Green Solutions for Food Safety: The Emerging Applications of Zearalenone-Degrading Enzymes
Abstract
1. Introduction
2. Enzymatic Characteristics of Lactone Hydrolase
2.1. Structural Characteristics and Catalytic Properties of Lactone Hydrolases
2.1.1. Structural Characteristics of Lactone Hydrolases
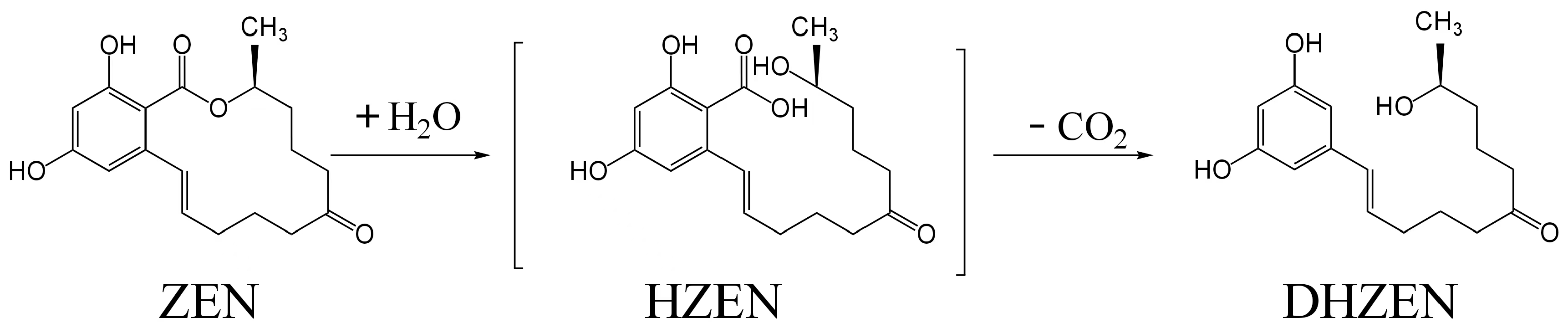
2.1.2. Catalytic Mechanism
2.2. Sources of Lactone Hydrolase, Catalytic Activity and Reaction Conditions
3. Molecular Modification and Optimization of Lactone Hydrolase
3.1. Directed Evolution Technology Strategy
3.2. Rationally Design Technical Strategies
3.3. Computer-Aided Rational Design Strategy
4. Application Research of Lactone Hydrolase
4.1. Direct Detoxification Application of Free Enzyme
4.1.1. Detoxification Applications in Food
4.1.2. Detoxification Application in Feed or Livestock and Poultry Animals
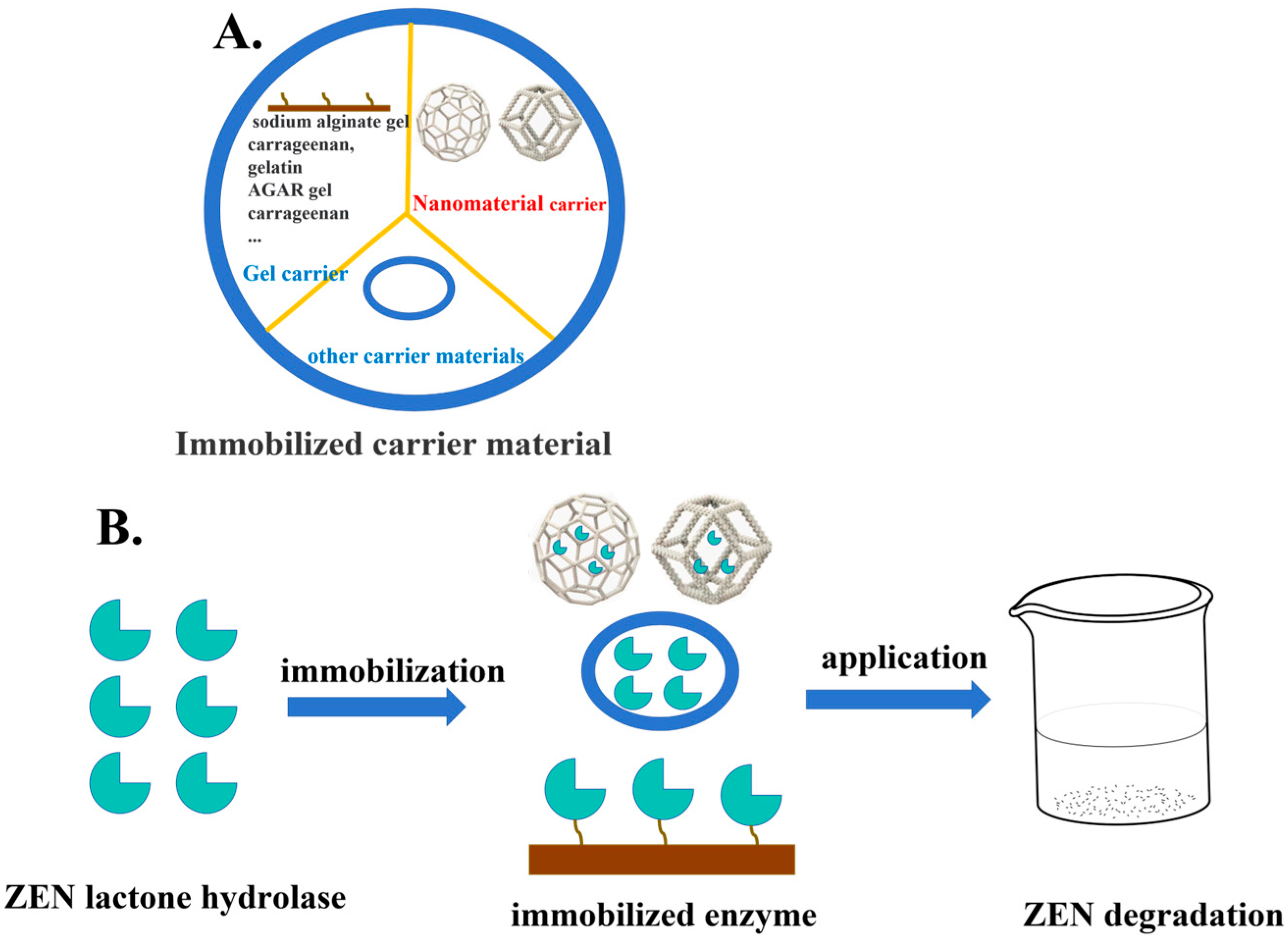
4.2. Application of Immobilized Enzymes for Detoxification
4.2.1. Application of Gel-Based Enzyme Immobilization Technology for ZEN Detoxification
4.2.2. Applications of Nano-Immobilized Enzyme Technology for ZEN Detoxification
4.2.3. Applications of Other Carrier Materials Technology for ZEN Detoxification
4.3. Current Applications and Cost Analysis of Enzymatic Detoxification Technologies
5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stob, M.; Baldwin, R.S.; Tuite, J.; Andrews, F.N.; Gillette, K.G. Isolation of an anabolic, uterotrophic compound from corn infected with Gibberella zeae. Nature 1962, 196, 1318. [Google Scholar] [CrossRef] [PubMed]
- Fruhauf, S.; Novak, B.; Nagl, V.; Hackl, M.; Hartinger, D.; Rainer, V.; Labudova, S.; Adam, G.; Aleschko, M.; Moll, W.D.; et al. Biotransformation of the Mycotoxin Zearalenone to its Metabolites Hydrolyzed Zearalenone (HZEN) and Decarboxylated Hydrolyzed Zearalenone (DHZEN) Diminishes its Estrogenicity In Vitro and In Vivo. Toxins 2019, 11, 481. [Google Scholar] [CrossRef]
- Fang, Y.; Zhang, Z.; Xu, W.; Zhang, W.; Guang, C.; Mu, W. Zearalenone lactonase: Characteristics, modification, and application. Appl. Microbiol. Biotechnol. 2022, 106, 6877–6886. [Google Scholar] [CrossRef]
- Rogowska, A.; Pomastowski, P.; Sagandykova, G.; Buszewski, B. Zearalenone and its metabolites: Effect on human health, metabolism and neutralisation methods. Toxicon 2019, 162, 46–56. [Google Scholar] [CrossRef]
- Karlovsky, P.; Suman, M.; Berthiller, F.; Meester, J.D.; Eisenbrand, G.; Perrin, I.; Oswald, I.P.; Speijers, G.; Chiodini, A.; Recker, T.; et al. Impact of food processing and detoxification treatments on mycotoxin contamination. Mycotoxin Res. 2016, 32, 179–205. [Google Scholar] [CrossRef]
- Kong, Q.; Shi, X.; Ma, W.; Zhang, F.; Yu, T.; Zhao, F.; Zhao, D.; Wei, C. Strategies to improve the adsorption properties of graphene-based adsorbent towards heavy metal ions and their compound pollutants: A review. J. Hazard. Mater. 2021, 415, 125690. [Google Scholar] [CrossRef] [PubMed]
- Murata, H.; Mitsumatsu, M.; Shimada, N. Reduction of feed-contaminating mycotoxins by ultraviolet irradiation: An in vitro study. Food Addit. Contam. 2008, 25, 1107–1110. [Google Scholar] [CrossRef]
- Alexandre, A.P.S.; Castanha, N.; Costa, N.S.; Santos, A.S.; Badiale-Furlong, E.; Augusto, P.E.D.; Calori-Domingues, M.A. Ozone technology to reduce zearalenone contamination in whole maize flour: Degradation kinetics and impact on quality. J. Sci. Food Agric. 2019, 99, 6814–6821. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Hossen, S.M.; Sago, Y.; Yoshida, M.; Nakagawa, H.; Nagashima, H.; Okadome, H.; Nakajima, T.; Kushiro, M. Effect of milling on the content of deoxynivalenol, nivalenol, and zearalenone in Japanese wheat. Food Control 2014, 40, 193–197. [Google Scholar] [CrossRef]
- Miklós, P.; Zelma, F.; Afshin, Z.; Tímea, B.; Beáta, L.; Sándor, K.-M.; Lajos, S. Removal of Zearalenone and Zearalenols from Aqueous Solutions Using Insoluble Beta-Cyclodextrin Bead Polymer. Toxins 2018, 10, 216. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Li, Y.; Luo, X.; Wang, R.; Zheng, R.; Wang, L.; Li, Y.; Yang, D.; Fang, W.; Chen, Z. Detoxification of zearalenone and ochratoxin A by ozone and quality evaluation of ozonised corn. Food Addit. Contaminants. Part A Chem. Anal. Control Expo. Risk Assess. 2016, 33, 1700–1710. [Google Scholar] [CrossRef] [PubMed]
- Inga, R.; Susanne, K.; Hana, V.; Sven, D. Hydrothermal treatment of naturally contaminated maize in the presence of sodium metabisulfite, methylamine and calcium hydroxide; effects on the concentration of zearalenone and deoxynivalenol. Mycotoxin Res. 2013, 29, 169–175. [Google Scholar] [CrossRef]
- Ilse, V.; Kris, A.; Leen, D.G. Biodegradation of Mycotoxins: Tales from Known and Unexplored Worlds. Front. Microbiol. 2016, 7, 561. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Huang, H. Engineering proteins for thermostability through rigidifying flexible sites. Biotechnol. Adv. 2014, 32, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhang, X.; Luan, H.; Zhang, Y.; Xu, W.; Feng, W.; Song, P. Bioenzymatic detoxification of mycotoxins. Front. Microbiol. 2024, 15, 1434987. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Xiao, J.; Chen, Y.; Yu, Y.; Xiao, X.; Yu, Y.; Wu, H. Secretory expression and characterization of a novel peroxiredoxin for zearalenone detoxification in Saccharomyces cerevisiae. Microbiol. Res. 2013, 168, 6–11. [Google Scholar] [CrossRef]
- Medyantseva, E.P.; Beilinson, R.M.; Nikolaenko, A.I.; Budnikov, H.C. Horseradish Peroxidase: Analytical Capabilities in the Determination of Zearalenone. J. Anal. Chem. 2022, 77, 671–680. [Google Scholar] [CrossRef]
- Zhang, L.; Zhu, J.; Li, K. Peroxidase activity based on Cu2SnS3 quantum dots for the degradation and visual detection of zearalenone. Mater. Today Commun. 2023, 35, 105871. [Google Scholar] [CrossRef]
- Guo, Y.; Tang, Y.; Zhang, L.; Liu, Y.; Ma, Q.; Zhao, L. Enzymatic characterization and application of soybean hull peroxidase as an efficient and renewable biocatalyst for degradation of zearalenone. Int. J. Biol. Macromol. 2024, 260, 129664. [Google Scholar] [CrossRef]
- Claus, H. Laccases: Structure, reactions, distribution. Micron 2003, 35, 93–96. [Google Scholar] [CrossRef]
- Qin, X.; Xin, Y.; Zou, J.; Su, X.; Wang, X.; Wang, Y.; Zhang, J.; Tu, T.; Yao, B.; Luo, H.; et al. Efficient Degradation of Aflatoxin B1 and Zearalenone by Laccase-like Multicopper Oxidase from Streptomyces thermocarboxydus in the Presence of Mediators. Toxins 2021, 13, 754. [Google Scholar] [CrossRef]
- Mwabulili, F.; Xie, Y.; Sun, S.; Ma, W.; Li, Q.; Yang, Y.; Jia, H.; Li, X. Thermo-Alkali-Tolerant Recombinant Laccase from Bacillus swezeyi and Its Degradation Potential against Zearalenone and Aflatoxin B1. J. Agric. Food Chem. 2024, 72, 13371–13381. [Google Scholar] [CrossRef]
- Huang, H.; Sun, F.; Yu, D.; Zhou, H.; Lin, H.; Yan, Z.; Wu, A. CotA laccase from Bacillus licheniformis ZOM-1 effectively degrades zearalenone, aflatoxin B1 and alternariol. Food Control 2023, 145, 109472. [Google Scholar]
- Yu, L.; Sun, B.; Ren, M.; Yan, X.; Liu, K.; Song, J.; Yue, L.; Wang, W.; Zhao, C. The Research on Zearalenone Lactonase and Its Application in Livestock and Poultry Diets. Acta Vet. Zootech. Sin. 2025, 1–13. Available online: https://link.cnki.net/urlid/11.1985.s.20250521.1647.007 (accessed on 24 June 2025).
- Wang, Y.; Chen, Y.; Jiang, L.; Huang, H. Improvement of the enzymatic detoxification activity towards mycotoxins through structure-based engineering. Biotechnol. Adv. 2022, 56, 107927. [Google Scholar] [CrossRef]
- Peng, W.; Ko, T.P.; Yang, Y.; Zheng, Y.; Chen, C.; Zhu, Z.; Huang, C.H.; Zeng, Y.; Huang, J.; Wang, A.; et al. Crystal structure and substrate-binding mode of the mycoestrogen-detoxifying lactonase ZHD from Clonostachys rosea. RSC Adv. 2014, 4, 62321–62325. [Google Scholar] [CrossRef]
- Rauwerdink, A.; Kazlauskas, R.J. How the Same Core Catalytic Machinery Catalyzes 17 Different Reactions: The Serine-Histidine-Aspartate Catalytic Triad of α/β-Hydrolase Fold Enzymes. ACS Catal. 2015, 5, 6153–6176. [Google Scholar] [CrossRef]
- Takahashi-Ando, N.; Shuichi, O.S.; Shibata, T.; Hamamoto, H.; Yamaguchi, I.; Kimura, M. Metabolism of zearalenone by genetically modified organisms expressing the detoxification gene from Clonostachys rosea. Appl. Environ. Microbiol. 2004, 70, 3239–3245. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhang, F.; Xiang, L.; Li, M.; Lu, Z.; Wu, P.; Sheng, X.; Zhou, J.; Zhang, G. Enhancing the activity of zearalenone lactone hydrolase toward the more toxic α-zearalanol via a single-point mutation. Appl. Environ. Microbiol. 2024, 90, e0181823. [Google Scholar] [CrossRef] [PubMed]
- Takahashi-Ando, N.; Kimura, M.; Kakeya, H.; Osada, H.; Yamaguchi, I. A novel lactonohydrolase responsible for the detoxification of zearalenone: Enzyme purification and gene cloning. Biochem. J. 2002, 365, 1–6. [Google Scholar] [CrossRef]
- Lin, M.; Tan, J.; Xu, Z.; Huang, J.; Tian, Y.; Chen, B.; Wu, Y.; Tong, Y.; Zhu, Y. Computational design of enhanced detoxification activity of a zearalenone lactonase from Clonostachys rosea in acidic medium. RSC Adv. 2019, 9, 31284–31295. [Google Scholar] [CrossRef] [PubMed]
- Qi, Q.; Yang, W.; Zhou, H.; Ming, D.; Sun, K.; Xu, T.; Hu, X.; Lv, H. The structure of a complex of the lactonohydrolase zearalenone hydrolase with the hydrolysis product of zearalenone at 1.60 Å resolution. Acta crystallographica. Sect. F Struct. Biol. Commun. 2017, 73, 376–381. [Google Scholar] [CrossRef]
- Christiane, G.; Johannes, F.; Barbara, D.; Markus, A.; Christian, S.; Andreas, H.; Karin, S.; Manuela, K.; Qendrim, Z.; Dian, S. Metabolism of Zearalenone in the Rumen of Dairy Cows with and without Application of a Zearalenone-Degrading Enzyme. Toxins 2021, 13, 84. [Google Scholar] [CrossRef]
- Christiane, G.; Manuela, K.; Andreas, H.; Roy, R.; Barbara, D.; Markus, A.; Heidi, S.; Oliver, G.; Zoran, M.; Marko, S.; et al. Enzymatic Degradation of Zearalenone in the Gastrointestinal Tract of Pigs, Chickens, and Rainbow Trout. Toxins 2023, 15, 48. [Google Scholar] [CrossRef]
- Yu, X.; Tu, T.; Luo, H.; Huang, H.; Su, X.; Wang, Y.; Wang, Y.; Zhang, J.; Bai, Y.; Yao, B. Biochemical Characterization and Mutational Analysis of a Lactone Hydrolase from Phialophora americana. J. Agric. Food Chem. 2020, 68, 2570–2577. [Google Scholar] [CrossRef]
- Bi, K.; Zhang, W.; Xiao, Z.; Zhang, D. Characterization, expression and application of a zearalenone degrading enzyme from Neurospora crassa. AMB Express 2018, 8, 194. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, B.; Zhang, W.; Guang, C.; Xu, W.; Mu, W. Identification and Modification of Enzymatic Substrate Specificity through Residue Alteration in the Cap Domain: A Thermostable Zearalenone Lactonase. J. Agric. Food Chem. 2023, 71, 18943–18952. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, X.M.; Zhang, Y.P.; Zhang, X.L.; Huang, H. Cloning and Characterization of Three Novel Enzymes Responsible for the Detoxification of Zearalenone. Toxins 2022, 14, 82. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Fang, Y.; Zhu, Y.; Tian, W.; Yu, J.; Tang, J. Biotransformation of zearalenone to non-estrogenic compounds with two novel recombinant lactonases from Gliocladium. BMC Microbiol. 2024, 24, 75. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wang, G.; Hou, M.; Du, S.; Han, J.; Yu, Y.; Gao, H.; He, D.; Shi, J.; Lee, Y.-W.; et al. New Hydrolase from Aeromicrobium sp. HA for the Biodegradation of Zearalenone: Identification, Mechanism, and Application. J. Agric. Food Chem. 2023, 71, 2411–2420. [Google Scholar] [CrossRef]
- Chen, S.R.; Pan, L.; Liu, S.Y.; Pan, L.J.; Li, X.J.; Wang, B. Recombinant expression and surface display of a zearalenone lactonohydrolase from Trichoderma aggressivum in Escherichia coli. Protein Expr. Purif. 2021, 187, 105933. [Google Scholar] [CrossRef]
- Shcherbakova, L.; Rozhkova, A.; Osipov, D.; Zorov, I.; Mikityuk, O.; Statsyuk, N.; Sinitsyna, O.; Dzhavakhiya, V.; Sinitsyn, A. Effective Zearalenone Degradation in Model Solutions and Infected Wheat Grain Using a Novel Heterologous Lactonohydrolase Secreted by Recombinant Penicillium canescens. Toxins 2020, 12, 475. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lu, Z.; Lin, X.; Wang, M.; Jiang, T.; Zhao, G.; La, X.; Xv, J.; Jiang, S.; Zhang, G. The N-terminal hydrophobicity modulates a distal structural domain conformation of zearalenone lacton hydrolase and its application in protein engineering. Enzym. Microb. Technol. 2023, 165, 110195. [Google Scholar] [CrossRef]
- Fruhauf, S.; Pühringer, D.; Thamhesl, M.; Fajtl, P.; Vekiru, E.K.; Gussl, A.H.; Schatzmayr, G.; Adam, G.; Damborsky, J.; Carugo, K.D.; et al. Bacterial Lactonases ZenA with Noncanonical Structural Features Hydrolyze the Mycotoxin Zearalenone. ACS Catal. 2024, 14, 3392–3410. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Zhang, N.; Wang, H.; Wang, M.; Jiang, S.; He, N.; Zhang, G. Enzymatic and structural characterization of a zearalenone lactone hydrolase ZHD11F from Phialophora attae. Acta Microbiol. Sin. 2022, 62, 4202–4212. [Google Scholar] [CrossRef]
- Fang, Y.; Huang, Z.; Xu, W.; Wang, C.; Sun, Y.; Zhang, W.; Guang, C.; Mu, W. Efficient elimination of zearalenone at high processing temperatures by a robust mutant of Gliocladium roseum zearalenone lactonase. Food Control 2022, 142, 109222. [Google Scholar] [CrossRef]
- Wang, M.; Yin, L.; Hu, H.; Selvaraj, J.; Zhou, Y.; Zhang, G. Expression, functional analysis and mutation of a novel neutral zearalenone-degrading enzyme. Int. J. Biol. Macromol. 2018, 118, 1284–1292. [Google Scholar] [CrossRef]
- Hu, X.; Liu, W.; Liu, W.; Zhan, X.; Guo, R.; Li, H.; Zheng, Y. Expression, purification and characterization of a novel zearalenone hydrolase from Rhinocladiella mackenziei. Microbiol. China 2018, 45, 2585–2591. [Google Scholar] [CrossRef]
- Gao, H.; Lu, D.; Xing, M.Y.; Xu, Q.; Xue, F. Excavation, expression, and functional analysis of a novel zearalenone-degrading enzyme. Folia Microbiol. 2022, 67, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xu, W.; Wu, H.; Zhang, W.; Mu, W. Identification of a Potent Enzyme for the Detoxification of Zearalenone. J. Agric. Food Chem. 2020, 68, 376–383. [Google Scholar] [CrossRef] [PubMed]
- Hui, R.; Hu, X.; Liu, W.; Liu, W.; Zheng, Y.; Chen, Y.; Guo, R.; Jin, J.; Chen, C. Characterization and crystal structure of a novel zearalenone hydrolase from Cladophialophora bantiana. Acta Crystallographica. Sect. F Struct. Biol. Commun. 2017, 73, 515–519. [Google Scholar] [CrossRef]
- Chai, C.; Chang, X.; Wang, N.; Wu, S.; Sun, C. Preliminary Study on Prokaryotic Expression and Its Degradation Activity of a New Zearalenone-Degradation Enzyme (mbZHD). J. Chin. Cereals Oils Assoc. 2017, 32, 29–33+38. [Google Scholar]
- Xiang, L.; Wang, Q.; Zhou, Y.; Yin, L.; Zhang, G.; Ma, Y. High-level expression of a ZEN-detoxifying gene by codon optimization and biobrick in Pichia pastoris. Microbiol. Res. 2016, 193, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Mwabulili, F.; Xie, Y.; Yang, Y.; Sun, S.; Li, Q.; Ma, W.; Jia, H. Characterization, Structural Analysis, and Thermal Stability Mutation of a New Zearalenone-Degrading Enzyme Mined from Bacillus subtilis. J. Agric. Food Chem. 2024, 72, 3025–3035. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Sun, C.; Wu, S.; Zhang, Y.; Wu, Z. Expression of Zearalenone Degrading Enzyme Gene from Gliocladium Roseum in Pichia Pastoris GS115. J. Chin. Cereals Oils Assoc. 2011, 26, 12–17. [Google Scholar]
- Sun, H.; He, Z.; Xiong, D.; Long, M. Mechanisms by which microbial enzymes degrade four mycotoxins and application in animal production: A review. Anim. Nutr. 2023, 15, 256–274. [Google Scholar] [CrossRef]
- Kohl, K.D.; Ashley, S.; Michal, S.-B.; Denise, D.M. Effects of anatomy and diet on gastrointestinal pH in rodents. J. Exp. Zool. Part A Ecol. Genet. Physiol. 2013, 319, 225–229. [Google Scholar] [CrossRef]
- Liu, Q.; Xun, G.; Feng, Y. The state-of-the-art strategies of protein engineering for enzyme stabilization. Biotechnol. Adv. 2019, 37, 530–537. [Google Scholar] [CrossRef] [PubMed]
- Rajendra, N.S. Directed Evolution Methods for Enzyme Engineering. Molecules 2021, 26, 5599. [Google Scholar] [CrossRef] [PubMed]
- Anshula, S.; Gaganjot, G.; Tawseef, A.; Sheikh, M.; Baljinder, K. Enzyme Engineering: Current Trends and Future Perspectives. Food Rev. Int. 2021, 37, 121–154. [Google Scholar]
- Fisher, A.K.; Freedman, B.G.; Bevan, D.R.; Senger, R.S. A review of metabolic and enzymatic engineering strategies for designing and optimizing performance of microbial cell factories. Comput. Struct. Biotechnol. J. 2014, 11, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Liu, Y.; Huang, Y.; Yu, P. Advances in the Directed Evolution of Computer-aided Enzymes. Curr. Top. Med. Chem. 2025, 25, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Xu, H.; Ji, Q.; Xu, R.; Zhu, M.; Dang, Y.; Shi, X.; Zhang, L.; Xia, Y. Mutation, food-grade expression, and characterization of a lactonase for zearalenone degradation. Appl. Microbiol. Biotechnol. 2023, 107, 5107–5118. [Google Scholar] [CrossRef] [PubMed]
- Dotsenko, A.; Sinelnikov, I.; Zorov, I.; Denisenko, Y.; Rozhkova, A.; Shcherbakova, L. The Protein Engineering of Zearalenone Hydrolase Results in a Shift in the pH Optimum of the Relative Activity of the Enzyme. Toxins 2024, 16, 540. [Google Scholar] [CrossRef]
- Guan, A.; Zhang, M.; Xu, F. Structure-guided engineering for improving the thermal stability of zearalenone hydrolase. Chin. J. Biotechnol. 2023, 39, 3336–3350. [Google Scholar] [CrossRef]
- Xing, X.; Chen, X.; You, X.; Huang, J.; Xue, D. Zearalenone degrading enzyme evolution to increase the hydrolysis efficiency under acidic conditions by the rational design. Food Chem. 2024, 456, 140088. [Google Scholar] [CrossRef]
- Xu, Z.; Liu, W.; Chen, C.; Li, Q.; Huang, J.; Ko, T.P.; Liu, G.; Liu, W.; Peng, W.; Cheng, Y.; et al. Enhanced α-Zearalenol Hydrolyzing Activity of a Mycoestrogen-Detoxifying Lactonase by Structure-Based Engineering. ACS Catal. 2016, 6, 7657–7663. [Google Scholar] [CrossRef]
- Xue, J.; Tehreem, S.; Rahim, K.; Wang, M.; Wu, P.; Zhang, G. Enhancing the thermal stability and activity of zearalenone lactone hydrolase to promote zearalenone degradation via semi-rational design. Enzym. Microb. Technol. 2024, 180, 110499. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Wang, M.; Li, X.; Wang, H.; Zhao, G.; Wu, P.; Lu, Z.; Zhang, G. The replacement of main cap domain to improve the activity of a ZEN lactone hydrolase with broad substrate spectrum. Biochem. Eng. J. 2022, 182, 108418. [Google Scholar] [CrossRef]
- Zheng, Y.; Liu, W.; Chen, C.; Hu, X.; Liu, W.; Ko, T.; Tang, X.; We, H.; Huang, J.; Guo, R. Crystal Structure of a Mycoestrogen-Detoxifying Lactonase from Rhinocladiella mackenziei: Molecular Insight into ZHD Substrate Selectivity. ACS Catal. 2018, 8, 4294–4298. [Google Scholar] [CrossRef]
- Xu, Z.; Liu, G.; Liu, W.; Guo, R.; Li, H.; Zheng, Y. Molecular Engineering of Mycoestrogen-Detoxifying Lactonase ZHD101 to Improve Enzyme Thermostability. J. Food Sci. Biotechnol. 2019, 38, 71–77. [Google Scholar]
- Chen, Q.; Lv, C.; Xv, F. Computation-aided design of the flexible region of zearalenone hydrolase improves its thermal stability. Chin. J. Biotechnol. 2021, 37, 4415–4429. [Google Scholar]
- Du, H.; Luo, S.; Adegoke, T.V.; Liu, Y.; Xing, F.; Gao, J.; Zheng, Y. Thermal stability modification of zearalenone detoxification enzyme ZLHY-6 based on thermolabile segment analysis and disulfide bond prediction. J. Food Saf. Qual. 2022, 13, 7150–7157. [Google Scholar] [CrossRef]
- Liu, X.; Wang, Y.; Fang, X.; Tang, Y.; Wang, G.; Guo, Y.; Yuan, J.; Zhao, L. Characteristics of a Novel Zearalenone Lactone Hydrolase ZHRnZ and Its Thermostability Modification. Int. J. Mol. Sci. 2024, 25, 9665. [Google Scholar] [CrossRef] [PubMed]
- Wijma, H.J.; Furst, M.J.L.J.; Janssen, D.B. A Computational Library Design Protocol for Rapid Improvement of Protein Stability: FRESCO. Methods Mol. Biol. 2018, 1685, 69–85. [Google Scholar] [PubMed]
- Zhang, Z.; Xu, W.; Wu, H. Zearalenone-detoxifying lactonase: Identification, modification and application. Food Ferment. Ind. 2021, 47, 285–291. [Google Scholar]
- Ranjith, A. Metabolites and degradation pathways of microbial detoxification of aflatoxins: A review. Mycotoxin Res. 2023, 40, 71–83. [Google Scholar] [CrossRef]
- Chang, X.; Liu, H.; Sun, J.; Wang, J.; Zhao, C.; Zhang, W.; Zhang, J.; Sun, C. Zearalenone Removal from Corn Oil by an Enzymatic Strategy. Toxins 2020, 12, 117. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Xie, P.; Jin, J.; Jin, Q.; Wang, X. Removal of Zearalenone from Degummed Corn Oil by Hydrolase on a Batch-Refining Unit. Foods 2022, 11, 3795. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Gu, X.; Li, G.; Xu, R.; Gu, Z.; Chen, B.; Lu, H. Optimized expression of a zearalenone degrading enzyme (ZHD101) and the application in detoxification of corn steep liquor. J. Hebei Agric. Univ. 2020, 43, 26–34. [Google Scholar] [CrossRef]
- Wang, R.; Zhou, J.; Hu, X.; Xu, T.; Lv, H. Expression of ZHD101 in Kluyveromyces marxianus and the Mutation Breeding of High-Field Strains. J. Fudan Univ. (Nat. Sci.) 2017, 56, 431–437. [Google Scholar] [CrossRef]
- Wang, X.; Sun, Y.; Ruan, C.; Zhang, Q.; Zhang, Y.; Hu, X. Expression of Zearalenone Degrading Enzyme in Bacillus subilis and Its Effects on Reproductive Performance of Sows. Chin. J. Anim. Nutr. 2017, 29, 4019–4025. [Google Scholar]
- Song, J.; Peng, Z.; Ning, Y.; Refaie, A.; Wang, C.; Liu, M.; Sun, L. A novel zearalenone lactonase can effectively mitigate zearalenone-induced reproductive toxicity in gilts. Toxicon Off. J. Int. Soc. Toxinol. 2025, 255, 108257. [Google Scholar] [CrossRef] [PubMed]
- Jothyswarupha, K.A.; Venkataraman, S.; Rajendran, D.S.; Shri, S.S.S.; Sivaprakasam, S.; Yamini, T.; Karthik, P.; Kumar, V.V. Immobilized enzymes: Exploring its potential in food industry applications. Food Sci. Biotechnol. 2024, 34, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.; Jiang, J.; Xiong, K.; Chen, Y.; Yao, Y.; Liu, L.; Liu, H.; Li, X. Enzyme Engineering: Performance Optimization, Novel Sources, and Applications in the Food Industry. Foods 2024, 13, 3846. [Google Scholar] [CrossRef]
- Wang, H.; Sun, J.; Chang, X.; Liu, H.; Sun, C. Study on the embedding and stability of zearalenone degrading enzyme ZLHY6. Sci. Technol. Cereals Oils Foods 2018, 26, 63–67. [Google Scholar] [CrossRef]
- Zheng, Y.; Dou, J.; Sun, C.; Liu, H.; Wu, W. A novel modified montmorillonite-based calcium alginate microsphere immobilized ZLHY6 enzyme for removal of zearalenone in maize gluten meal. Int. J. Biol. Macromol. 2025, 307, 142168. [Google Scholar] [CrossRef] [PubMed]
- Bi, K. The Expression of A Recombined Zearalenone Degrading Enzyme. Master’s Thesis, Qilu University of Technology, Jinan China, June 2019. [Google Scholar]
- Fu, X.; Xu, M.; Li, T.; Li, Y.; Zhang, H.; Zhang, C. The Improved Expression and Stability of Zearalenone Lactonohydrolase from Escherichia coli BL21 (DE3). Appl. Biochem. Microbiol. 2021, 57, 79–85. [Google Scholar] [CrossRef]
- Mozhgan, R.; Ahmad, H.; Fabio, V.; Taha, A.; Tanvi, S.; Kumar, N.A.; Roberto, S.; Muhammad, B.; Iqbal, H.M. Industrial applications of immobilized nano-biocatalysts. Bioprocess Biosyst. Eng. 2021, 45, 1–20. [Google Scholar] [CrossRef]
- Wu, Y.; Xu, W.; Jiao, L.; Gu, W.; Du, D.; Hu, L.; Lin, Y.; Zhu, C. Nanobiocatalysis: A materials science road to biocatalysis. Chem. Soc. Rev. 2022, 51, 6948–6964. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Lu, T.; Dai, X.; Ding, P.; Xiong, Y.; Ge, J.; Li, X. Co-Immobilization of Enzymes and Metals on the Covalent-Organic Framework for the Efficient Removal of Mycotoxins. ACS Appl. Mater. Interfaces 2023, 15, 6859–6867. [Google Scholar] [CrossRef]
- Zhou, C.; He, N.; Lin, X.; Liu, H.; Lu, Z.; Zhang, G. Site-directed display of zearalenone lactonase on spilt-intein functionalized nanocarrier for green and efficient detoxification of zearalenone. Food Chem. 2024, 446, 138804. [Google Scholar] [CrossRef] [PubMed]
- Ayub, J.; Saeed, M.U.; Hussain, N.; Zulfiqar, I.; Mehmood, T.; Iqbal, H.M.N.; Bilal, M. Designing robust nano-biocatalysts using nanomaterials as multifunctional carriers-expanding the application scope of bio-enzymes. Top. Catal. 2023, 66, 625–648. [Google Scholar] [CrossRef]
- Dong, Z.; Tan, J.; Pinelo, M.; Zhang, H.; Wan, Y.; Luo, J. Engineering Mussel-Inspired Coating on Membranes for Green Enzyme Immobilization and Hyperstable Reuse. Ind. Eng. Chem. Res. 2022, 61, 5042–5053. [Google Scholar] [CrossRef]
- He, M.; Li, Y.; Pi, F.; Ji, J.; He, X.; Zhang, Y.; Sun, X. A novel detoxifying agent: Using rice husk carriers to immobilize zearalenone-degrading enzyme from Aspergillus niger FS10. Food Control 2016, 68, 271–279. [Google Scholar] [CrossRef]
- Zhao, C. Enzymatic Degradation of Zearalenone for Production of Zero Trans Fatty Acid Corn Oil. Ph.D. Thesis, Jiangnan University, Wuxi, China, June 2023. [Google Scholar]
- Sun, M.; Wang, M.; Li, Y.; Yuan, B.; Chen, C.; Wang, L.; Chen, H.; Sun, M. Application and research progress of artificial intelligence in enzyme engineering. Chem. Life 2025, 45, 252–264. [Google Scholar]

| Source Microorganism | Gene Name | Substrate | Expression Host | Optimum pH | Optimum Temperature/°C | Enzyme Activity Value |
|---|---|---|---|---|---|---|
| Clonostachys rosea IFO 7063 | ZHD101 | ZEN | S. pombe and E. coli | 9.0–10.0 | 37–45 | NR [30] |
| Cladophialophora bantiana | CLA | ZEN | E. coli BL21 | 7.0 | 40 | 114.8 U/mg a [38] |
| Exophiala aquamarina | EXO | ZEN | E. coli BL21 | 9.0 | 40 | 459.0 U/mg a [38] |
| Trichoderma aggressivum | TRI | ZEN | E. coli BL21 | 9.5 | 40 | 239.8 U/mg a [38] |
| Gliocladium spp. | ZHDR52, ZHDP83 | ZEN, α/β-ZEL α/β-ZAL | E. coli BL21 (DE3) | 9.0 | 45 | ZEN 196.11 U/mg a, 229.64 U/mg a >90% removal of α/β-ZEL and α/β-ZAL in 6 h [39] |
| Exophiala aquamarina CBS 119918 | ZHDAY3 | ZEN, α/β-ZEL α/β-ZAL | E. coli BL21 (DE3) | 9.5 | 40 | ZEN 157.5 U/mg a α-ZEL 79.6 U/mg a α-ZAL 115 U/mg a β-ZEL 71.7 U/mg a β-ZAL 53.0 U/mg a [29] |
| Aeromicrobium strain | ZenH | ZEN | E. coli | 7.0 | 55 | The specific activity of ZenH against ZEN is 7.05 U/mg a [40] |
| Trichoderma aggressivum | ZHD-P | ZEN | E. coli BL21 (DE3) | 7.5–9.0 | 45 | 191.94 U/mg a [41] |
| Clonostachys rosea strain GrZ7 | PR-ZHD | ZEN | E. coli Rosetta TM (DE3) | 8.5 | 30 | Complete degradation [42] |
| Fonsecaea multimorphosa CBS 102226 | ZHD11C | ZEN, α/β-ZEL α/β-ZAL | E. coli DH5α and BL21 (DE3) | NR | 45 | ZEN 55.8 U/mg a α-ZEL 21.6 U/mg a α-ZAL 37.3 U/mg a β-ZEL 9.3 U/mg a β-ZAL 6.3 U/mg a [43] |
| Rhodococcus erythropolis PFAD 8-1 | ZENA | ZEN | NR | 8.2 | 50–60 | NR [44] |
| Phialophora attae | ZHD11F | ZEN | E. coli BL21 (DE3) | 8.0 | 35 | 40.04 U/mg a [45] |
| Monosporascus sp. GIB2. | ZENM | ZEN, α/β-ZEL, α-ZAL | E. coli BL21 (DE3) and E. coli DH5α | 9.0 | 60 | ZEN 333 U/mg a α-ZEL 316 U/mg a α-ZAL 300 U/mg a β-ZEL 210 U/mg a [37] |
| Gliocladium roseum | S162P/S220R | ZEN | E. coli JM109 and E. coli BL21 (DE3) | 7.0 | 35 | 371 U/mg a 224 U/mg a [46] |
| Rhinocladiella mackenziei | ZHD518 | ZEN, α/β-ZEL α/β-ZAL | E. coli BL21 (DE3) | 8.0 | 40 | ZEN 207.0 U/mg a α-ZEL 23.0 U/mg a α-ZAL 119.8 U/mg a β-ZEL 64.7 U/mg a β-ZAL 66.5 U/mg a [47] |
| Rhinocladiella mackenziei CBS 650.93 | RmZHD | ZEN | E. coli BL21 (DE3) | 8.6 | 45 | 1.27 U/mg c [48] |
| Exophiala spinifera CBS 89,968 | ZHD_LD | ZEN | E. coli | 9.0 | 50 | 1.15 ± 0.04 U/mg c [49] |
| Glocladium roseum | ZENG | ZEN, α-ZEL α-ZAL | E. coli BL21 (DE3) | 7.0 | 38 | ZEN 315.0 U/mg a α-ZEL 187 U/mg a α-ZAL 117 U/mg a [50] |
| Glodophialophora bantiana | CbZHD | ZEN | E. coli BL21 (DE3) | 8.0 | 35 | 0.688 U/mg c [51] |
| Marasonina brannaea | mbZHD | ZEN | E. coli BL21 (DE3) | NR | NR | 200 U/mg d [52] |
| Neurospora crassa | ZENC | ZEN | P. pastoris | 8.0 | 45 | 530.4 U/mg a [36] |
| Glocladium roseum | ZHD101 | ZEN, α/β-ZEL | P. pastoris GS115 | 9.5 | 37 | ZEN 4976.5 U/mg α-ZEL1257.1 U/mg β-ZEL 780.9 U/mL (15 min) [53] |
| Phialophora americana | ZHD607 | ZEN | P. pastoris | 8.0 | 35 | 4940 U/mg b [35] |
| Bacillus subtilis YT-4 | ZENY | ZEN | E. coli DH5α and P. pastoris GS115 | 8.0 | 37 | 60% degradation in 6 h. 95% degradation in 36 h [54] |
| Glocladium roseum | zlhy-6 | ZEN | P. pastoris GS115 | NR | NR | 10.0 U/mL a [55] |
| Strain Name | Enzyme | Transformed Strategy | Optimization Method | Active Effect |
|---|---|---|---|---|
| Clonostachys rosea | ZHD101 | Directed evolution | Combination mutation (Screen out the optimal mutant V153H-V158F, zhd101.1) | Its specific activity is 1.1 times higher than that of the wild type, and it has better thermal stability and pH stability [63] |
| Clonostachys rosea | ZHD | Directed evolution | site-directed mutagenesis | The pKa values of the residues E126 and H242 catalyzed by ZHD enzyme were reduced [64] |
| Trichoderma aggressivum | ZHD-P | Directed evolution | The E. coli cell surface display system of ZHD-P and gene screening were constructed | Intracellular ZHD-P remained 100% active after incubation at 25–40 °C for 1 h. The surface displayed ZHD-P showed high activity against ZEN and remained 80% active after incubation at pH 5.0–11.0 for 12 h [41] |
| Clonostachys rosea | ZHD101 | Rational design | site-directed mutagenesis (There are 9 mutants, represented by S220R and S220W.) | The thermal melting temperature (Tm) of the 9 mutants increased by 0.4–5.6 °C. Among them, S220RandS220W showed the best thermal stability, with Tm increasing by 5.6 °Cand 4.0 °C, respectively. The thermal half-inactivation time at 45 °C was extended by 15.4 times and3.1 times, respectively. The relative enzyme activities were 70.6% and 57.3% of those of the wild type, respectively [65] |
| Gliocladium roseum | ZHD101 | Rational design | mutation | Under the conditions of pH 4.2 and 37 °C, the degrading enzyme activity of 8 mutant sites increased from 7.69 U/mg to 38.67 U/mg. After evolution, Km decreased from 283.61 μM to 75.33 μM [66] |
| Clonostachys rosea | ZHD101 | Rational design | site-directed mutagenesis (V153H) | The activity of V153H against ZEN remained unchanged, but its specific activity against α-ZOL increased by 3.7 times, the affinity for substrates decreased by 2.7 times, but the conversion rate increased by 5.2 times [67] |
| Gliocladium roseum | ZENG | Rational design -structure -based modification. | site-directed mutagenesis (H134F/S136F, H134I/S13 6I, H134L/S136L) | The three mutants retained 40% of their activity after being incubated at 48 °C for more than 7 min [50] |
| Phialophora macrospora | ZHD11A | Rational design | site-directed mutagenesis (I160Y-G242S) | The mutant can still maintain about 40% residual activity at 55 °C for 10 min. Compared with ZHD11A, the specific activity of I160Y-G242S is also increased by 2 times, from 220 U/mg to 450 U/mg [68] |
| Fonsecae monophora | Zhd11B | Rational design -structure -based modification. | site-directed mutagenesis (T158H) | The relative activity of the T158H mutant against α-ZOL increased by 1.3 times [69] |
| Phialophora americana | ZHD607 | Rational design | site-directed mutagenesis (ZHDM1 and I160Y) | The degradation activity of the mutant ZHDM1 and I160Y for ZEN was 2.9 times and3.4 times that of ZHD607, respectively [35] |
| Rhinocladiella mackenziei | Zhd518 | Rational design | site-directed mutagenesis (N156H) | The degradation efficiency of α-ZOL is 3.3 times that of the wild type [47] |
| Rhinocladiella mackenziei | RmZHD | Rational design | site-directed mutagenesis (V153H and Y160A) | The degradation efficiency of V153H for α-ZOL has increased by 3.17 times, and the hydrolysis activity of Y160A for α-ZOL has increased by 70% [70] |
| Rhodococcus erythropolis PFA D8-1 | ZENA | Rational design | site-directed mutagenesis (D264A, D264L, D264N) | The catalytic triplet was identified and defined as Ser-128-His303-Asp-153, and it had degradation activity [44] |
| Monosporascus sp. GIB2 | ZENM | Rational design | site-directed mutagenesis (G163S) | The catalytic activity of the mutant against α-ZOL (kcat/Km 0.223 min−1 μM−1) is higher than that against ZEN (kcat/Km 0.191 min−1 μM−1), and α-ZOL is its optimal substrate. The mutant can change the substrate specificity of lactone hydrolase [37] |
| Bacillus subtilis YT-4 | ZENY | Rational design | Site-directed mutagenesis (N∆11 and N5V) | The first 11 amino acids at the N-terminal were replaced by the first 13 amino acids in the N-terminal region of ZHD11C, especially the fifth residue n was replaced by V, and a 25% stability improvement was achieved at 45 °C [54] |
| Clonostachys rosea | ZHD101 | Rational design | introduce disulfide bonds (D143C/P18 1C) | The residual activity after treatment at 50 °C for 2 min is approximately twice that of the wild type [71] |
| Fonsecaea multimorphosa CBS 102226 | ZHD11C | Rational design | gene selection | After incubation at 45 °C for 1 h, it retained nearly 90% of its activity. After incubation at pH6.5 to 9.0 for 12 h, it still retained over 12% of its activity and was capable of hydrolyzing α-ZAL, α-ZOL, β-ZAL, and β-ZOL [43] |
| Fonsecae monophora | Zhd11B | Rational design -structure -based modification | Hat domain swap (V131-L172 replaces the corresponding area of Zhd518) | The activities against ZEN and α-ZAL and β-ZAL were increased by 1.5, 1.6 and 2.9 times, respectively [69] |
| Clonostachys rosea | ZHD101 | Computer-aided rational design-Computational design of pH-activity profiles for enzymes | site-directed mutagenesis (D157K and E171K) | M2 (D157K) and M9 (E171K) moderately enhanced the catalytic efficiency of ZHD101 under acidic conditions. The kcat/Km of the two mutants was 1.34 and 2.06 times that of the wild type, respectively [31] |
| Exophiala aquamarina CBS 119918 | ZHDAY3 | Computer-aided rational design | site-directed mutagenesis (N153H) | The hydrolytic activity increased from 115.1 ± 2.1 U/mg to 253.3 ± 4.3 U/mg [29] |
| Clonostachys rosea | ZHD101 | Computer-aided rational design-Virtual saturation mutation based on flexible regions | site-directed mutagenesis (N156F, S194T and T259F) | The enzyme activities of the three mutants were 95.8%, 131.6% and 169.0%, respectively, compared with the wild type. The Tm of the double mutant TIN156F/S194T and the triple mutant N156F/S194T/T259F increased by 6.7 °C and 6.1 °C, respectively [72] |
| Gliocladium roseum | ZENG | Computer-aided rational design-MD simulations | site-directed mutagenesis (S162P/S220R) | S162P/S220R mutant under 55 °C half-life (t1/2) is 36.8 times higher than wild-type enzyme, Tm significantly increased by 8.2 °C [46] |
| Glocladium roseum | ZLHY6 | Computer-aided rational design -Computer virtual saturation mutation | site-directed mutagenesis (H134 W) | After heat treatment at 45 °C for 20 min, the relative enzymatic activity of mutant H134W was 10 times that of the wild type, and it retained a certain activity against ZEN (41.33%) [73] |
| Glocladium roseum | ZLHY6 | Computer-aided rational design -Computer virtual saturation mutation | introduce disulfide bonds (Q45C/A253 C) | After heat treatment at 45 °C for 20 min, the relative enzymatic activities of Q45C/A253C were 3.1 times that of the wild type, respectively, and they retained a certain activity (41.33%) against ZEN [73] |
| Rosellinia necatrix | ZHRnZ | Computer-aided rational design | site-directed mutagenesis (E122R) | The catalytic efficiency of E122R is 1.3 times higher than that of the wild type [74] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Ren, X.; Xu, B.; Fan, L.; Guo, C.; Zhang, B.; Ning, M. Green Solutions for Food Safety: The Emerging Applications of Zearalenone-Degrading Enzymes. Foods 2025, 14, 3010. https://doi.org/10.3390/foods14173010
Zhang Y, Ren X, Xu B, Fan L, Guo C, Zhang B, Ning M. Green Solutions for Food Safety: The Emerging Applications of Zearalenone-Degrading Enzymes. Foods. 2025; 14(17):3010. https://doi.org/10.3390/foods14173010
Chicago/Turabian StyleZhang, Yawei, Xianfeng Ren, Baocheng Xu, Lixia Fan, Changying Guo, Bingchun Zhang, and Mingxiao Ning. 2025. "Green Solutions for Food Safety: The Emerging Applications of Zearalenone-Degrading Enzymes" Foods 14, no. 17: 3010. https://doi.org/10.3390/foods14173010
APA StyleZhang, Y., Ren, X., Xu, B., Fan, L., Guo, C., Zhang, B., & Ning, M. (2025). Green Solutions for Food Safety: The Emerging Applications of Zearalenone-Degrading Enzymes. Foods, 14(17), 3010. https://doi.org/10.3390/foods14173010





