest dilution of the cell suspension of each sample in each lot. A minimum of three colonies were collected if the macroscopic morphology appeared similar. These colonies were cultured for 3 d at 25 °C in test tubes containing yeast peptone dextrose (YPD) broth (Condalab, Torrejón de Ardo, Spain) and purified by streaking on the same agar medium. The purity of the isolates was verified microscopically. Pure colonies (n = 54) were then stored at −80 °C in 20% (v/v) glycerol YPD broth for further characterization, for a maximum of 12 months. The axenic yeast isolates were initially differentiated by observing the following characteristics after growth on Wallerstein Laboratory (WL) nutrient agar (25 °C, 5 days): colony colour, shape, rim, opacity, surface, and consistency [40,41,42]. Genomic DNA was extracted from all isolates using the same method as for bacterial isolates. The extracted DNA was amplified for 5.8S rRNA using ITS1 and ITS4 primers [43]. Preliminary species identification was achieved through Restriction Fragment Length Polymorphism (RFLP) analysis by generating restriction profiles through DNA digestion with the enzymes CfoI, HaeIII, and HinfI (Thermo Fisher Scientific, Monza, Italy) following the methodology of Sinacori et al. [44]. Band visualization was performed as described by Alfonzo et al. [45]. Isolates with identical RFLP profiles were further distinguished into strains using DNA (RAPD)-PCR analysis with M13 primer [46], following the amplification conditions, band visualization, and RAPD profile analysis reported by Alfonzo et al. [45]. To confirm the identification at species level determined by RFLP analysis, each yeast strain was subjected to sequencing of the D1/D2 region of the 26S rRNA gene with primers NL1 and NL4, following the methodology reported by Kurtzman and Robnett [47]. Sequence verification was manually performed using Chromas 2.6.6 software (Technelysium Pty Ltd., Brisbane, Australia) after DNA sequencing at BMR Genomics (Padova, Italy). The sequence of each yeast strain was entered into the BLAST search engine to compare the percentage similarity with sequences in the GenBank database (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 13 November 2024).
2.6. Extraction of the DNA and Preparation of the MiSeq Library
Total DNA was extracted from each table olive sample by separating the fruit from the brine using the NucleoSpin Food kit (Macherey-Nagel GmbH & Co. KG, Düren, Germany) following the manufacturer’s protocol. For each sample (both fruit and brine), a 464 bp fragment of the V3–V4 region [48,49] of the 16S rRNA gene (corresponding to Escherichia coli positions 341 to 805) was amplified from the extracted DNA. PCR amplification of each sample was performed in 25 μL of reaction volume, with 12.5 μL of 2× KAPA Hifi HotStart Ready Mix (Kapa Biosystems Ltd., London, UK), 1 μM of each primer, 2 μL of DNA (10 ng/μL), and 9.5 μL of ddH20. All PCR reactions were carried out using a Verity™ 96-well Thermal Cycler, according to the following protocol: 95 °C for 5 min and 25 cycles of 95 °C for 30 s, 55 °C for 30 s, 72 °C for 40 s, with a final elongation step of 72 °C for 5 min. PCR products were checked by gel electrophoresis and cleaned using an Agencourt AMPure XP system (Beckman Coulter, Brea, CA, USA), following the manufacturer’s instructions. After seven PCR cycles (16S metataxonomic Sequencing Library Preparation, Illumina), Illumina adaptors were attached (Illumina Nextera XT Index Primer). Libraries were purified using Agencourt AMPure XP (Beckman Coulter, Brea, CA, USA) and then sequenced on an Illumina® MiSeq (Run Chemistry: 2 × 300 PE) platform (MiSeq Control Software 2.0.5 and Real-Time Analysis software 1.16.18, Illumina, San Diego, CA, USA). Amplicon library preparation, quality and quantification of pooled libraries and high-throughput sequencing by Illumina technology were performed at the Sequencing Platform, Fondazione Edmund Mach (FEM, San Michele all’ Adige, TN, Italy).
2.7. Illumina Data Analysis and Sequences Identification
The raw paired-end FASTQ files were demultiplexed using idemp (available at https://github.com/yhwu/idemp/blob/master/idemp.cpp, accessed on 13 November 2024). Subsequently, they were imported into Quantitative Insights Into Microbial Ecology (Qiime2, version 2018.2). The sequences underwent quality filtering, trimming, de-noising, and merging using DADA2 [50]. Briefly: forward and reverse reads with a total number of expected errors of >2 were discarded. Next, the DADA2 workflow was used to trim quality-controlled reads to specific lengths, to identify exact sequences with single-nucleotide resolution, and to filter de novo chimeras with consensus method.
Taxonomic and compositional analyses for bacteria were conducted using the feature-classifier plugin (available at https://github.com/qiime2/q2-feature-classifier, accessed on 13 November 2024) [51]. A pre-trained Naive Bayes classifier, based on the Greengenes 13_8 99% Operational Taxonomic Units (OTUs) database (previously trimmed to the V3–V4 region of 16S rDNA and bound by the 341F/805R primer pair), was used to generate taxonomy tables from paired-end sequence reads. The MiSeq Illumina sequencing data have been deposited in the NCBI Sequence Read Archive (SRA) and are accessible under Accession Number PRJNA1185664 [52].
2.8. Statistical Analysis
The microbial count data were subjected to analysis of variance (ANOVA) and compared pairwise using Tukey’s post hoc test. Statistical significance was set at p ≤ 0.001. The software used for statistical data processing was XLStat software ver. 2019.2.2 (Addinsoft, New York, NY, USA).
3. Results
3.1. pH Values of Brine Samples
The pH values measured in the brine samples at the conclusion of the storage period were 4.38 ± 0.04 (S1), 4.34 ± 0.06 (S2), 4.30 ± 0.03 (S3), and 4.35 ± 0.02 (S4), respectively. These values indicate the extent of acidification achieved during the fermentation process across the four production batches.
3.2. Microbial Counts
The results of the microbial count (Table 1) revealed the presence of key microbial groups characteristic of table olives.

Table 1.
Microbial counts of different groups of microorganisms determined on 4 different batches of Sant’Agostino olives.
Variations were noted among the different samples regarding total mesophilic microorganism count, yeasts, rod LAB, and staphylococci. Conversely, no significant differences were observed for cocci-shaped LAB. Pseudomonads, Enterobacteriaceae and coliforms were not detected in any of the samples by direct enumeration, while the absence of Salmonella spp. and Shigella spp. was confirmed through enrichment-based methods. Higher levels of total mesophilic microorganisms were found in the brines (S2, S3, and S4), ranging from 6.8 to 6.9 log CFU/mL. Similarly, yeast counts were higher in the brine samples (5.6–5.9 log CFU/mL) compared to the fruit samples (4.5–4.9 log CFU/g). Sample P4 had the lactobacilli count (7.3 log CFU/g), while S1 had the lowest (6.0 log CFU/mL). There was low variability between brine and fruit samples for LAB cocci populations, with counts ranging from 5.8 log CFU/mL (S1) to 6.7 log CFU/g (P4). Spore-forming bacteria were only found in brine samples (S1, S2, S3, and S4), with microbial concentrations of approximately 2.5 log cycles. Staphylococcaceae levels were higher in brine samples, ranging from 5.2 to 5.9 log CFU/mL, whereas fruit samples showed values between 2.7 and 4.4 log CFU/g.
3.3. Phenotypic Grouping and Genetic Identification
3.3.1. Lactic Acid Bacteria
Colonies with different macroscopic characteristics in Petri dishes used to enumerate rod-shaped (MRS) and coccus-shaped (M17) LAB in both brine and olive fruit were subjected to purification and phenotypic grouping. A total of 62 isolates, characterised by Gram positivity and catalase negativity, were identified as presumptive LAB. Microscopic observations revealed that the most common cell morphology was coccus, observed in 58 isolates, with short chains present in 22 isolates. Only four isolates exhibited a rod-shaped morphology. Preliminary physiological and biochemical characterisation (Table 2) enabled differentiation of the coccus-shaped isolates into four distinct groups, while the rod-shape morphology was represented by a single phenotypic group.

Table 2.
Phenotypic grouping of bacteria isolated from Sant’Agostino table olives.
The presumptive LAB were subjected to RAPD-PCR analysis to differentiate them at the strain level. A comparison of the polymorphic profiles allowed the differentiation of 10 strains (Figure 1), which were subsequently identified to the species level by sequencing the 16S rRNA gene.
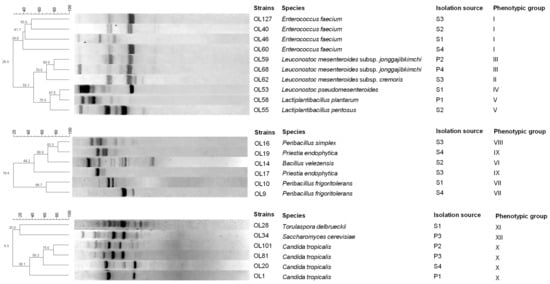
Figure 1.
Dendrogram obtained for LAB, spore-forming bacteria and yeast strains isolated from the brine and fruit of Sant’Agostino table olives using RAPD-PCR. The top line indicates the percentage of similarity. Acronyms: S1–S4, brine from 4 production batches; P1–P4, fruit from 4 different production batches.
Species-specific and multiplex PCRs were also used to analyse the species within the L. plantarum group. In total, six species were identified (Table 3): Enterococcus faecium (n = 4); Leuconostoc mesenteroides subsp. jonggajibkimchii (n = 2); Leuconostoc mesenteroides subsp. cremoris (n = 1); Leuconostoc pseudomesenteroides (n = 1); L. plantarum (n = 1); and L. pentosus (n = 1).

Table 3.
Molecular identification of bacteria strains in Sant’Agostino table olives through PCR-amplified products of 16S rDNA.
3.3.2. Spore-Forming Bacteria
Isolates of spore-forming bacteria found only in the brine samples were positive for Gram stain and catalase tests and exclusively exhibited rod cell morphology. Based on their physiological and biochemical characteristics (Table 2), they were divided into four groups. RAPD-PCR analysis differentiated the 18 isolates into six strains (Figure 1). Each strain was then identified at the species level by sequencing the 16S rRNA region: Peribacillus simplex (n = 1); Priestia endophytica (n = 2); Bacillus velezensis (n = 1); and Peribacillus frigoritolerans (n = 1).
3.3.3. Yeasts
Fifty-four yeast isolates were classified into three phenotypic groups (Table 4) based on the macroscopic characteristics of the colonies.

Table 4.
Macroscopic characters and phenotypic groups of yeasts isolated from Sant’Agostino table olives.
Group VI had the largest number of isolates (n = 50), followed by group VIII (n = 3), and group VII (n = 1). Common characteristics included a rounded colony shape and a butyrous consistency. Colony colour facilitated phenotypic discrimination of the groups, with additional differences observed in edge, opacity, and surface characteristics. RFLP analysis presumptively identified the yeast isolates as belonging to three species: Candida tropicalis, Torulaspora delbrueckii, and Saccharomyces cerevisiae (Table 5).

Table 5.
Molecular identification of yeasts isolated from Sant’Agostino table olives.
RAPD-PCR analysis further differentiated isolates with the same RFLP profile at the strain level. The 54 yeast isolates comprised six strains: four of C. tropicalis, and one each of T. delbrueckii and S. cerevisiae. Sequencing of the D1/D2 region of the 26S rRNA gene and comparison with the sequences in Genbank confirmed the species identifications obtained by RFLP analysis.
3.4. Cultivable LAB, Spore-Forming Bacteria and Yeast Species Distribution
The species distributions of microorganisms isolated from the brine and olive fruit samples are reported in Table 6.

Table 6.
Distributions of cultivable LAB, spore-forming bacteria and yeast species among Sant’Agostino table olives.
The highest number of microbial species was observed in the brine samples S1 and S3 (six species), while the lowest number of species was recorded in fruit sample P1 (three species). Both brine and fruit samples revealed the presence of several LAB species, including E. faecium (P2–P4, S1–S4), L. plantarum (P1, S3), Ln. mesenteroides subsp. cremoris (P1, P3 and S1–S3), and Ln. mesenteroides subsp. jonggajibkimchii (P2–P4 and S4). LAB species found only in brine samples were L. pentosus (S2) and Ln. pseudomesenteroides (S1). Spore-forming bacteria were detected only in the brine samples. Specifically, P. frigoritolerans and Pr. endophytica were found in S1 and S4, and S3 and S4, respectively. Bacillus velezensis was present in S2 sample, and P. simplex in S3.
Candida tropicalis was confirmed in all brine and fruit samples. In contrast, S. cerevisiae was identified only in three fruit samples (P2, P3, and P4), and T. delbrueckii was isolated solely from brine sample S1.
3.5. Characterisation of the Illumina Data and Taxonomic Analysis of the Bacterial Community
Illumina technology enabled identification of non-culturable (dormant and/or viable) bacterial populations in the brine and olive fruit samples. The total DNA extracted from the eight samples was successfully amplified targeting the bacterial V3–V4 region of the 16S rRNA gene. The distribution of bacterial taxa in the samples is shown in Figure 2. Eight bacterial taxa were characterised in fruit and brine samples: Actinobacteria, Bacteroidetes, Enterococcus, Lactobacillus, Leuconostoc, Alphaproteobacteria, Enterobacteriaceae, and other Gammaproteobacteria.
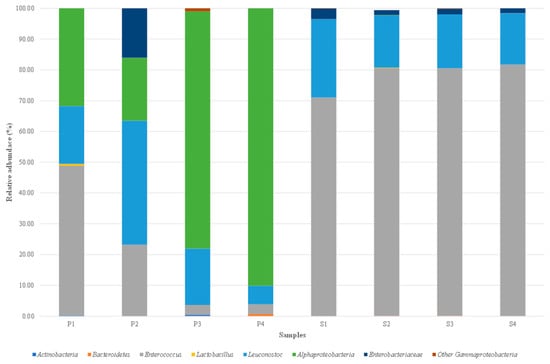
Figure 2.
Relative abundance (%) of bacterial genera identified by MiSeq Illumina on Sant’Agostino table olives aromatised with Foeniculum vulgare. Only genera with abundance > 0.1% in at least one sample were included. Acronyms: S1–S4, brine from 4 production batches; P1–P4, fruit from 4 different production batches.
The LAB group genera Enterococcus and Leuconostoc were present in all brine and olive fruit samples, with relative abundances varying according to sample type. For Enterococcus, percentages in the fruit ranged from 3.19% (P4) to 48.69% (P1), and higher percentages in the brine ranged from 71.01% (S1) to 81.67% (S4). For Leuconostoc, percentages ranged from 6.05 to 40.33% in the fruit and 16.72 to 25.51% in the brine. The genus Lactobacillus was found in one fruit sample (P1) and one brine sample (S2), but exhibited lower relative abundance than Enterococcus and Leuconostoc, ranging from 0.11% in S2 to 0.54% in P1. The Alphaproteobacteria class was highly representative in all the olive fruit samples, with relative abundances from 20.49% (P2) to 90.12% (P4), but was low in the brine sample S2 (0.11%). Enterobacteriaceae family was present in all brine samples, with percentages from 1.55% (S4) to 3.41 (S1), and was only found in fruit sample P2 (16.03%). Other bacterial taxa were present in low relative abundances and were occasionally detected in various fruit and brine samples. Specifically, Gammaproteobacteria had the highest percentages in P3 (0.98%), Bacteroidetes in P4 (0.65%), and Actinobacteria in P3 (0.42%).
4. Discussion
This research assessed the microbial diversity of bacterial populations in Sant’Agostino green table olives flavoured with F. vulgare using both culture-dependent and culture-independent methods. While DNA-based independent approaches have recently been employed to study the microbial communities in table olives [53], combining these with culture-dependent methods enabled the assessment of the viability of the identified taxa. The cultivable microbiota of table olives mainly consists of lactic acid bacteria and yeast populations [54]. However, the microbial groups characterizing spontaneously fermented table olives are diverse and influenced by several factors. These include olive variety [55], processing location [56], pH, salinity, the amount of fermentable sugars, fermentation temperature, water activity, and oxygen availability [57], and the processing style [58].
The Sant’Agostino cultivar is well-suited to table olive processing due to its firm fruit, low oil content, and sufficient fermentable sugars [59]. Numerous studies have examined the microbiota of various table olive varieties such as Bella di Cerignola, Nocellara Etnea, Tonda di Cagliari, Cipriota, Kalamata, Picual, Gordal, Hojiblanca, and Manzanilla [7,55,58,60,61,62]. However, information on the microbiota cultivar Sant’Agostino is limited. To better understand the microorganisms associated with Sant’Agostino table olives, a study was conducted to examine the ecology of microbial groups characterizing these olives, processed following a traditional Apulian protocol. Microbiological counts on the final product identified the microbial groups present in Sant’Agostino table olives flavoured with F. vulgare. LAB and yeasts were the dominant groups, consistent with other table olive varieties processed differently [63,64,65]. Notably, all four production batches’ brines and pulp samples were free of pseudomonads, typically associated with microbial spoilage. Pseudomonas spp. exhibit proteolytic activity, causing a decrease in brine acidity and drupe swelling [66]. Although commonly associated with Greek and Seville table olives [66] and other methods like Castelvetrano [45], their absence may be due to the addition of F. vulgare during processing. Extracts or essential oils from wild fennel seeds and plant parts released into the brine during immersion have antimicrobial activity against various pathogenic and spoilage bacteria, including pseudomonads [67,68,69]. Although fresh wild fennel was added after fermentation was complete, its antimicrobial properties may have contributed to shaping the final microbial composition. The consistent absence of pseudomonads and other spoilage microorganisms across all samples could be partially attributed to the addition of wild fennel. However, as wild fennel was introduced post-fermentation and no control batches without fennel were included in the study, its specific impact cannot be conclusively determined. Further investigations comparing batches with and without fennel are required to clarify its role in modulating the microbiota of the final product.
The absence of Enterobacteriaceae and coliforms in the final product is attributable to the regular acidification process of the brine, which creates an unfavourable environment for their proliferation; in fact, the pH values of the brine in the different samples varied between 4.30 (S3) and 4.38 (S1). This results from the combination of low pH values and adequate salt content. Enterobacteriaceae and coliforms are commonly present in the early stages of the process, coinciding with high pH levels. However, during fermentation, as the brine’s pH decreases, their concentrations drop below detectable levels [70]. Elevated levels of Enterobacteriaceae and coliforms early in fermentation can irreversibly affect the quality of the final product, leading to off odours [71]. The absence of Shigella spp. and Salmonella spp. is also indicative of a regular fermentation process. Additionally, the presence of phenolic and oleosidic compounds in the brine, which have antibacterial effects, combined with a low pH and adequate salt content, creates conditions hostile to these foodborne pathogens [72]. Staphylococcaceae were the only spoilage/pathogenic bacteria present in the four production batches due to their tolerance to acidic pH and high salinity [73]. The microbial levels for Staphylococcaceae are comparable to those observed in other studies on green olives, such as Aloreña de Málaga [74], Manzanilla and Gordal [56]. However, the absence of Staphylococcus aureus colonies indicates that processing was carried out with appropriate hygienic conditions [75].
Investigations into the microbial groups revealed the presence of L. plantarum and L. pentosus among LAB, frequently isolated from various table olive varieties [64]. Strains of both species have been characterized and successfully employed as starter cultures to enhance the fermentation process in Nocellara del Belice [76], Cobrançosa [77], Campiñesa [78], Nocellara Etnea [79] and Manzanilla [80] table olives. However, E. faecium is the most prevalent LAB species in the four production batches of Sant’Agostino table olive. A study of LAB from Cypriot green table olives [81] found that the enterococci group was predominant, with all identified strains belonging to E. faecium. These strains demonstrate potential for application in the food industry due to their good resistance to low pH values and bile salts. El Issaoui et al. [82] identified several E. faecium strains from Moroccan table olives that grew at low temperatures, tolerated acidic pH and salinity, and exhibited antimicrobial activity against common pathogens. Other identified LAB species belonged to the genus Leuconostoc. Portilha-Cunha et al. [10] describe how Leuconostoc, along with Lactobacillus, Pediococcus, Enterococcus, and Streptococcus, is commonly found in table olives. Using Ln. mesenteroides as a starter culture, in combination with different yeast species, has shortened the brine acidification period and improved the sensory quality of the final product [83]. Maoloni et al. [84] used Ln. pseudomesenteroides (strain PB288) in a multi-strain starter culture to optimize the fermentation of Ascolana Tenera table olives flavoured with sea fennel (Crithmum maritimum L.). Leuconostoc mesenteroides subsp. jonggajibkimchii, a dominant LAB species found in kimchi [85], has not previously been reported in fermented table olives. To our knowledge, this is the first documented presence of this subspecies in such products. Additional research and repeated isolations will be necessary to clarify its ecological role and persistence in this food matrix. However, it has also been reported as the dominant fermenting agent in the Turkish vegetable drink Şalgam [86]. Leuconostoc mesenteroides subsp. cremoris is typically associated with the fermentation of dairy products, for which it is used as a starter culture [87]. Its presence in table olives has already been documented by Kumral et al. [88], and the species has been isolated from olive fruit. The present study adds to the existing evidence of its presence in fermented vegetables by detecting it in wild fennel-flavoured Sant’Agostino olives, thereby supporting the hypothesis of its adaptability to different fermentation environments.
In the group of spore-forming bacteria found exclusively in the brine samples of all four batches, none of the identified species have been previously associated with table olives. Peribacillus frigoritolerans was isolated from leaf samples in Andalusian olive groves [26]. Priestia endophytica is a spore-forming endophytic bacterium known to colonise various agricultural plants [89,90,91]. Its presence in fermented table olives has not previously been documented, but it may be associated with the use of wild fennel or water sources. Hussein et al. [92] identified P. endophytica strains with plant-growth-promoting (PGP) properties from barley seeds, which, when inoculated into maize plantations, improved productivity [93]. Similarly, P. simplex, a spore-forming bacterium that was previously isolated from olive tree environments in Spain [26], is known for its PGP properties in various crops [94]. This is the first report of its presence in fermented table olives, suggesting possible transfer from plant material, such as wild fennel, or from water used during processing.
Additionally, B. velezensis is a well-researched biocontrol agent effective against several fungal diseases, including verticilliosis, fusariosis, and root rot in olive trees [95,96,97]. The presence of these spore-forming bacterial species in the brine of Sant’Agostino table olives may be attributed to the washing water used to remove soda residues or the water used to produce the brine. Alternatively, their endophytic nature suggests their presence in the stems, leaves, or seeds of wild fennel used as a flavouring agent [91]. However, none of these spore-forming bacteria has been identified as a spoilage agent of fermented plant products. Priestia endophytica is rarely associated with infections in immunocompromised patients or those with orthopedic complications [98].
The predominant yeast species was C. tropicalis, frequently found in both Seville table olives [99] and Greek-style table olives [100] that undergo “spontaneous” fermentation. While the presence of yeasts in table olives was once seen negatively [101], this perspective has evolved. Selected yeast strains are currently employed, often in combination with LAB, to improve the fermentation process [102,103]. Some C. tropicalis strains from fermenting table olives have shown significant probiotic potential, suggesting their use in producing probiotic table olives [104,105]. Saccharomyces cerevisiae, known for its fermentative capacity, is used as a starter in bakery and alcoholic beverage production [106]. Its presence in table olives has been detected in several spontaneously fermented products [107,108]. Tufariello et al. [109] used a S. cerevisiae strain as a starter in Manzanilla, Picual, and Kalamàta table olives, reducing fermentation time and standardizing sensory and nutritional qualities. Other studies highlight its probiotic potential and application in new probiotic food products [104,105]. Combining S. cerevisiae with olive leaf extracts in Spanish-style table olives has enhanced fermentation performance and increased hydroxytyrosol levels, an antioxidant with anti-inflammatory, anticarcinogenic, and neuroprotective properties [110]. Unlike C. tropicalis and S. cerevisiae, T. delbrueckii has been occasionally isolated in Sant’Agostino table olives flavored with wild fennel. Psani and Kotzekidou [111] identified T. delbrueckii in Greek-style black table olives, which could degrade oleuropein and showed antimicrobial activity against Listeria monocytogenes, Bacillus cereus and Salmonella Typhimurium. Recently, Mujdeci and Ozbas [112] isolated T. delbrueckii from salted Gemlik black table olives, along with Pichia membranifaciens, Candida sorbosivorans, Citeromyces nyonsensis, Candida etchelsii, Wickerhamomyces subpelliculosus, Candida apicola, Wickerhamomyces anomalus, and Candida versatilis, which were dominant in this production process. However, no studies to date have investigated T. delbrueckii as a starter or probiotic strain in table olives.
Next-generation sequencing (NGS) using Illumina technology was employed to survey the microbial diversity in the production of Sant’Agostino green olives flavoured with wild fennel. NGS tools allowed for the detection of viable non-culturable microorganisms and dead cells, provided the DNA was accessible [113]. Specifically, the bacterial composition of samples from the four production batches was analyzed in both brine and fruit by amplifying the V3-V4 rRNA gene region. The Enteroccoccus genus present in all fruit and brine samples, similar to other table olive productions where its abundance varied with fermentation times. Notably, Enterococcus was found in Sevillian-style processed Manzanilla [7] and Hojiblanca [114] olives. For the genus Leuconostoc, despite lower relative abundance, its presence was noted in Greek or natural-style olives of the Cypriot, Kalamata, and Picual cultivars that underwent “spontaneous” fermentation [55]. The difference observed between the culture-dependent and culture-independent results, particularly regarding the dominance of Enterococcus faecium in culture-based analyses versus the co-dominance of Enterococcus and Leuconostoc in the MiSeq Illumina data, reflects the methodological distinctions between the two approaches. Similar discrepancies have been reported in studies on table olives, where culture-based methods tend to favour the recovery of viable and fast-growing microorganisms, while metataxonomic techniques provide a broader overview of the microbial community, including viable but non-culturable (VBNC) organisms and DNA from dead cells [73]. These differences highlight the complementarity of the two approaches, with culture-dependent methods offering insights into the technological potential of isolates, and sequencing-based methods enhancing our understanding of microbial diversity and structure. Contrary to several publications, the genus Lactobacillus showed lower relative abundance in various Greek- and Seville-style olive varieties [7,55,115,116]. Alphaproteobacteria were present in all fruit samples with an average relative abundance of 55%, compared to lower percentages found by Michailidou et al. [117] and Mougiou et al. [53] in Greek-style table olives. However, the NGS approach did not detect spore-forming bacteria in the brine samples from different production batches. It is plausible that these bacteria were present in low abundance, and the DNA amount was insufficient for NGS detection, favouring the most abundant taxa [118].
5. Conclusions
This study aimed to characterise both the cultivable and non-cultivable microbiota of Sant’Agostino table olives, subjected to a traditional fermentation process, with wild fennel added as a flavouring agent at the final stage. The final product, resulting from spontaneous fermentation, exhibited considerable microbial diversity. The predominant LAB species was E. faecium, while C. tropicalis was the most frequently isolated yeast. The presence of Enterococcus in all samples was corroborated by culture-independent analyses. The absence of viable pathogenic microorganisms indicates that, despite the spontaneous nature of the fermentation and the traditional production protocol, the four tested batches are unlikely to pose a microbiological risk to consumers. Although the number of production batches analysed was limited, further studies are warranted to comprehensively assess the microbiota associated with this specific table olive production process. The selection of starter and/or probiotic cultures of lactic acid bacteria and yeasts could further enhance microbiological control and standardise the fermentation process of Sant’Agostino olives flavoured with wild fennel.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/foods14213689/s1, Figure S1: Ingredients and finished product of the traditional processing of Sant’Agostino table olives flavoured with wild fennel. (a) Wild fennel (Foeniculum vulgare) used as a natural flavouring; (b) Sant’Agostino cultivar table olives before processing, harvested at different stages of maturation; (c) glass jar containing olives processed according to the traditional Apulian method with the addition of wild fennel.
Author Contributions
Conceptualization: R.G. and L.S.; methodology: R.G., E.F., G.G., R.P. and A.P.; software: R.G. and E.F.; validation: A.A. and L.S.; formal analysis: A.A., D.A., E.F., G.P., R.P. and V.N.; investigation: E.F., A.P. and L.S.; resources: N.F. and G.M.; data curation: A.A., D.A., E.F., G.P., G.G., R.P. and V.N.; writing—original draft: A.A., N.F., G.M. and L.S.; writing—review & editing: A.A., N.F., G.M. and L.S.; visualization: G.G.; supervision: A.A., N.F., G.M. and L.S.; project administration: N.F. and G.M.; funding acquisition: N.F. and G.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by distinct funding sources, each allocated to separate components of the study to support and develop different and separate parts of total research. There was no overlap in the use of funds for the same activity. The funding were: (i) European Union-FESR or FSE, PON Research and Innovation 2014–2020-DM 1062/2021. Ministry of University and Research in Italy, Grant number: B7521002300001; (ii) European Union—Project PRIMA MEDIET4ALL—“Transnational Movement to Support the Sustainable Transition towards a Healthy and Eco-friendly Agri-Food System through the Promotion of MEDIET and its Lifestyle in Modern Society.”–Transnational call PRIMA Partnership for Research and Innovative in the Mediterranean Area—Call 2022, Thematic Area 3-Food value chain: Topic 2.3.1-2022 (RIA) Enabling the transition to healthy and sustainable dietary behaviour. Grant number: B73C23000060001.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
Acknowledgments
We acknowledge the contributions and support from the Research Infrastructure MIRRI-IT.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- International Olive Council. Word Table Olive Figures: Production. Available online: https://www.internationaloliveoil.org/wp-content/uploads/2023/12/OT-W901-13-12-2023-P.pdf (accessed on 16 February 2024).
- International Olive Council. EU Table Olive Figures: Production. Available online: https://www.internationaloliveoil.org/wp-content/uploads/2023/12/OT-CE-901-13-12-2023-P.pdf (accessed on 16 February 2024).
- International Olive Council. World Table Olive Figures: Consumptions. Available online: https://www.internationaloliveoil.org/wp-content/uploads/2023/12/OT-W901-13-12-2023-C.pdf (accessed on 16 February 2024).
- Conte, P.; Fadda, C.; Del Caro, A.; Urgeghe, P.P.; Piga, A. Table olives: An overview on effects of processing on nutritional and sensory quality. Foods 2020, 9, 514. [Google Scholar] [CrossRef] [PubMed]
- Bouranta, M.; Papakitsou, K.; Papakitsos, E. Inquiring quality assurance of the table olive products. Glob. Acad. J. Agric. Biosci. 2023, 5, 29–37. [Google Scholar] [CrossRef]
- Anagnostopoulos, D.A.; Tsaltas, D. Current status, recent advances, and main challenges on table olive fermentation: The present meets the future. Front. Microbiol. 2022, 12, 797295. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Barba, J.L.; Sánchez, A.H.; López-López, A.; Cortés-Delgado, A.; Montaño, A. Microbial and chemical characterization of natural-style green table olives from the Gordal, Hojiblanca and Manzanilla cultivars. Foods 2023, 12, 2386. [Google Scholar] [CrossRef]
- Rokni, Y.; Abouloifa, H.; Bellaouchi, R.; Hasnaoui, I.; Gaamouche, S.; Lamzira, Z.; Salah, R.B.; Saalaoui, E.; Ghabbour, N.; Asehraou, A. Characterization of β-glucosidase of Lactobacillus plantarum FSO1 and Candida pelliculosa L18 isolated from traditional fermented green olive. J. Genet. Eng. Biotechnol. 2021, 19, 117. [Google Scholar] [CrossRef]
- Ramírez, E.; Lara, E.; Valero, A.; Rodríguez-Gómez, F. Proposal for technological adaptation of small-sized green olives to Spanish-style processing. Food Control 2021, 126, 108067. [Google Scholar] [CrossRef]
- Portilha-Cunha, M.F.; Macedo, A.C.; Malcata, F.X. A review on adventitious lactic acid bacteria from table olives. Foods 2020, 9, 948. [Google Scholar] [CrossRef]
- Perez, R.A.; Navarro, T.; Lorenzo, C.D. HS–SPME analysis of the volatile compounds from spices as a source of flavour in “Campo Real” table olive preparations. Flavour Fragr. J. 2007, 22, 265–273. [Google Scholar] [CrossRef]
- Pires-Cabral, P.; Barros, T.; Mateus, T.; Prata, J.; Quintas, C. The effect of seasoning with herbs on the nutritional, safety and sensory properties of reduced-sodium fermented Cobrançosa cv. table olives. AIMS Agric. Food 2018, 3, 521–534. [Google Scholar] [CrossRef]
- Papapostolou, M.; Mantzouridou, F.T.; Tsimidou, M.Z. Flavored olive oil as a preservation means of reduced salt Spanish style green table olives (Cv. Chalkidiki). Foods 2021, 10, 392. [Google Scholar] [CrossRef]
- Noreen, S.; Tufail, T.; Badar Ul Ain, H.; Awuchi, C.G. Pharmacological, nutraceutical, functional and therapeutic properties of fennel (Foeniculum vulgare). Int. J. Food Prop. 2023, 26, 915–927. [Google Scholar] [CrossRef]
- Gharby, S.; Oubannin, S.; Ait Bouzid, H.; Bijla, L.; Ibourki, M.; Gagour, J.; Koubachi, J.; Sakar, E.H.; Majourhat, K.; Lee, L.-H.; et al. An Overview on the Use of Extracts from Medicinal and Aromatic Plants to Improve Nutritional Value and Oxidative Stability of Vegetable Oils. Foods 2022, 11, 3258. [Google Scholar] [CrossRef]
- Auchtung, J.M.; Hallen-Adams, H.E.; Hutkins, R. Microbial interactions and ecology in fermented food ecosystems. Nat. Rev. Microbiol. 2025, 23, 622–634. [Google Scholar] [CrossRef]
- Gautam, A.; Sharma, R.; Singh, P.; Kumari, S.; Thakur, N.; Kumar, A.; Singh, R. Ecological factors that drive microbial communities in culturally diverse fermented foods. BMC Microbiol. 2025, 25, 655. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Kumar, S. Exploring the functionality of microbes in fermented foods: Technological advancements and future directions. Fermentation 2025, 11, 300. [Google Scholar] [CrossRef]
- Moumen, B.E.; Bouzoubaa, A.; Drioiche, A.; Eddahmouny, M.; Al Kamaly, O.; Shahat, A.A.; Touijer, H.; Hadi, N.; Kharchouf, S.; Cherrat, A.; et al. Unveiling the Chemical Composition, Antioxidant, and Antimicrobial Potentials of Foeniculum vulgare Mill: A Combined In Vitro and In Silico Approach. Int. J. Mol. Sci. 2025, 26, 4499. [Google Scholar] [CrossRef] [PubMed]
- Elkiran, O.; Telhuner, O. Chemical profiles and antimicrobial activities of essential oil from different plant parts of Foeniculum vulgare Mill. Food Sci. Nutr. 2025, 13, e70307. [Google Scholar] [CrossRef]
- Tofalo, R.; Schirone, M.; Perpetuini, G.; Angelozzi, G.; Suzzi, G.; Corsetti, A. Microbiological and chemical profiles of naturally fermented table olives and brines from different Italian cultivars. Antonie Van Leeuwenhoek 2012, 102, 121–131. [Google Scholar] [CrossRef]
- Martins, F.; Rodrigues, N.; Pereira, J.A.; Baptista, P.; Ramalhosa, E. Effect of the cleaning and disinfection methods on the hygienic conditions of fermentation tanks of table olives (Olea europaea L.) Negrinha de Freixo cultivar. Food Microbiol. 2024, 119, 104425. [Google Scholar] [CrossRef]
- Galli, V.; Venturi, M.; Mari, E.; Guerrini, S.; Granchi, L. Selection of Yeast and Lactic Acid Bacteria Strains, Isolated from Spontaneous Raw Milk Fermentation, for the Production of a Potential Probiotic Fermented Milk. Fermentation 2022, 8, 407. [Google Scholar] [CrossRef]
- Martorana, A.; Alfonzo, A.; Settanni, L.; Corona, O.; La Croce, F.; Caruso, T.; Moschetti, G.; Francesca, N. An innovative method to produce green table olives based on pied de cuve technology. Food Microbiol. 2015, 50, 126–140. [Google Scholar] [CrossRef] [PubMed]
- Benincasa, C.; Muccilli, S.; Amenta, M.; Perri, E.; Romeo, F.V. Phenolic trend and hygienic quality of green table olives fermented with Lactobacillus plantarum starter culture. Food Chem. 2015, 186, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Manetsberger, J.; Caballero Gómez, N.; Benomar, N.; Christie, G.; Abriouel, H. Characterization of the culturable sporobiota of Spanish olive groves and its tolerance toward environmental challenges. Microbiol. Spectr. 2023, 11, e04013-22. [Google Scholar] [CrossRef] [PubMed]
- Restivo, I.; Sciurba, L.; Indelicato, S.; Allegra, M.; Lino, C.; Garofalo, G.; Bongiorno, D.; Davino, S.; Avellone, G.; Settanni, L. Repurposing olive oil mill wastewater into a valuable ingredient for functional bread production. Foods 2025, 14, 1945. [Google Scholar] [CrossRef]
- Busetta, G.; Garofalo, G.; Barbera, M.; Di Trana, A.; Claps, S.; Lovallo, C.; Franciosi, E.; Gaglio, R.; Settanni, L. Metagenomic, microbiological, chemical and sensory profiling of Caciocavallo Podolico Lucano cheese. Food Res. Int. 2023, 169, 112926. [Google Scholar] [CrossRef]
- Aponte, M.; Ventorino, V.; Blaiotta, G.; Volpe, G.; Farina, V.; Avellone, G.; Lanza, C.M.; Moschetti, G. Study of green Sicilian table olive fermentations through microbiological, chemical and sensory analyses. Food Microbiol. 2010, 27, 162–170. [Google Scholar] [CrossRef]
- Paray, A.; Singh, M.; Mir, M. Gram staining: A brief review. Int. J. Res. Rev. 2023, 10, 336–341. [Google Scholar] [CrossRef]
- Rahmawati, N.; Syukri, M.; Darmawi, D.; Zachreini, I.; Yusuf, M.; Idroes, R. Identification of lactic acid bacteria from Etawa goat milk Kopelma Darussalam Village, Banda Aceh. IOP Conf. Ser. Earth Environ. Sci. 2021, 667, 012022. [Google Scholar] [CrossRef]
- Gaglio, R.; Cirlincione, F.; Di Miceli, G.; Franciosi, E.; Di Gerlando, R.; Francesca, N.; Settanni, L.; Moschetti, G. Microbial dynamics in durum wheat kernels during aging. Int. J. Food Microbiol. 2020, 324, 108631. [Google Scholar] [CrossRef]
- Vashist, H.; Sharma, D.; Gupta, A. A review on commonly used biochemical test for bacteria. Innovare J. Life Sci. 2013, 1, 1–7. [Google Scholar]
- Fancello, F.; Multineddu, C.; Santona, M.; Deiana, P.; Zara, G.; Mannazzu, I.; Budroni, M.; Dettori, S.; Zara, S. Bacterial biodiversity of extra virgin olive oils and their potential biotechnological exploitation. Microorganisms 2020, 8, 97. [Google Scholar] [CrossRef] [PubMed]
- Gaglio, R.; Francesca, N.; Di Gerlando, R.; Mahony, J.; De Martino, S.; Stucchi, C.; Moschetti, G.; Settanni, L. Enteric bacteria of food ice and their survival in alcoholic beverages and soft drinks. Food Microbiol. 2017, 67, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Zotta, T.; Giavalisco, M.; Parente, E.; Picariello, G.; Siano, F.; Ricciardi, A. Selection of Lactiplantibacillus strains for the production of fermented table olives. Microorganisms 2022, 10, 625. [Google Scholar] [CrossRef] [PubMed]
- Alfonzo, A.; Francesca, N.; Naselli, V.; Gaglio, R.; Corona, O.; Seminerio, V.; Settanni, L.; La Croce, F.; Moschetti, G. Effect of glucose and inactivated yeast additions on the fermentation performances of Lactiplantibacillus pentosus OM13 during the production of Nocellara del Belice table olives. Fermentation 2023, 9, 634. [Google Scholar] [CrossRef]
- Allioui, N.; Driss, F.; Dhouib, H.; Jlail, L.; Tounsi, S.; Frikha-Gargouri, O. Two novel Bacillus strains (subtilis and simplex species) with promising potential for the biocontrol of Zymoseptoria tritici, the causal agent of septoria tritici blotch of wheat. BioMed Res. Int. 2021, 2021, 6611657. [Google Scholar] [CrossRef]
- Torriani, S.; Felis, G.E.; Dellaglio, F. Differentiation of Lactobacillus plantarum, L. pentosus, and L. paraplantarum by recA gene sequence analysis and multiplex PCR assay with recA gene-derived primers. Appl. Environ. Microbiol. 2001, 67, 3450–3454. [Google Scholar] [CrossRef]
- Alfonzo, A.; Sicard, D.; Di Miceli, G.; Guezenec, S.; Settanni, L. Ecology of yeasts associated with kernels of several durum wheat genotypes and their role in co-culture with Saccharomyces cerevisiae during dough leavening. Food Microbiol. 2021, 94, 103666. [Google Scholar] [CrossRef]
- Cavazza, A.; Grando, M.S.; Zini, C. Rilevazione della flora microbica di mosti e vini. Vignevini 1992, 9, 17–20. [Google Scholar]
- Pallmann, C.L.; Brown, J.A.; Olineka, T.L.; Cocolin, L.; Mills, D.A.; Bisson, L.F. Use of WL medium to profile native flora fermentations. Am. J. Enol. Vitic. 2001, 52, 198–203. [Google Scholar] [CrossRef]
- Parafati, L.; Palmeri, R.; Pitino, I.; Restuccia, C. Killer yeasts isolated from olive brines: Technological and probiotic aptitudes. Food Microbiol. 2022, 103, 103950. [Google Scholar] [CrossRef]
- Sinacori, M.; Francesca, N.; Alfonzo, A.; Cruciata, M.; Sannino, C.; Settanni, L.; Moschetti, G. Cultivable microorganisms associated with honeys of different geographical and botanical origin. Food Microbiol. 2014, 38, 284–294. [Google Scholar] [CrossRef]
- Alfonzo, A.; Alongi, D.; Prestianni, R.; Pirrone, A.; Naselli, V.; Viola, E.; De Pasquale, C.; La Croce, F.; Gaglio, R.; Settanni, L.; et al. Enhancing the quality and safety of Nocellara del Belice green table olives produced using the Castelvetrano method. Food Microbiol. 2024, 120, 104477. [Google Scholar] [CrossRef]
- Porru, C.; Rodríguez-Gómez, F.; Benítez-Cabello, A.; Jiménez-Díaz, R.; Zara, G.; Budroni, M.; Mannazzu, I.; Arroyo-López, F.N. Genotyping, identification and multifunctional features of yeasts associated to Bosana naturally black table olive fermentations. Food Microbiol. 2018, 69, 33–42. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Robnett, C.J. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 1998, 73, 331–371. [Google Scholar] [CrossRef]
- Baker, G.C.; Smith, J.J.; Cowan, D.A. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 2003, 55, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Claesson, M.J.; Wang, Q.; O’Sullivan, O.; Greene-Diniz, R.; Cole, J.R.; Ross, R.P.; O’Toole, P.W. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 2010, 38, e200. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Caporaso, J.G. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2′s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Licata, A.G.; Zoppi, M.; Dossena, C.; Rossignoli, F.; Rizzo, D.; Marra, M.; Gargari, G.; Mantegazza, G.; Guglielmetti, S.; Bergamaschi, L.; et al. QIIME2 enhances multi-amplicon sequencing data analysis: A standardized and validated open-source pipeline for comprehensive 16S rRNA gene profiling. Microbiol. Spectr. 2025, 13, e01673-25. [Google Scholar] [CrossRef]
- Mougiou, N.; Tsoureki, A.; Didos, S.; Bouzouka, I.; Michailidou, S.; Argiriou, A. Microbial and biochemical profile of different types of Greek table olives. Foods 2023, 12, 1527. [Google Scholar] [CrossRef]
- Speranza, B.; Sinigaglia, M.; Corbo, M.R.; D’Errico, N.; Bevilacqua, A. A preliminary approach to define the microbiological profile of naturally fermented Peranzana Alta Daunia table olives. Foods 2022, 11, 2100. [Google Scholar] [CrossRef]
- Kamilari, E.; Anagnostopoulos, D.A.; Tsaltas, D. Fermented table olives from Cyprus: Microbiota profile of three varieties from different regions through metabarcoding sequencing. Front. Microbiol. 2023, 13, 1101515. [Google Scholar] [CrossRef]
- Lucena-Padrós, H.; Ruiz-Barba, J.L. Microbial biogeography of Spanish-style green olive fermentations in the province of Seville, Spain. Food Microbiol. 2019, 82, 259–268. [Google Scholar] [CrossRef]
- Benítez-Cabello, A.; Delgado, A.M.; Quintas, C. Main challenges expected from the impact of climate change on microbial biodiversity of table olives: Current status and trends. Foods 2023, 12, 3712. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Barba, J.L.; Sánchez, A.H.; López-López, A.; Cortés-Delgado, A.; Montaño, A. Microbial community and volatilome changes in brines along the spontaneous fermentation of Spanish-style and natural-style green table olives (Manzanilla cultivar). Food Microbiol. 2023, 113, 104286. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, E.; Lorusso, G.; Lamparelli, F. A study of floral biology and the technological features of seven olive cultivars of different origins. Acta Hortic. 1999, 474, 279–284. [Google Scholar] [CrossRef]
- Comunian, R.; Ferrocino, I.; Paba, A.; Daga, E.; Campus, M.; Di Salvo, R.; Cauli, E.; Piras, F.; Zurru, R.; Cocolin, L. Evolution of microbiota during spontaneous and inoculated Tonda di Cagliari table olives fermentation and impact on sensory characteristics. LWT—Food Sci. Technol. 2017, 84, 64–72. [Google Scholar] [CrossRef]
- De Angelis, M.; Campanella, D.; Cosmai, L.; Summo, C.; Rizzello, C.G.; Caponio, F. Microbiota and metabolome of un-started and started Greek-type fermentation of Bella di Cerignola table olives. Food Microbiol. 2015, 52, 18–30. [Google Scholar] [CrossRef]
- Randazzo, C.L.; Todaro, A.; Pino, A.; Pitino, I.; Corona, O.; Caggia, C. Microbiota and metabolome during controlled and spontaneous fermentation of Nocellara Etnea table olives. Food Microbiol. 2017, 65, 136–148. [Google Scholar] [CrossRef]
- Campus, M.; Değirmencioğlu, N.; Comunian, R. Technologies and trends to improve table olive quality and safety. Front. Microbiol. 2018, 9, 617. [Google Scholar] [CrossRef]
- Martins, F.; Rodrigues, N.; Ramalhosa, E. A Review of the Microbial Dynamics of Natural and Traditional Fermentations of Table Olive. Appl. Microbiol. 2025, 5, 52. [Google Scholar] [CrossRef]
- Tassou, C.C.; Panagou, E.Z.; Nychas, G.J. Microbial colonization of naturally fermented olives. In Olives and Olive Oil in Health and Disease Prevention; Preedy, V.R., Watson, R.R., Eds.; Academic Press: Cambridge, MA, USA, 2010; pp. 397–406. [Google Scholar]
- Campaniello, D.; Bevilacqua, A.; D’Amato, D.; Corbo, M.R.; Altieri, C.; Sinigaglia, M. Microbial characterization of table olives processed according to Spanish and natural styles. Food Technol. Biotechnol. 2005, 43, 289–294. [Google Scholar]
- Barrahi, M.; Esmail, A.; Elhartiti, H.; Chahboun, N.; Benali, A.; Amiyare, R.; Lakhrissi, B.; Rhaiem, N.; Zarrouk, A.; Ouhssine, M. Chemical composition and evaluation of antibacterial activity of fennel (Foeniculum vulgare Mill) seed essential oil against some pathogenic bacterial strains. Caspian J. Environ. Sci. 2020, 18, 295–307. [Google Scholar]
- Milenković, A.; Ilić, Z.; Stanojević, L.; Milenković, L.; Šunić, L.; Lalević, D.; Stanojević, J.; Danilović, B.; Cvetković, D. Essential Oil Yield, Composition, Antioxidant and Microbial Activity of Wild Fennel (Foeniculum vulgare Mill.) from Monte Negro Coast. Horticulturae 2022, 8, 1015. [Google Scholar] [CrossRef]
- Miraj, S.; Kiani, S. Study of antibacterial, antimycobacterial, antifungal, and antioxidant activities of Foeniculum vulgare: A review. Pharm. Lett. 2016, 8, 200–205. [Google Scholar]
- Yahyaoui, M.; Moumnassi, S.; Bentouhami, N.; Houmy, N.; Ed-Daoui, A.; Bellaouchi, R.; Taibi, M.; Haddou, M.; Bouzidi, A.; Brasca, M.; et al. Enhancing the fermentation of unsalted Moroccan Picholine green olives through heat-shock treatment, Lactiplantibacillus plantarum S61 inoculation and orange peel addition. CyTA–J. Food 2024, 22, 2384610. [Google Scholar] [CrossRef]
- Sab, C.; Romero, C.; Brenes, M.; Montaño, A.; Ouelhadj, A.; Medina, E. Industrial processing of Algerian table olive cultivars elaborated as Spanish style. Front. Microbiol. 2021, 12, 729436. [Google Scholar] [CrossRef]
- Medina, E.; Brenes, M.; Romero, C.; Ramírez, E.; de Castro, A. Survival of foodborne pathogenic bacteria in table olive brines. Food Control 2013, 34, 719–724. [Google Scholar] [CrossRef]
- Benítez-Cabello, A.; Romero-Gil, V.; Medina-Pradas, E.; Garrido-Fernández, A.; Arroyo-López, F.N. Exploring bacteria diversity in commercialized table olive biofilms by metataxonomic and compositional data analysis. Sci. Rep. 2020, 10, 11381. [Google Scholar] [CrossRef]
- Medina, E.; Ruiz-Bellido, M.A.; Romero-Gil, V.; Rodríguez-Gómez, F.; Montes-Borrego, M.; Landa, B.B.; Arroyo-López, F.N. Assessment of the bacterial community in directly brined Aloreña de Málaga table olive fermentations by metagenetic analysis. Int. J. Food Microbiol. 2016, 236, 47–55. [Google Scholar] [CrossRef]
- Kazou, M.; Tzamourani, A.; Panagou, E.Z.; Tsakalidou, E. Unraveling the microbiota of natural black cv. Kalamata fermented olives through 16S and ITS metataxonomic analysis. Microorganisms 2020, 8, 672. [Google Scholar] [CrossRef]
- Aponte, M.; Blaiotta, G.; La Croce, F.; Mazzaglia, A.; Farina, V.; Settanni, L.; Moschetti, G. Use of selected autochthonous lactic acid bacteria for Spanish-style table olive fermentation. Food Microbiol. 2012, 30, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Coimbra-Gomes, J.; Reis, P.J.; Tavares, T.G.; Silva, A.A.; Mendes, E.; Casal, S.; Malcata, F.X.; Macedo, A.C. Cobrançosa table olive fermentation as per the Portuguese traditional method, using potentially probiotic Lactiplantibacillus pentosus i106 upon alternative inoculation strategies. Fermentation 2022, 9, 12. [Google Scholar] [CrossRef]
- Rodríguez-Gómez, F.; Valero, A.; Vives Lara, E.; Marín, A.; Ramírez, E.M. LP309 a new strain of Lactiplantibacillus pentosus that improves the lactic fermentation of Spanish-style table olives. J. Food Sci. 2023, 88, 5191–5202. [Google Scholar] [CrossRef] [PubMed]
- Vaccalluzzo, A.; Celano, G.; Pino, A.; Calabrese, F.M.; Foti, P.; Caggia, C.; Randazzo, C. Metagenetic and volatilomic approaches to elucidate the effect of Lactiplantibacillus plantarum starter cultures on Sicilian table olives. Front. Microbiol. 2022, 12, 771636. [Google Scholar] [CrossRef]
- Benítez-Cabello, A.; Calero-Delgado, B.; Rodríguez-Gómez, F.; Bautista-Gallego, J.; Garrido-Fernández, A.; Jiménez-Díaz, R.; Arroyo-López, F.N. The use of multifunctional yeast-lactobacilli starter cultures improves fermentation performance of Spanish-style green table olives. Food Microbiol. 2020, 91, 103497. [Google Scholar]
- Anagnostopoulos, D.A.; Bozoudi, D.; Tsaltas, D. Enterococci isolated from Cypriot green table olives as a new source of technological and probiotic properties. Fermentation 2018, 4, 48. [Google Scholar] [CrossRef]
- El Issaoui, K.; Senhaji, N.S.; Wieme, A.; Abrini, J.; Khay, E.O. Probiotic properties and physicochemical potential of lactic acid bacteria isolated from Moroccan table olives. J. Food Qual. Hazards Control 2022, 9, 169–178. [Google Scholar] [CrossRef]
- Tufariello, M.; Durante, M.; Ramires, F.A.; Grieco, F.; Tommasi, L.; Perbellini, E.; Falco, V.; Tsaioula-Margari, M.; Logrieco, A.F.; Mita, G.; et al. New process for production of fermented black table olives using selected autochthonous microbial resources. Front. Microbiol. 2015, 6, 1007. [Google Scholar] [CrossRef]
- Maoloni, A.; Cardinali, F.; Milanović, V.; Osimani, A.; Garofalo, C.; Ferrocino, I.; Corvaglia, M.R.; Cocolin, L.; Aquilanti, L. Microbial dynamics and key sensory traits of laboratory-scale co-fermented green olives (Olea europaea L. cv. Ascolana tenera) and sea fennel (Crithmum maritimum L.). Food Biosci. 2022, 50, 102077. [Google Scholar]
- Lee, J.; Lee, M.; Jung, M.; Kim, Y.; Roh, S.; Ryu, B.; Jeon, C.; Choi, H.; Whon, T.; Lee, S. Unravelling the key factors for the dominance of Leuconostoc starters during kimchi fermentation. NPJ Sci. Food 2025, 9, 15. [Google Scholar] [CrossRef]
- Agirman, B.; Settanni, L.; Erten, H. Effect of different mineral salt mixtures and dough extraction procedure on the physical, chemical and microbiological composition of Şalgam: A black carrot fermented beverage. Food Chem. 2021, 344, 128618. [Google Scholar] [CrossRef]
- Ruppitsch, W.; Nisic, A.; Hyden, P.; Cabal, A.; Sucher, J.; Stöger, A.; Allerberger, F.; Martinović, A. Genetic Diversity of Leuconostoc mesenteroides Isolates from Traditional Montenegrin Brine Cheese. Microorganisms 2021, 9, 1612. [Google Scholar] [CrossRef]
- Kumral, A.Y.; Basoglu, F.; Sahin, I. Effect of the use of different lactic starters on the microbiological and physicochemical characteristics of naturally black table olives of Gemlik cultivar. J. Food Process. Preserv. 2009, 33, 651–664. [Google Scholar] [CrossRef]
- Kharkhota, M.; Kharchuk, M.; Duplij, V.; Brindza, J.; Avdieieva, L.; Matvieieva, N. Effect of Priestia endophytica on the metabolites accumulation in chicory and lettuce plants cultivated in vitro. Prep. Biochem. Biotechnol. 2023, 53, 1137–1142. [Google Scholar] [CrossRef]
- Matvieieva, N.; Duplij, V.; Kharkhota, M.; Avdeeva, L. Priestia endophytica bacteria stimulate Rhodiola rosea L. in vitro growth. Agrobiodivers. Improv. Nutr. Health Life Qual. 2022, 6, 149–155. [Google Scholar] [CrossRef]
- Shurigin, V.; Li, L.; Alaylar, B.; Egamberdieva, D.; Liu, Y.H.; Li, W.J. Plant beneficial traits of endophytic bacteria associated with fennel (Foeniculum vulgare Mill.). AIMS Microbiol. 2024, 10, 449. [Google Scholar] [CrossRef] [PubMed]
- Hussein, W.; Ramadan, W.A.; Ibrahim, H.F. Isolation and identification of associated endophytic bacteria from barley seeds harbour non-ribosomal peptides and enhance tolerance to salinity stress. Beni-Suef Univ. J. Basic Appl. Sci. 2024, 13, 27. [Google Scholar] [CrossRef]
- Agunbiade, V.F.; Fadiji, A.E.; Agbodjato, N.A.; Babalola, O.O. Isolation and characterization of plant-growth-promoting, drought-tolerant rhizobacteria for improved maize productivity. Plants 2024, 13, 1298. [Google Scholar] [CrossRef] [PubMed]
- Manetsberger, J.; Caballero Gómez, N.; Soria-Rodríguez, C.; Benomar, N.; Abriouel, H. Simply versatile: The use of Peribacillus simplex in sustainable agriculture. Microorganisms 2023, 11, 2540. [Google Scholar] [CrossRef]
- Azabou, M.C.; Gharbi, Y.; Medhioub, I.; Ennouri, K.; Barham, H.; Tounsi, S.; Triki, M.A. The endophytic strain Bacillus velezensis OEE1: An efficient biocontrol agent against Verticillium wilt of olive and a potential plant growth promoting bacteria. Biol. Control 2020, 142, 104168. [Google Scholar]
- Castro, D.; Torres, M.; Sampedro, I.; Martínez-Checa, F.; Torres, B.; Béjar, V. Biological control of Verticillium wilt on olive trees by the salt-tolerant strain Bacillus velezensis XT1. Microorganisms 2020, 8, 1080. [Google Scholar] [CrossRef] [PubMed]
- Cheffi, M.; Chenari Bouket, A.; Alenezi, F.N.; Luptakova, L.; Belka, M.; Vallat, A.; Rateb, M.E.; Tounsi, S.; Triki, M.A.; Belbahri, L. Olea europaea L. root endophyte Bacillus velezensis OEE1 counteracts oomycete and fungal harmful pathogens and harbours a large repertoire of secreted and volatile metabolites and beneficial functional genes. Microorganisms 2019, 7, 314. [Google Scholar] [CrossRef] [PubMed]
- Covington, A.L.; Cerqueira, F.M.; Pavia, J.E.; Reynoso, D.; Ren, P. Complex trauma sequelae: Mycobacterium goodii and Priestia endophytica hardware infection in a patient with Ehlers-Danlos syndrome. BMC Infect. Dis. 2024, 24, 1064. [Google Scholar] [CrossRef] [PubMed]
- Bautista-Gallego, J.; Rodríguez-Gómez, F.; Barrio, E.; Querol, A.; Garrido-Fernández, A.; Arroyo-López, F.N. Exploring the yeast biodiversity of green table olive industrial fermentations for technological applications. Int. J. Food Microbiol. 2011, 147, 89–96. [Google Scholar] [CrossRef]
- Pereira, E.L.; Ramalhosa, E.; Borges, A.; Pereira, J.A.; Baptista, P. Yeast dynamics during the natural fermentation process of table olives (Negrinha de Freixo cv.). Food Microbiol. 2015, 46, 582–586. [Google Scholar] [CrossRef]
- Bencresciuto, G.; Mandalà, C.; Migliori, C.; Cortellino, G.; Vanoli, M.; Bardi, L. Assessment of starters of lactic acid bacteria and killer yeasts: Selected strains in lab-scale fermentations of table olives (Olea europaea L.) cv. Leccino. Fermentation 2023, 9, 182. [Google Scholar] [CrossRef]
- Alongi, D.; Pirrone, A.; Naselli, V.; Prestianni, R.; Monte, M.; Gaglio, R.; De Pasquale, C.; Settanni, L.; Alfonzo, A.; Moschetti, G.; et al. Co-inoculation approach combining lactic acid bacteria and yeasts to enhance the production of Nocellara del Belice green split table olives. Food Biosci. 2024, 61, 104816. [Google Scholar] [CrossRef]
- Chytiri, A.; Tasioula-Margari, M.; Bleve, G.; Kontogianni, V.G.; Kallimanis, A.; Kontominas, M.G. Effect of different inoculation strategies of selected yeast and LAB cultures on Conservolea and Kalamàta table olives considering phenol content, texture, and sensory attributes. J. Sci. Food Agric. 2020, 100, 926–935. [Google Scholar] [CrossRef]
- Oliveira, T.; Ramalhosa, E.; Nunes, L.; Pereira, J.A.; Colla, E.; Pereira, E.L. Probiotic potential of indigenous yeasts isolated during the fermentation of table olives from Northeast of Portugal. Innov. Food Sci. Emerg. Technol. 2017, 44, 167–172. [Google Scholar] [CrossRef]
- Simões, L.A.; de Souza, A.C.; Ferreira, I.; Melo, D.S.; Lopes, L.A.A.; Magnani, M.; Schwan, R.F.; Dias, D.R. Probiotic properties of yeasts isolated from Brazilian fermented table olives. J. Appl. Microbiol. 2021, 131, 1983–1997. [Google Scholar] [CrossRef]
- Parapouli, M.; Vasileiadis, A.; Afendra, A.S.; Hatziloukas, E. Saccharomyces cerevisiae and its industrial applications. AIMS Microbiol. 2020, 6, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Ciafardini, G.; Venditti, G.; Zullo, B. Yeast dynamics in the black table olives processing using fermented brine as starter. J. Food Sci. 2021, 5, 92–106. [Google Scholar] [CrossRef] [PubMed]
- Perpetuini, G.; Prete, R.; Garcia-Gonzalez, N.; Khairul Alam, M.; Corsetti, A. Table olives more than a fermented food. Foods 2020, 9, 178. [Google Scholar] [CrossRef] [PubMed]
- Tufariello, M.; Anglana, C.; Crupi, P.; Virtuosi, I.; Fiume, P.; Di Terlizzi, B.; Moselhy, N.; Attay, H.A.G.; Pati, S.; Logrieco, A.F.; et al. Efficacy of yeast starters to drive and improve Picual, Manzanilla and Kalamàta table olive fermentation. J. Sci. Food Agric. 2019, 99, 2504–2512. [Google Scholar] [CrossRef]
- Schaide, T.; Cabrera-Bañegil, M.; Pérez-Nevado, F.; Esperilla, A.; Martín-Vertedor, D. Effect of olive leaf extract combined with Saccharomyces cerevisiae in the fermentation process of table olives. J. Food Sci. Technol. 2019, 56, 3001–3013. [Google Scholar] [CrossRef]
- Psani, M.; Kotzekidou, P. Technological characteristics of yeast strains and their potential as starter adjuncts in Greek-style black olive fermentation. World J. Microbiol. Biotechnol. 2006, 22, 1329–1336. [Google Scholar] [CrossRef]
- Mujdeci, G.N.; Ozbas, Z.Y. Technological and enzymatic characterization of the yeasts isolated from natural fermentation media of Gemlik olives. J. Appl. Microbiol. 2021, 131, 801–818. [Google Scholar] [CrossRef]
- Settanni, L.; Barbaccia, P.; Bonanno, A.; Ponte, M.; Di Gerlando, R.; Franciosi, E.; Di Grigoli, A.; Gaglio, R. Evolution of indigenous starter microorganisms and physicochemical parameters in spontaneously fermented beef, horse, wild boar and pork salamis produced under controlled conditions. Food Microbiol. 2020, 87, 103385. [Google Scholar] [CrossRef]
- Correa-Galeote, D.; Ghomari, I.; Asehraou, A.; González-López, J. Revealing the bacterial abundance and diversity in brines from started Spanish-style green table olives. LWT—Food Sci. Technol. 2022, 160, 113212. [Google Scholar] [CrossRef]
- Anagnostopoulos, D.A.; Kamilari, E.; Tsaltas, D. Evolution of bacterial communities, physicochemical changes and sensorial attributes of natural whole and cracked Picual table olives during spontaneous and inoculated fermentation. Front. Microbiol. 2020, 11, 1128. [Google Scholar] [CrossRef]
- Tzamourani, A.P.; Kasimati, A.; Karagianni, E.; Manthou, E.; Panagou, E.Z. Exploring microbial communities of Spanish-style green table olives of Conservolea and Halkidiki cultivars during modified atmosphere packaging in multi-layered pouches through culture-dependent techniques and metataxonomic analysis. Food Microbiol. 2022, 107, 104063. [Google Scholar] [CrossRef]
- Michailidou, S.; Trikka, F.; Pasentsis, K.; Petrovits, G.E.; Kyritsi, M.; Argiriou, A. Insights into the evolution of Greek style table olives microbiome stored under modified atmosphere: Biochemical implications on the product quality. Food Control 2021, 130, 108286. [Google Scholar] [CrossRef]
- Kallastu, A.; Malv, E.; Aro, V.; Meikas, A.; Vendelin, M.; Kattel, A.; Nahku, R.; Kazantseva, J. Absolute quantification of viable bacteria abundances in food by next-generation sequencing: Quantitative NGS of viable microbes. Curr. Res. Food Sci. 2023, 6, 100443. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).