Antimicrobial Peptides from Porcine Blood Cruor Hydrolysates as a Promising Source of Antifungal Activity
Abstract
1. Introduction
2. Materials and Methods
2.1. Peptide Sequences
2.2. Strain Collection and Mantainance
2.3. Antimicrobial Activities
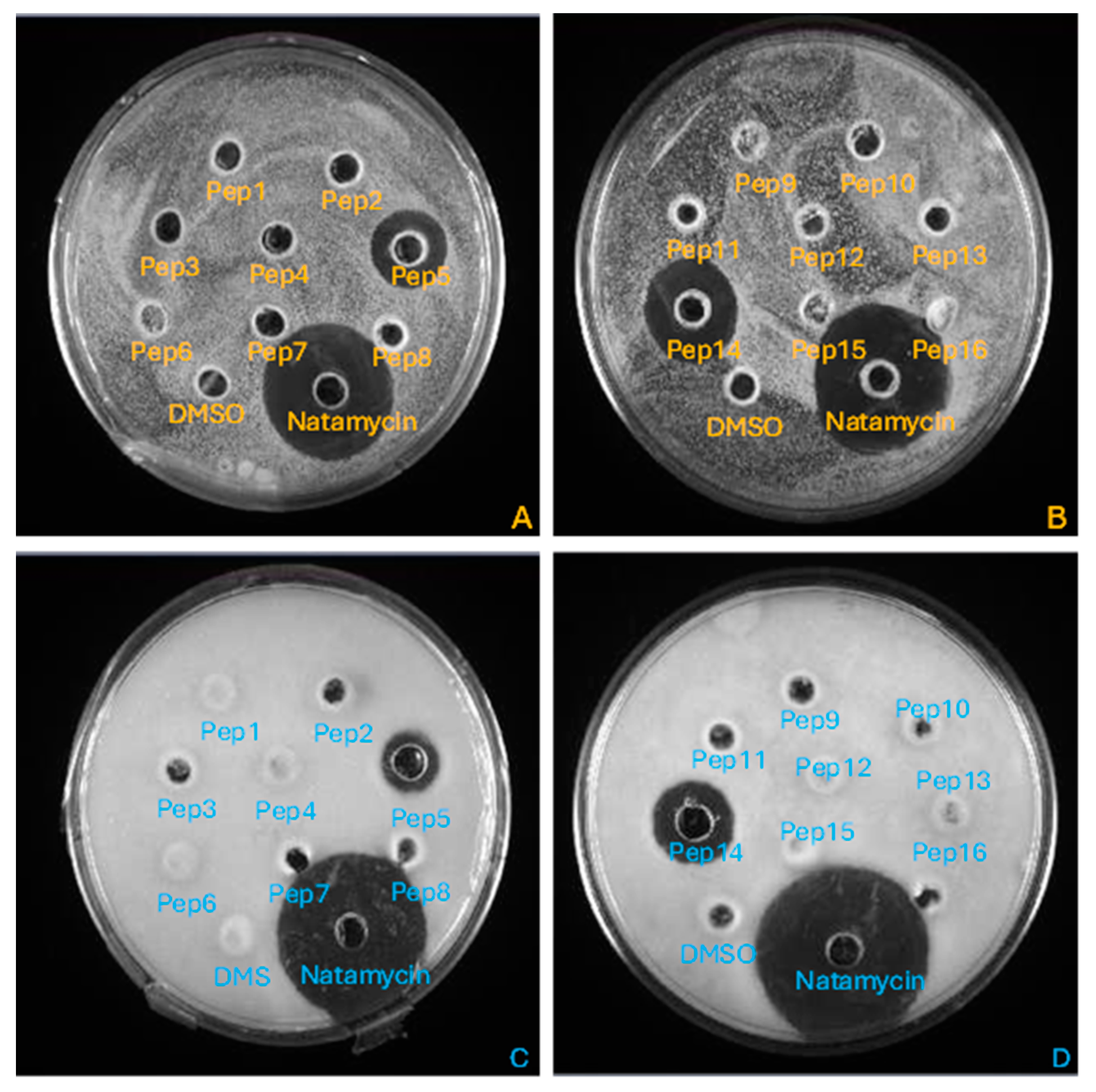
2.3.1. Agar Well Diffusion
2.3.2. Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal/Fungicidal Concentration (MBC/MFC)
2.3.3. Checkerboard Assay
3. Results
3.1. Antimicrobial Activity of the Synthetized Peptides
3.2. Synergistic Effect Between Pep4 and Pep5
3.3. Pep5 3D Structure
4. Discussion and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Góchez, D.; Raicek, M.; Pinto Ferreira, J.; Jeannin, M.; Moulin, G.; Erlacher-Vindel, E. OIE Annual Report on Antimicrobial Agents Intended for Use in Animals: Methods Used. Front. Vet. Sci. 2019, 6, 317. [Google Scholar] [CrossRef] [PubMed]
- Morrison, L.; Zembower, T.R. Antimicrobial resistance. Gastrointest. Endosc. Clin. N. Am. 2020, 30, 619–635. [Google Scholar] [CrossRef]
- Serna, C.; Gonzalez-Zorn, B. Antimicrobial resistance and One Health. Rev. Esp. Quim. 2022, 35 (Suppl. S3), 37–40. [Google Scholar] [CrossRef]
- Reygaert, W.C. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018, 3, 482. [Google Scholar] [CrossRef] [PubMed]
- Ben-Ami, R.; Kontoyiannis, D.P. Resistance to Antifungal Drugs. Infect. Dis. Clin. North. Am. 2021, 35, 279–311. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.C.; Alastruey-Izquierdo, A.; Berman, J.; Bicanic, T.; Bignell, E.M.; Bowyer, P.; Bromley, M.; Brüggemann, R.; Garber, G.; Cornely, O.A.; et al. Tackling the emerging threat of antifungal resistance to human health. Nat. Rev. Microbiol. 2022, 20, 557–571. [Google Scholar] [CrossRef]
- Revie, N.M.; Iyer, K.R.; Robbins, N.; Cowen, L.E. Antifungal drug resistance: Evolution, mechanisms and impact. Curr. Opin. Microbiol. 2018, 45, 70–76. [Google Scholar] [CrossRef]
- Lee, Y.; Robbins, N.; Cowen, L.E. Molecular mechanisms governing antifungal drug resistance. NPJ Antimicrob. Resist. 2023, 1, 5. [Google Scholar] [CrossRef]
- McEwen, S.A.; Collignon, P.J. Antimicrobial Resistance: A One Health Perspective. Microbiol. Spectr. 2018, 6, 521–547. [Google Scholar] [CrossRef] [PubMed]
- Cook, M.A.; Wright, G.D. The past, present, and future of antibiotics. Sci. Transl. Med. 2022, 14, eabo7793. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Sharma, S. Role of alternatives to antibiotics in mitigating the antimicrobial resistance crisis. Indian. J. Med. Res. 2022, 156, 464–477. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Li, J.; Yao, Z.; Li, M. A review on the alternatives to antibiotics and the treatment of antibiotic pollution: Current development and future prospects. Sci. Total Environ. 2024, 926, 171757. [Google Scholar] [CrossRef]
- Berger, S.; El Chazli, Y.; Babu, A.F.; Coste, A.T. Azole Resistance in Aspergillus fumigatus: A Consequence of Antifungal Use in Agriculture? Front. Microbiol. 2017, 8, 1024. [Google Scholar] [CrossRef] [PubMed]
- Thery, T.; Lynch, K.M.; Arendt, E.K. Natural Antifungal Peptides/Proteins as Model for Novel Food Preservatives. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1327–1360. [Google Scholar] [CrossRef]
- Qiu, M.; Wang, Y.; Sun, L.; Deng, Q.; Zhao, J. Fatty Acids and Oxylipins as Antifungal and Anti-Mycotoxin Agents in Food: A Review. Toxins 2021, 13, 852. [Google Scholar] [CrossRef]
- Shori, A.B.; Albloushi, S. Antifungal Activity of Lactobacillus plantarum and Sage Extract on Aspergillus Fumigatus in Yogurt. Am. J. Biomed. Life Sci. 2018, 6, 37–42. [Google Scholar] [CrossRef]
- Luo, Y.; Song, Y. Mechanism of Antimicrobial Peptides: Antimicrobial, Anti-Inflammatory and Antibiofilm Activities. Int. J. Mol. Sci. 2021, 22, 11401. [Google Scholar] [CrossRef] [PubMed]
- Bin Hafeez, A.; Jiang, X.; Bergen, P.J.; Zhu, Y. Antimicrobial Peptides: An Update on Classifications and Databases. Int. J. Mol. Sci. 2021, 22, 11691. [Google Scholar] [CrossRef] [PubMed]
- Boparai, J.K.; Sharma, P.K. Mini Review on Antimicrobial Peptides, Sources, Mechanism and Recent Applications. Protein Pept. Lett. 2020, 27, 4–16. [Google Scholar] [CrossRef]
- Zhang, R.; Xu, L.; Dong, C. Antimicrobial Peptides: An Overview of their Structure, Function and Mechanism of Action. Protein Pept. Lett. 2022, 29, 641–650. [Google Scholar] [CrossRef]
- Erdem Büyükkiraz, M.; Kesmen, Z. Antimicrobial peptides (AMPs): A promising class of antimicrobial compounds. J. Appl. Microbiol. 2022, 132, 1573–1596. [Google Scholar] [CrossRef]
- Cournoyer, A.; Thibodeau, J.; Ben Said, L.; Sanchez-Reinoso, Z.; Mikhaylin, S.; Fliss, I.; Bazinet, L. How Discoloration of Porcine Cruor Hydrolysate Allowed the Identification of New Antifungal Peptides. Foods 2022, 11, 4035. [Google Scholar] [CrossRef]
- Toldrà, M.; Lynch, S.A.; Couture, R.; Álvarez, C. Blood proteins as functional ingredients. In Sustainable Meat Production and Processing; Academic Press: Cambridge, MA, USA, 2019; pp. 85–101. [Google Scholar] [CrossRef]
- Bah, C.S.; Bekhit, A.E.D.A.; Carne, A.; McConnell, M.A. Slaughterhouse blood: An emerging source of bioactive compounds. Compr. Rev. Food Sci. Food Saf. 2013, 12, 314–331. [Google Scholar] [CrossRef]
- Zouari, O.; Przybylski, R.; Hannioui, M.; Sion, L.; Dhulster, P.; Nedjar-Arroume, N. High added-value co-product: The porcine cruor is an attractive source of active peptides. J. Nutr. Health Food Sci. 2020, 7, 1–9. [Google Scholar] [CrossRef]
- Hedhili, K.; Dimitrov, K.; Vauchel, P.; Sila, A.; Chataigné, G.; Dhulster, P.; Nedjar, N. Valorization of cruor slaughterhouse by-product by enzymatic hydrolysis for the production of antibacterial peptides: Focus on α 1-32 family peptides mechanism and kinetics modeling. Bioprocess. Biosyst. Eng. 2015, 38, 1867–1877. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, V.T.; Karelin, A.A.; Philippova, M.M.; Nazimov, I.V.; Pletnev, V.Z. Hemoglobin as a source of endogenous bioactive peptides: The concept of tissue-specific peptide pool. Biopolymers 1997, 43, 171–188. [Google Scholar] [CrossRef]
- Outman, A.; Deracinois, B.; Flahaut, C.; Diab, M.A.; Dhaouefi, J.; Gressier, B.; Eto, B.; Nedjar, N. Comparison of the Bioactive Properties of Human and Bovine Hemoglobin Hydrolysates Obtained by Enzymatic Hydrolysis: Antimicrobial and Antioxidant Potential of the Active Peptide α137-141. Int. J. Mol. Sci. 2023, 24, 13055. [Google Scholar] [CrossRef]
- Cournoyer, A.; Daigle, G.; Thibodeau, J.; Perreault, V.; Bazinet, L. Effects of applied current modes on the migration selectivity of peptides from a porcine cruor hydrolysate during electrodialysis with ultrafiltration membrane. Sep. Purif. Technol. 2024, 338, 126280. [Google Scholar] [CrossRef]
- Cournoyer, A.; Bazinet, M.; Clément, J.-P.; Plante, P.-L.; Fliss, I.; Bazinet, L. How peptide migration and fraction bioactivity are modulated by applied electrical current conditions during electromembrane process separation: A comprehensive machine learning-based peptidomic approach. Food Res. Int. 2024, in press. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.E.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server; Humana Press: Clifton, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Sanchez-Reinoso, Z.; Todeschini, S.; Thibodeau, J.; Ben Said, L.; Fliss, I.; Bazinet, L.; Mikhaylin, S. Biocontrol Strategy of Listeria monocytogenes in Ready-to-Eat Pork Cooked Ham Using Peptic Hydrolysates of Porcine Hemoglobin. Foods 2024, 13, 2394. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed]
- Pirtskhalava, M.; Amstrong, A.A.; Grigolava, M.; Chubinidze, M.; Alimbarashvili, E.; Vishnepolsky, B.; Gabrielian, A.; Rosenthal, A.; Hurt, D.E.; Tartakovsky, M. DBAASP v3: Database of antimicrobial/cytotoxic activity and structure of peptides as a resource for development of new therapeutics. Nucleic Acids Res. 2021, 49, D288–D297. [Google Scholar] [CrossRef]
- Lai, Z.; Yuan, X.; Chen, W.; Chen, H.; Li, B.; Bi, Z.; Lyu, Y.; Shan, A. Design of Proteolytic-Resistant Antifungal Peptides by Utilizing Minimum d-Amino Acid Ratios. Med. Chem. 2024, 67, 10891–10905. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res 2022, 50, W276–W279. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Zhang, C.; Li, Y.; Pearce, R.; Bell, E.W.; Zhang, Y. Folding non-homologous proteins by coupling deep-learning contact maps with I-TASSER assembly simulations. Cell Rep. Methods 2021, 1, 100014. [Google Scholar] [CrossRef]
- Zhou, X.; Zheng, W.; Li, Y.; Pearce, R.; Zhang, C.; Bell, E.W.; Zhang, G.; Zhang, Y. I-TASSER-MTD: A deep-learning-based platform for multi-domain protein structure and function prediction. Nat. Protoc. 2022, 17, 2326–2353. [Google Scholar] [CrossRef] [PubMed]
- Hernández, A.; Pérez-Nevado, F.; Ruiz-Moyano, S.; Serradilla, M.J.; Villalobos, M.C.; Martín, A.; Córdoba, M.G. Spoilage yeasts: What are the sources of contamination of foods and beverages? Int. J. Food Microbiol. 2018, 286, 98–110. [Google Scholar] [CrossRef]
- García-Vela, S.; Guay, L.D.; Rahman, M.R.T.; Biron, E.; Torres, C.; Fliss, I. Antimicrobial Activity of Synthetic Enterocins A, B, P, SEK4, and L50, Alone and in Combinations, Against Clostridium perfringens. Int. J. Mol. Sci. 2024, 25, 1597. [Google Scholar] [CrossRef]
- Soltani, S.; Biron, E.; Ben Said, L.; Subirade, M.; Fliss, I. Bacteriocin-Based Synergetic Consortia: A Promising Strategy to Enhance Antimicrobial Activity and Broaden the Spectrum of Inhibition. Microbiol. Spectr. 2022, 10, e0040621. [Google Scholar] [CrossRef] [PubMed]
- Borselli, D.; Lieutaud, A.; Thefenne, H.; Garnotel, E.; Pagès, J.M.; Brunel, J.M.; Bolla, J.M. Polyamino-Isoprenic Derivatives Block Intrinsic Resistance of P. aeruginosa to Doxycycline and Chloramphenicol In Vitro. PLoS ONE 2016, 11, e0154490. [Google Scholar] [CrossRef] [PubMed]
- Przybylski, R.; Firdaous, L.; Châtaigné, G.; Dhulster, P.; Nedjar, N. Production of an antimicrobial peptide derived from slaughterhouse by-product and its potential application on meat as preservative. Food Chem. 2016, 211, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Rizzello, C.G.; Cassone, A.; Coda, R.; Gobbetti, M. Antifungal activity of sourdough fermented wheat germ used as an ingredient for bread making. Food Chem. 2011, 127, 952–959. [Google Scholar] [CrossRef]
- Venegas-Ortega, M.G.; Flores-Gallegos, A.C.; Aguilar, C.N.; Rodríguez-Herrera, R.; Martínez-Hernández, J.L.; Nevárez-Moorillón, G.V. Multi-Functional Potential of Presumptive Lactic Acid Bacteria Isolated from Chihuahua Cheese. Foods 2020, 9, 276. [Google Scholar] [CrossRef]
- Gasu, E.N.; Ahor, H.S.; Borquaye, L.S. Peptide Extract from Olivancillaria hiatula Exhibits Broad-Spectrum Antibacterial Activity. Biomed. Res. Int. 2018, 2018, 6010572. [Google Scholar] [CrossRef] [PubMed]
- Nedjar-Arroume, N.; Dubois-Delval, V.; Adje, E.Y.; Traisnel, J.; Krier, F.; Mary, P.; Kouach, M.; Briand, G.; Guillochon, D. Bovine hemoglobin: An attractive source of antibacterial peptides. Peptides 2008, 29, 969–977. [Google Scholar] [CrossRef] [PubMed]
- Abou-Diab, M.; Thibodeau, J.; Fliss, I.; Dhulster, P.; Bazinet, L.; Nedjar, N. Production of Demineralized Antibacterial, Antifungal and Antioxidant Peptides from Bovine Hemoglobin Using an Optimized Multiple-Step System: Electrodialysis with Bipolar Membrane. Membranes 2022, 12, 512. [Google Scholar] [CrossRef] [PubMed]
- Outman, A.; Deracinois, B.; Flahaut, C.; Diab, M.A.; Gressier, B.; Eto, B.; Nedjar, N. Potential of Human Hemoglobin as a Source of Bioactive Peptides: Comparative Study of Enzymatic Hydrolysis with Bovine Hemoglobin and the Production of Active Peptide α137-141. Int. J. Mol. Sci. 2023, 24, 11921. [Google Scholar] [CrossRef] [PubMed]
- Odeyemi, O.A.; Alegbeleye, O.O.; Strateva, M.; Stratev, D. Understanding spoilage microbial community and spoilage mechanisms in foods of animal origin. Compr. Rev. Food Sci. Food Saf. 2020, 19, 311–331. [Google Scholar] [CrossRef] [PubMed]
- Axel, C.; Zannini, E.; Arendt, E.K. Mold spoilage of bread and its biopreservation: A review of current strategies for bread shelf life extension. Crit. Rev. Food Sci. Nutr. 2017, 57, 3528–3542. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.M.; Yap, Y.Y.; Cheong, J.W.; Ng, F.M.; Lau, Q.Y.; Barkham, T.; Teo, J.W.; Hill, J.; Chia, C.S. Antifungal peptides: A potential new class of antifungals for treating vulvovaginal candidiasis caused by fluconazole-resistant Candida albicans. J. Pept. Sci. 2017, 23, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Raman, N.; Lee, M.R.; Lynn, D.M.; Palecek, S.P. Antifungal Activity of 14-Helical β-Peptides against Planktonic Cells and Biofilms of Candida Species. Pharmaceuticals 2015, 8, 483–503. [Google Scholar] [CrossRef]
- Schmidtchen, A.; Pasupuleti, M.; Malmsten, M. Effect of hydrophobic modifications in antimicrobial peptides. Adv. Colloid Interface Sci. 2014, 205, 265–274. [Google Scholar] [CrossRef]
- Le Huy, B.; Phuong, H.B.T.; Thanh, B.N.T.; Van, Q.T.; Dinh, H.V.; Xuan, H.L. Influence of hydrophobicity on the antimicrobial activity of helical antimicrobial peptides: A study focusing on three mastoparans. Mol. Divers. 2024. [Google Scholar] [CrossRef] [PubMed]
- Hollmann, A.; Martínez, M.; Noguera, M.E.; Augusto, M.T.; Disalvo, A.; Santos, N.C.; Semorile, L.; Maffía, P.C. Role of amphipathicity and hydrophobicity in the balance between hemolysis and peptide-membrane interactions of three related antimicrobial peptides. Colloids Surf. B Biointerfaces 2016, 141, 528–536. [Google Scholar] [CrossRef]
- Decker, A.P.; Mechesso, A.F.; Wang, G. Expanding the Landscape of Amino Acid-Rich Antimicrobial Peptides: Definition, Deployment in Nature, Implications for Peptide Design and Therapeutic Potential. Int. J. Mol. Sci. 2022, 23, 12874. [Google Scholar] [CrossRef]
- Dong, N.; Ma, Q.; Shan, A.; Lv, Y.; Hu, W.; Gu, Y.; Li, Y. Strand length-dependent antimicrobial activity and membrane-active mechanism of arginine- and valine-rich β-hairpin-like antimicrobial peptides. Antimicrob. Agents Chemother. 2012, 56, 2994–3003. [Google Scholar] [CrossRef]
- Koehbach, J.; Craik, D.J. The Vast Structural Diversity of Antimicrobial Peptides. Trends Pharmacol. Sci. 2019, 40, 517–528. [Google Scholar] [CrossRef]
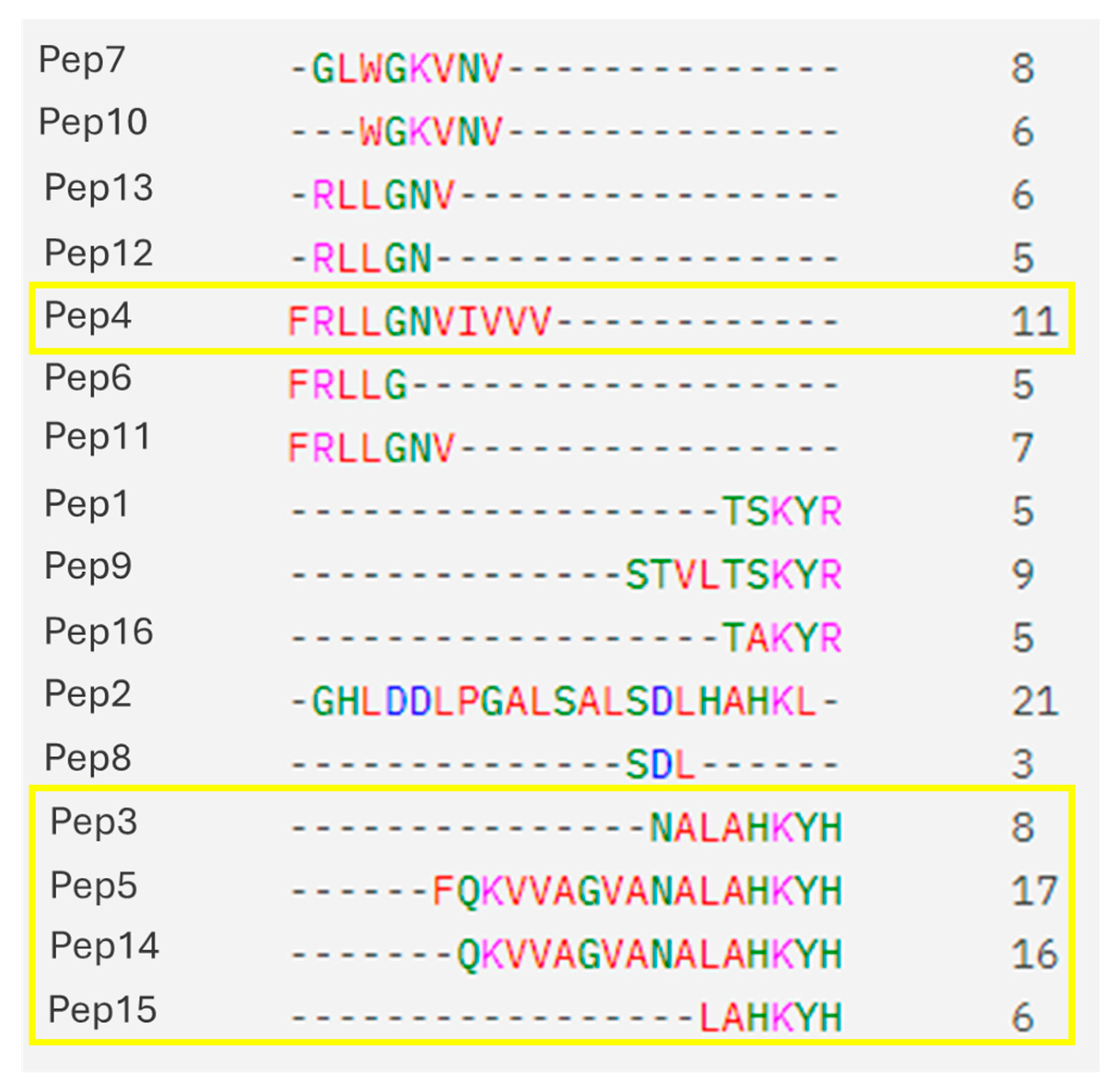

| Name | Sequence | Length | Molecular Weight (g/mol) | Isoelectric Point | GRAVY |
|---|---|---|---|---|---|
| Pep1 | TSKYR | 5 | 653.74 | 9.99 | −2.24 |
| Pep2 | GHLDDLPGALSALSDLHAHKL | 21 | 2180.45 | 5.71 | 0.01 |
| Pep3 | NALAHKYH | 8 | 953.07 | 8.61 | −0.96 |
| Pep4 | FRLLGNVIVVV | 11 | 1228.54 | 9.75 | 2.12 |
| Pep5 | FQKVVAGVANALAHKYH | 17 | 1853.16 | 9.70 | 0.20 |
| Pep6 | FRLLG | 5 | 604.75 | 9.75 | 1.10 |
| Pep7 | GLWGKVNV | 8 | 872.03 | 8.75 | 0.39 |
| Pep8 | SDL | 3 | 333.341 | 4.05 | −0.17 |
| Pep9 | STVLTSKYR | 9 | 1054.21 | 9.99 | −0.52 |
| Pep10 | WGKVNV | 6 | 701.82 | 8.75 | −0.05 |
| Pep11 | FRLLGNV | 7 | 817.99 | 9.75 | 0.88 |
| Pep12 | RLLGN | 5 | 571.68 | 9.75 | −0.16 |
| Pep13 | RLLGNV | 6 | 670.81 | 9.75 | 0.56 |
| Pep14 | QKVVAGVANALAHKYH | 16 | 1705.98 | 9.70 | 0.04 |
| Pep15 | LAHKYH | 6 | 767.89 | 8.61 | −1.00 |
| Pep16 | TAKYR | 5 | 637.74 | 9.99 | −1.72 |
| Name | Activity Against R. mucilaginosa | Activity Against Paecilomyces spp. | ||||
|---|---|---|---|---|---|---|
| MIC | MFC | MFC/MIC | MIC | MFC | MFC/MIC | |
| Pep1 | NI | NI | ND | NI | NI | ND |
| Pep2 | NI | NI | ND | NI | NI | ND |
| Pep3 | 0.625 ± 0.0 | 0.625 ± 0.0 | 1 | 0.625 ± 0.0 | 1.25 ± 0.0 | 2 |
| Pep4 | 0.375 ± 0.0 | NI | ND | 0.625 ± 0.0 | 0.625 ± 0.0 | 1 |
| Pep5 | 0.039 ± 0.0 | 0.078 ± 0.0 | 2 | 0.009 ± 0.0 | 0.009 ± 0.0 | 1 |
| Pep6 | NI | NI | ND | NI | NI | ND |
| Pep7 | NI | NI | ND | NI | NI | ND |
| Pep8 | NI | NI | ND | NI | NI | ND |
| Pep9 | NI | NI | ND | NI | NI | ND |
| Pep10 | NI | NI | ND | NI | NI | ND |
| Pep11 | NI | NI | ND | NI | NI | ND |
| Pep12 | NI | NI | ND | NI | NI | ND |
| Pep13 | NI | NI | ND | NI | NI | ND |
| Pep14 | 0.078 ± 0.0 | 0.078 ± 0.0 | 1 | 0.078 ± 0.0 | 0.078 ± 0.0 | 1 |
| Pep15 | 1.25 ± 0.0 | 1.25 ± 0.0 | 1 | 1.25 ± 0.0 | NI | ND |
| Pep16 | NI | NI | ND | NI | NI | ND |
| Indicator Fungal Strains | Values | Pep4 | Pep5 |
|---|---|---|---|
| Candida guilliermondii | MIC | NI | 0.312 ± 0.0 |
| MFC | NI | 0.625 ± 0.0 | |
| MFC/MIC | ND | 2 | |
| Candida parasilopsis | MIC | NI | NI |
| MFC | NI | NI | |
| MFC/MIC | ND | ND | |
| Debaryomyces hansenii | MIC | 0.312 ± 0.0 | 0.156 ± 0.0 |
| MFC | NI | 0.156 ± 0.0 | |
| MFC/MIC | ND | 1 | |
| Aspergillus versicolor | MIC | 0.312 ± 0.0 | 0.039 ± 0.0 |
| MFC | NI | 0.078 ± 0.0 | |
| MFC/MIC | ND | 2 | |
| Aspergillus niger | MIC | NI | NI |
| MFC | NI | NI | |
| MFC/MIC | ND | ND | |
| Penicillium commune | MIC | NI | 0.312 ± 0.0 |
| MFC | NI | 0.312 ± 0.0 | |
| MFC/MIC | ND | 1 | |
| Penicillium chrysogenum | MIC | 0.156 ± 0.0 | 0.312 ± 0.0 |
| MFC | NI | 0.156 ± 0.0 | |
| MFC/MIC | ND | 2 | |
| Eurotium rubrum | MIC | 0.312 ± 0.0 | 0.039 ± 0.0 |
| MFC | NI | 0.078 ± 0.0 | |
| MFC/MIC | ND | 2 | |
| Mucor racemosus | MIC | NI | 0.312 ± 0.0 |
| MFC | NI | 0.312 ± 0.0 | |
| MFC/MIC | ND | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Vela, S.; Cournoyer, A.; Sánchez-Reinoso, Z.; Bazinet, L. Antimicrobial Peptides from Porcine Blood Cruor Hydrolysates as a Promising Source of Antifungal Activity. Foods 2025, 14, 8. https://doi.org/10.3390/foods14010008
García-Vela S, Cournoyer A, Sánchez-Reinoso Z, Bazinet L. Antimicrobial Peptides from Porcine Blood Cruor Hydrolysates as a Promising Source of Antifungal Activity. Foods. 2025; 14(1):8. https://doi.org/10.3390/foods14010008
Chicago/Turabian StyleGarcía-Vela, Sara, Aurore Cournoyer, Zain Sánchez-Reinoso, and Laurent Bazinet. 2025. "Antimicrobial Peptides from Porcine Blood Cruor Hydrolysates as a Promising Source of Antifungal Activity" Foods 14, no. 1: 8. https://doi.org/10.3390/foods14010008
APA StyleGarcía-Vela, S., Cournoyer, A., Sánchez-Reinoso, Z., & Bazinet, L. (2025). Antimicrobial Peptides from Porcine Blood Cruor Hydrolysates as a Promising Source of Antifungal Activity. Foods, 14(1), 8. https://doi.org/10.3390/foods14010008







