Metabolomics on Apple (Malus domestica) Cuticle—Search for Authenticity Markers
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Chemicals
2.3. Methods
2.3.1. Sample Preparation
2.3.2. UHPLC-HRMS/MS Non-Target Screening
2.3.3. Data Processing
2.3.4. Statistical Analysis
2.3.5. Marker Identification
3. Results and Discussion
3.1. Selection of Extraction Solvent/Mixture
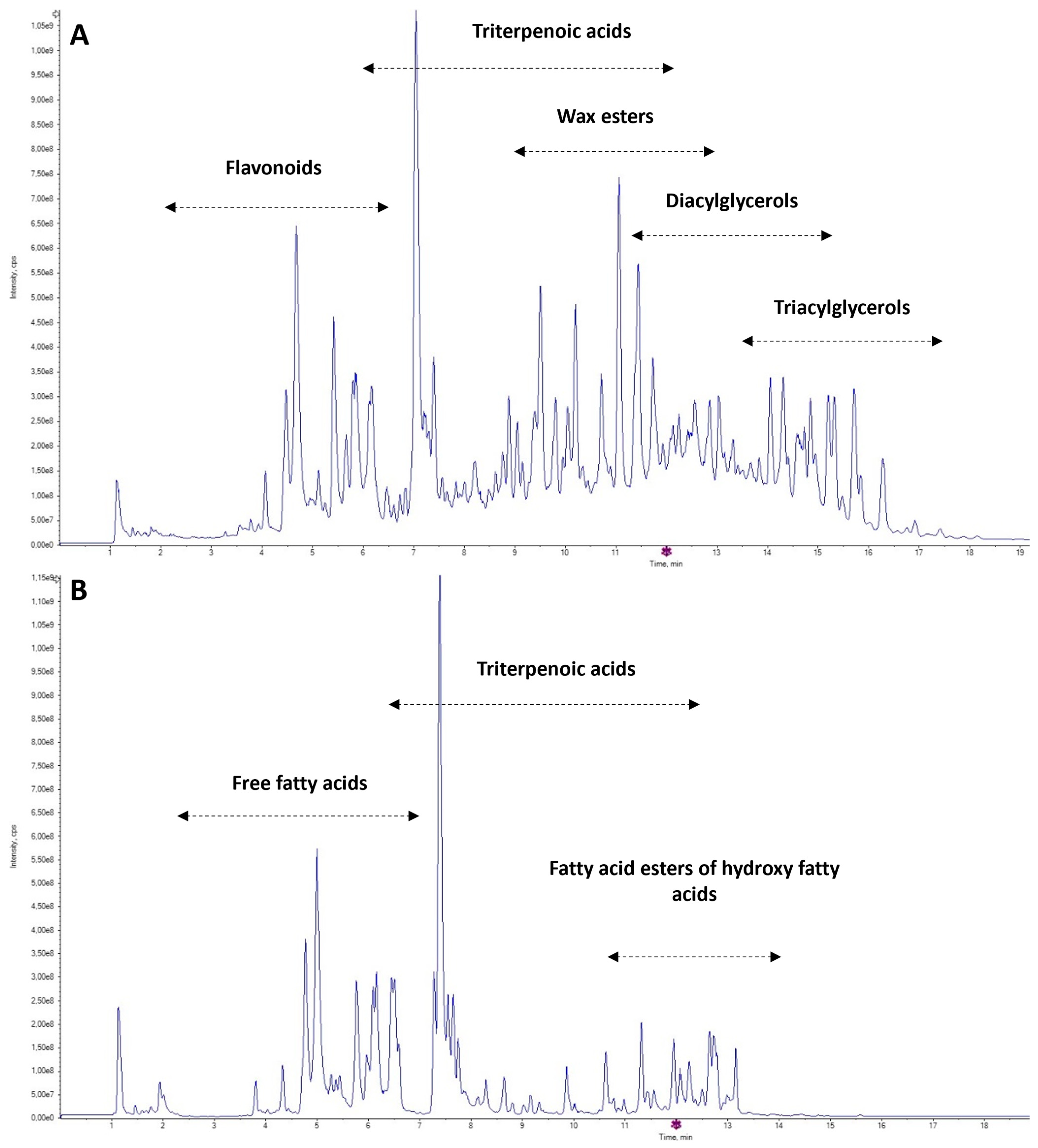
3.2. UHPLC-HRMS/MS Analysis
3.3. Chemometric Analysis
3.3.1. Data Overview
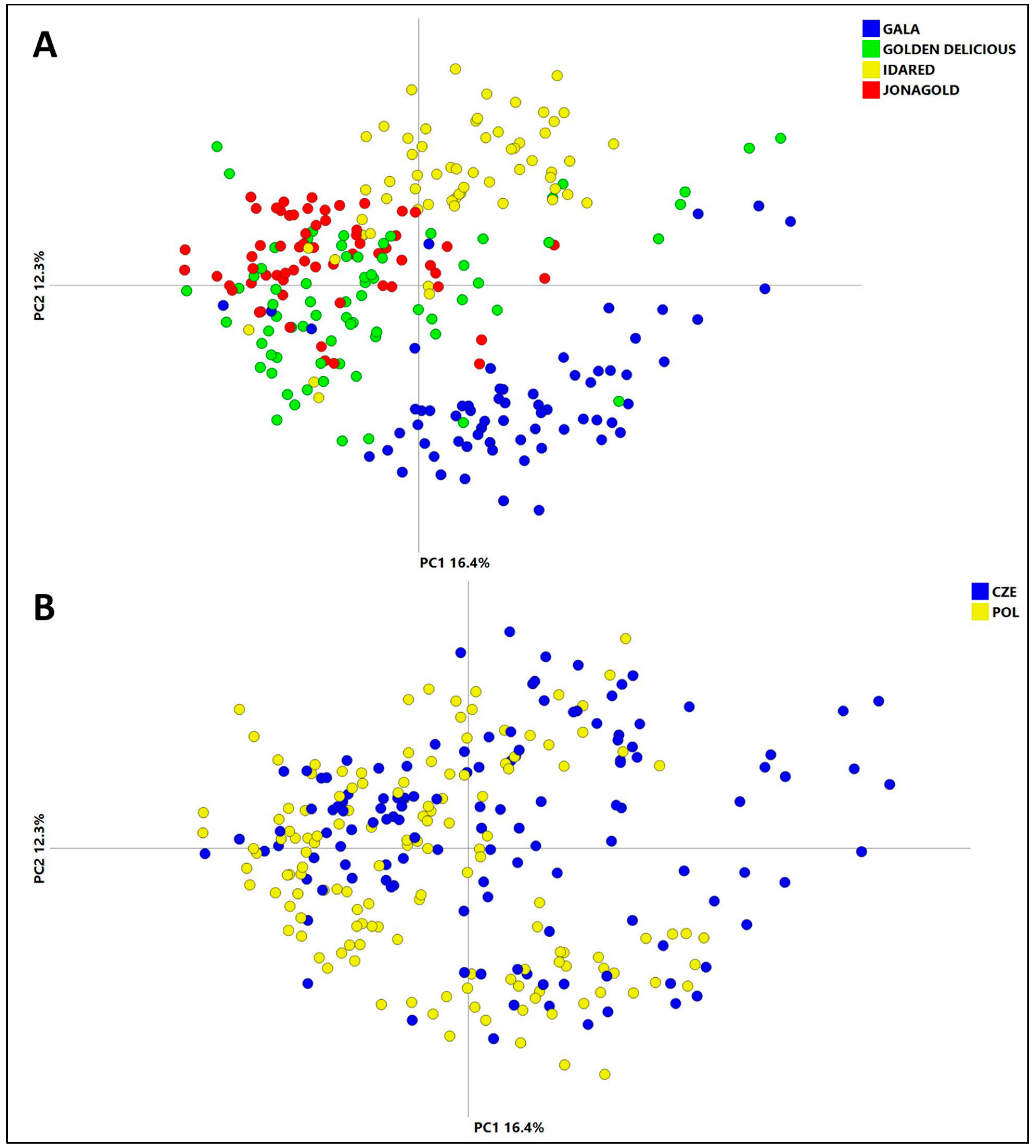
3.3.2. Apple Cultivars Classification
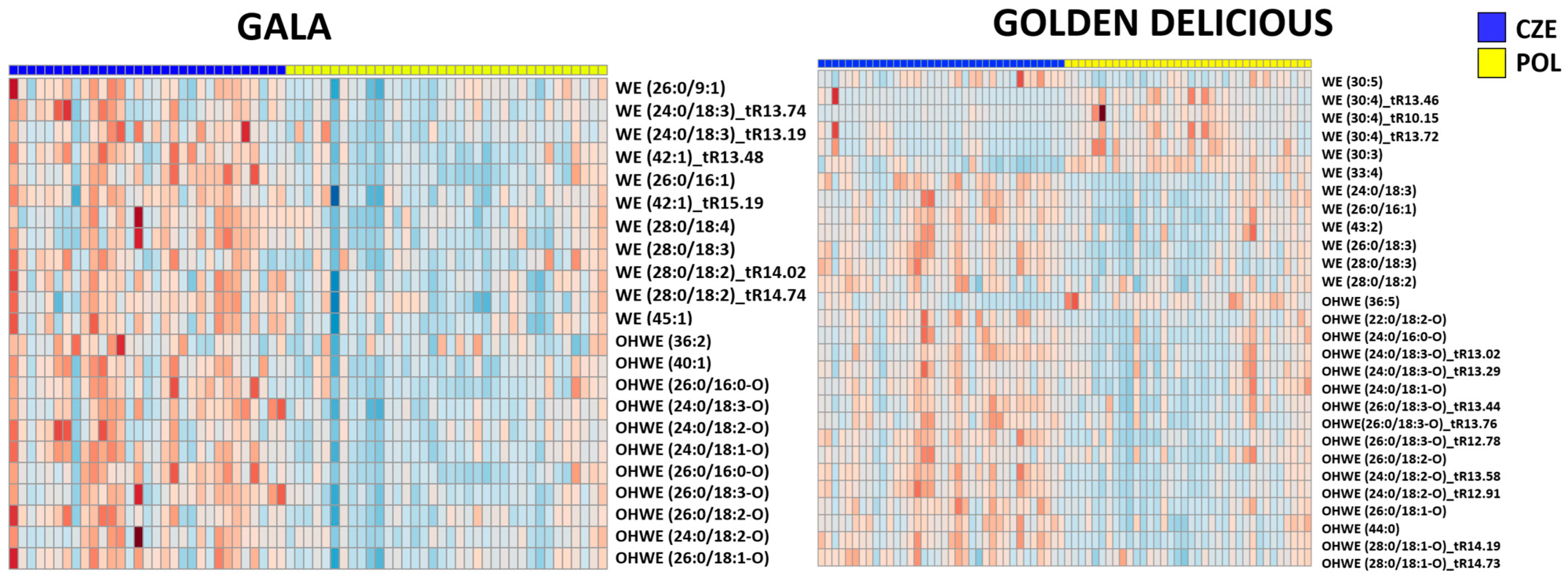
3.3.3. Classification of Apple Geographical Origin
4. Conclusions
- PCA showed a more pronounced cultivar impact on the metabolites occurring in the apple cuticle compared to that of geographical origin.
- The created PLS-DA models enabled reliable apple cultivar classification; 13 markers encompassing mainly waxes and triterpenoids were identified,
- The created OPLS-DA models enabled the safe classification of geographical origins of “Gala”, “Golden Delicious” and “Idared” cultivars; however, for “Jonagold”, it was unsuccessful.
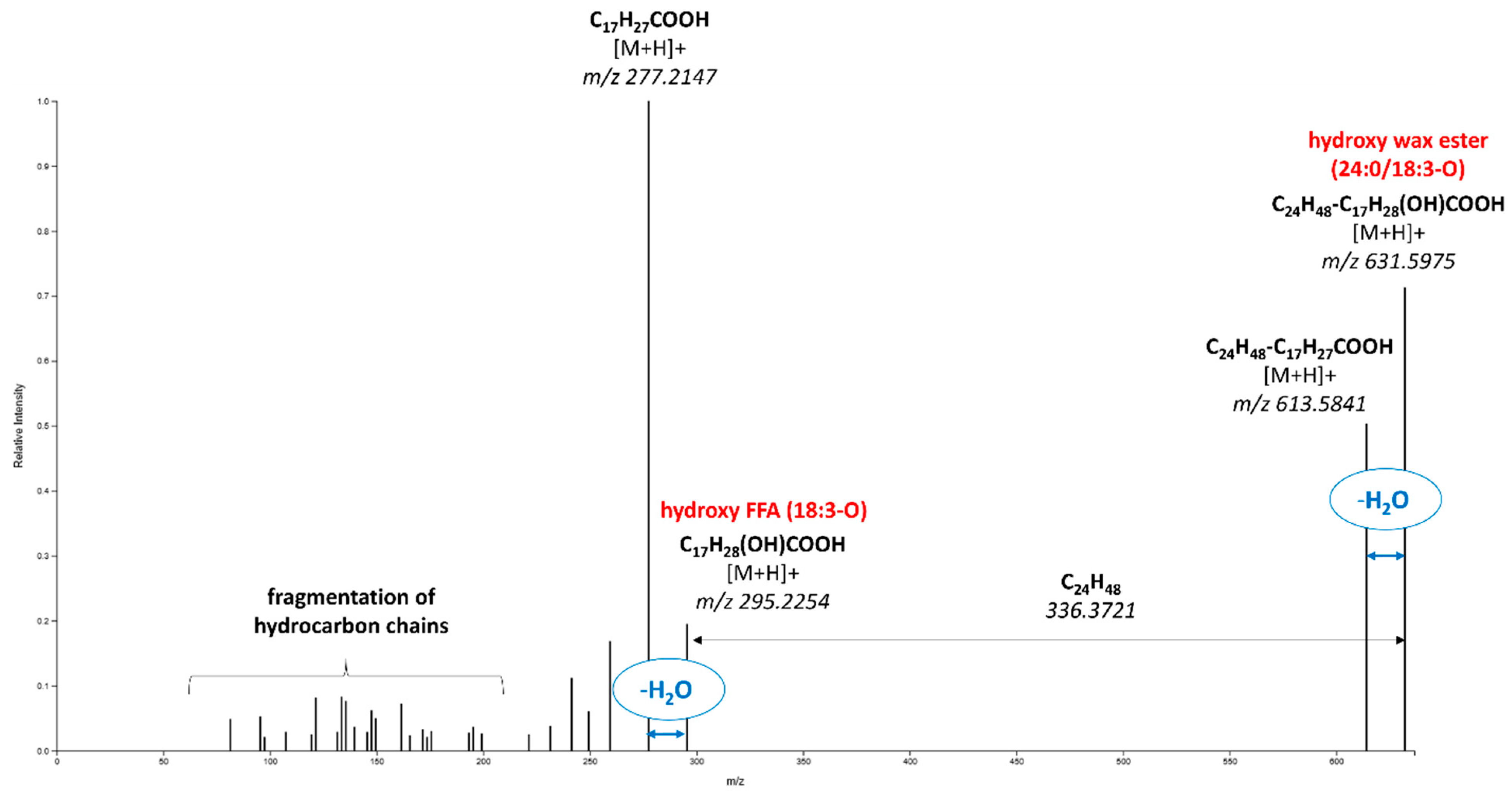
- Wax esters, including those with bound hydroxy fatty acids (reported for the first time in apple cuticular wax), represented a significant group of identified markers, the amount of which in “Golden Delicious” and “Gala” cultivars was higher (upregulated) in samples from the Czech Republic compared those from Poland.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shanbandeh, M. Apple Production Worldwide in 2022. Available online: https://www.statista.com/statistics/237605/production-of-apples-worldwide-by-region-2007/ (accessed on 29 February 2024).
- Bat, K.; Vidrih, R.; Nečemer, M.; Mozetič Vodopivec, B.; Mulič, I.; Kump, P.; Ogrinc, N. Characterization of Slovenian apples with respect to their botanical and geographical origin and agricultural production practice. Food Technol. Biotechnol. 2012, 50, 107–116. [Google Scholar]
- Wu, H.; Yue, T.; Yuan, Y. Authenticity Tracing of Apples According to Variety and Geographical Origin Based on Electronic Nose and Electronic Tongue. Food Anal. Methods 2018, 11, 522–532. [Google Scholar] [CrossRef]
- Li, C.; Li, L.; Wu, Y.; Lu, M.; Yang, Y.; Li, L. Apple Variety Identification Using Near-Infrared Spectroscopy. J. Spectrosc. 2018, 2018, e6935197. [Google Scholar] [CrossRef]
- Bian, H.; Sheng, L.; Yao, H.; Ji, R.; Yu, Y.; Chen, R.; Wei, D.; Han, Y. Application of fluorescence spectroscopy in classifying apple juice according to the variety. Optik 2021, 231, 166361. [Google Scholar] [CrossRef]
- Guo, J.; Yue, T.; Yuan, Y. Feature selection and recognition from nonspecific volatile profiles for discrimination of apple juices according to variety and geographical origin. J. Food Sci. 2012, 77, C1090–C1096. [Google Scholar] [CrossRef] [PubMed]
- Medina, S.; Perestrelo, R.; Santos, R.; Pereira, R.; Câmara, J.S. Differential volatile organic compounds signatures of apple juices from Madeira Island according to variety and geographical origin. Microchem. J. 2019, 150, 104094. [Google Scholar] [CrossRef]
- Mimmo, T.; Camin, F.; Bontempo, L.; Capici, C.; Tagliavini, M.; Cesco, S.; Scampicchio, M. Traceability of different apple varieties by multivariate analysis of isotope ratio mass spectrometry data. Rapid Commun. Mass Spectrom. 2015, 29, 1984–1990. [Google Scholar] [CrossRef] [PubMed]
- Brombin, V.; Mistri, E.; Bianchini, G. Multi stable isotope ratio analysis for the traceability of northern Italian apples. Food Chem. X 2022, 16, 100514. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhao, Y.; Mu, J.; Zhang, J.; Zhang, A. Determination of geographical origin of concentrated apple juice through analysis of stable isotopic and mineral elemental fingerprints: Preliminary results. J. Sci. Food Agric. 2021, 101, 3795–3803. [Google Scholar] [CrossRef]
- Belding, R.D.; Blankenship, S.M.; Young, E.; Leidy, R.B. Composition and Variability of Epicuticular Waxes in Apple Cultivars. J. Am. Soc. Hort. Sci. 1998, 123, 348–356. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, B.; Zhang, J.; Wang, C.; Liu, C.; Liu, Y.; Zhu, X.; Ren, X. Relationships between cuticular waxes and skin greasiness of apples during storage. Postharv. Biol. Technol. 2017, 131, 55–67. [Google Scholar] [CrossRef]
- Wu, W.; Jiang, B.; Liu, R.; Han, Y.; Fang, X.; Mu, H.; Farag, M.; Simal-Gandara, J.; Prieto, M.A.; Chen, H.; et al. Structures and functions of cuticular wax in postharvest fruit and its regulation: A comprehensive review with future perspectives. Engineering 2023, 23, 118–129. [Google Scholar] [CrossRef]
- Leide, J.; Xavier de Souza, A.; Papp, I.; Riederer, M. Specific characteristics of the apple fruit cuticle: Investigation of early and late season cultivars ‘Prima’ and ‘Florina’ (Malus domestica Borkh.). Sci. Hortic. 2018, 229, 137–147. [Google Scholar] [CrossRef]
- Shepherd, T.; Wynne Griffiths, D. The effects of stress on plant cuticular waxes. New Phytol. 2006, 171, 469–499. [Google Scholar] [CrossRef]
- Fernandez-Moreno, J.-P.; Malitsky, S.; Lashbrooke, J.; Biswal, A.K.; Racovita, R.C.; Mellerowicz, E.J.; Jetter, R.; Orzaez, D.; Aharoni, A.; Granell, A. An efficient method for medium throughput screening of cuticular wax composition in different plant species. Metabolomics 2016, 12, 73. [Google Scholar] [CrossRef]
- Lara, I.; Belge, B.; Goulao, L.F. A Focus on the Biosynthesis and Composition of Cuticle in Fruits. J. Agric. Food Chem. 2015, 63, 4005–4019. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Min, D.; Song, B.; Shao, S.; Zhang, X. Ethylene effects on apple fruit cuticular wax composition and content during cold storage. Postharv. Biol. Technol. 2017, 134, 98–105. [Google Scholar] [CrossRef]
- Li, F.; Min, D.; Ren, C.; Dong, L.; Shu, P.; Cui, X.; Zhang, X. Ethylene altered fruit cuticular wax, the expression of cuticular wax synthesis-related genes and fruit quality during cold storage of apple (Malus domestica Borkh. C.v. Starkrimson) fruit. Postharv. Biol. Technol. 2019, 149, 58–65. [Google Scholar] [CrossRef]
- Chen, J.; Green, K.B.; Nichols, K.K. Characterization of Wax Esters by Electrospray Ionization Tandem Mass Spectrometry: Double Bond Effect and Unusual Product Ions. Lipids 2015, 50, 821–836. [Google Scholar] [CrossRef]
- Tada, A.; Jin, Z.-L.; Sugimoto, N.; Sato, K.; Yamazaki, T.; Tanamoto, K. Analysis of the constituents in jojoba wax used as a food additive by LC/MS/MS. Shokuhin Eiseigaku Zasshi. J. Food Hyg. Soc. Jpn 2005, 46, 198–204. [Google Scholar] [CrossRef]
- McGhie, T.K.; Hudault, S.; Lunken, R.C.M.; Christeller, J.T. Apple peels, from seven cultivars, have lipase-inhibitory activity and contain numerous ursenoic acids as identified by LC-ESI-QTOF-HRMS. J. Agric. Food Chem. 2012, 60, 482–491. [Google Scholar] [CrossRef] [PubMed]
- Poirier, B.C.; Buchanan, D.A.; Rudell, D.R.; Mattheis, J.P. Differential Partitioning of Triterpenes and Triterpene Esters in Apple Peel. J. Agric. Food Chem. 2018, 66, 1800–1806. [Google Scholar] [CrossRef] [PubMed]
- Andre, C.M.; Greenwood, J.M.; Walker, E.G.; Rassam, M.; Sullivan, M.; Evers, D.; Perry, N.B.; Laing, W.A. Anti-inflammatory procyanidins and triterpenes in 109 apple varieties. J. Agric. Food Chem. 2012, 60, 10546–10554. [Google Scholar] [CrossRef] [PubMed]
- Andre, C.M.; Larsen, L.; Burgess, E.J.; Jensen, D.J.; Cooney, J.M.; Evers, D.; Zhang, J.; Perry, N.B.; Laing, W.A. Unusual immuno-modulatory triterpene-caffeates in the skins of russeted varieties of apples and pears. J. Agric. Food Chem. 2013, 61, 2773–2779. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Kind, T.; Nakabayashi, R.; Yukihira, D.; Tanaka, W.; Cajka, T.; Saito, K.; Fiehn, O.; Arita, M. Hydrogen Rearrangement Rules: Computational MS/MS Fragmentation and Structure Elucidation Using MS-FINDER Software. Anal. Chem. 2016, 88, 7946–7958. [Google Scholar] [CrossRef] [PubMed]
- Dührkop, K.; Fleischauer, M.; Ludwig, M.; Aksenov, A.A.; Melnik, A.V.; Meusel, M.; Dorrestein, P.C.; Rousu, J.; Böcker, S. SIRIUS 4: A rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 2019, 16, 4. [Google Scholar] [CrossRef] [PubMed]
- Dührkop, K.; Nothias, L.-F.; Fleischauer, M.; Reher, R.; Ludwig, M.; Hoffmann, M.A.; Petras, D.; Gerwick, W.H.; Rousu, J.; Dorrestein, P.C.; et al. Systematic classification of unknown metabolites using high-resolution fragmentation mass spectra. Nat. Biotechnol. 2021, 39, 4. [Google Scholar] [CrossRef] [PubMed]
- Dührkop, K.; Shen, H.; Meusel, M.; Rousu, J.; Böcker, S. Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proc. Natl. Acad. Sci. USA 2015, 112, 12580–12585. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.W.; Wang, M.; Leber, C.A.; Nothias, L.-F.; Reher, R.; Kang, K.B.; van der Hooft, J.J.J.; Dorrestein, P.C.; Gerwick, W.H.; Cottrell, G.W. NPClassifier: A Deep Neural Network-Based Structural Classification Tool for Natural Products. J. Nat. Prod. 2021, 84, 2795–2807. [Google Scholar] [CrossRef]
- Djoumbou Feunang, Y.; Eisner, R.; Knox, C.; Chepelev, L.; Hastings, J.; Owen, G.; Fahy, E.; Steinbeck, C.; Subramanian, S.; Bolton, E.; et al. ClassyFire: Automated chemical classification with a comprehensive, computable taxonomy. J. Cheminform. 2016, 8, 61. [Google Scholar] [CrossRef]
- Karp, P.D.; Billington, R.; Caspi, R.; Fulcher, C.A.; Latendresse, M.; Kothari, A.; Keseler, I.M.; Krummenacker, M.; Midford, P.E.; Ong, Q.; et al. The BioCyc collection of microbial genomes and metabolic pathways. Brief. Bioinform. 2019, 20, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
- Sorokina, M.; Merseburger, P.; Rajan, K.; Yirik, M.A.; Steinbeck, C. COCONUT online: Collection of open natural products database. J. Cheminform. 2021, 13, 2. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Guo, A.C.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef] [PubMed]
- Reisdorph, N.A.; Walmsley, S.; Reisdorph, R. A Perspective and Framework for Developing Sample Type Specific Databases for LC/MS-Based Clinical Metabolomics. Metabolites 2019, 10, 8. [Google Scholar] [CrossRef] [PubMed]
- Blaženović, I.; Kind, T.; Ji, J.; Fiehn, O. Software Tools and Approaches for Compound Identification of LC-MS/MS Data in Metabolomics. Metabolites 2018, 8, 31. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Bylesjö, M.; Rantalainen, M.; Cloarec, O.; Nicholson, J.K.; Holmes, E.; Trygg, J. OPLS discriminant analysis: Combining the strengths of PLS-DA and SIMCA classification. J. Chemom. 2006, 20, 341–351. [Google Scholar] [CrossRef]
- Triba, M.N.; Le Moyec, L.; Amathieu, R.; Goossens, C.; Bouchemal, N.; Nahon, P.; Rutledge, D.N.; Savarin, P. PLS/OPLS models in metabolomics: The impact of permutation of dataset rows on the K-fold cross-validation quality parameters. Mol. BioSyst. 2015, 11, 13–19. [Google Scholar] [CrossRef]
- Cocchi, M.; Biancolillo, A.; Marini, F. Chapter Ten—Chemometric Methods for Classification and Feature Selection. In Comprehensive Analytical Chemistry; Jaumot, J., Bedia, C., Tauler, R., Eds.; Data Analysis for Omic Sciences: Methods and Applications; Elsevier: Amsterdam, The Netherlands, 2018; Volume 82, pp. 265–299. [Google Scholar] [CrossRef]
- Pennington, J.A.T. Food Composition Databases for Bioactive Food Components. J. Food Comp. Anal. 2002, 15, 419–434. [Google Scholar] [CrossRef]
- Ramírez-Ambrosi, M.; López-Márquez, D.M.; Abad-García, B.; Dapena, E.; Berrueta, L.Á.; Gallo, B. Comparative study of phenolic profile of fruit and juice samples of a progeny of ‘Meana’ × ‘Florina’ from an Asturian cider apple breeding program. Eur. Food Res. Technol. 2015, 241, 769–784. [Google Scholar] [CrossRef]
- Sud, M.; Fahy, E.; Cotter, D.; Brown, A.; Dennis, E.A.; Glass, C.K.; Merrill Jr, A.H.; Murphy, R.C.; Raetz, C.R.H.; Russell, D.W.; et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007, 35, D527–D532. [Google Scholar] [CrossRef] [PubMed]
- Veraverbeke, E.A.; Lammertyn, J.; Saevels, S.; Nicolaï, B.M. Changes in chemical wax composition of three different apple (Malus domestica Borkh.) cultivars during storage. Postharv. Biol. Technol. 2001, 23, 197–208. [Google Scholar] [CrossRef]
- Klein, B.; Falk, R.B.; Thewes, F.R.; Anese, R.d.O.; dos Santos, I.D.; Ribeiro, S.R.; Donadel, J.Z.; Brackmann, A.; Barin, J.S.; Cichoski, A.J.; et al. Dynamic controlled atmosphere: Effects on the chemical composition of cuticular wax of ‘Cripps Pink’ apples after long-term storage. Postharv. Biol. Technol. 2020, 164, 111170. [Google Scholar] [CrossRef]
- Morice, I.M.; Shorland, F.B. Composition of the surface waxes of apple fruits and changes during storage. J. Sci. Food Agric. 1973, 24, 1331–1339. [Google Scholar] [CrossRef] [PubMed]
- Urbanova, K.; Vrkoslav, V.; Valterova, I.; Haková, M.; Cvacka, J. Structural characterization of wax esters by electron ionization mass spectrometry. J. Lipid Res. 2011, 53, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Vrkoslav, V.; Urbanová, K.; Cvacka, J. Analysis of wax ester molecular species by high performance liquid chromatography/atmospheric pressure chemical ionisation mass spectrometry. J. Chromatogr. A 2010, 1217, 4184–4194. [Google Scholar] [CrossRef] [PubMed]
- Vrkoslav, V.; Urbanova, K.; Haková, M.; Cvacka, J. Analysis of wax esters by silver-ion high-performance liquid chromatography–tandem mass spectrometry. J. Chromatogr. A 2013, 1302, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Sekosan, G. LC-MS Identification of Wax Esters in Cloudy Canola Oil. LCGC Suppl. 2014, 12, 16–21. [Google Scholar]
- Iven, T.; Herrfurth, C.; Hornung, E.; Heilmann, M.; Hofvander, P.; Stymne, S.; Feussner, I. Wax ester profiling of seed oil by nano-electrospray ionization tandem mass spectrometry. Plant Methods 2013, 9, 24. [Google Scholar] [CrossRef]
- Liebisch, G.; Fahy, E.; Aoki, J.; Dennis, E.A.; Durand, T.; Ejsing, C.S.; Fedorova, M.; Feussner, I.; Griffiths, W.J.; Köfeler, H.; et al. Update on LIPID MAPS classification, nomenclature, and shorthand notation for MS-derived lipid structures. J. Lipid Res. 2020, 61, 1539–1555. [Google Scholar] [CrossRef]
- Liberati-Čizmek, A.-M.; Biluš, M.; Brkić, A.L.; Barić, I.C.; Bakula, M.; Hozić, A.; Cindrić, M. Analysis of Fatty Acid Esters of Hydroxyl Fatty Acid in Selected Plant Food. Plant Foods Hum. Nutr. 2019, 74, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Mandrekar, J.N. Receiver Operating Characteristic Curve in Diagnostic Test Assessment. J. Thorac. Oncol. 2010, 5, 1315–1316. [Google Scholar] [CrossRef] [PubMed]
- Westerhuis, J.A.; Hoefsloot, H.C.J.; Smit, S.; Vis, D.J.; Smilde, A.K.; van Velzen, E.J.J.; van Duijnhoven, J.P.M.; van Dorsten, F.A. Assessment of PLSDA cross validation. Metabolomics 2008, 4, 81–89. [Google Scholar] [CrossRef]
- Spickett, C.M.; Pitt, A.R. Oxidative Lipidomics Coming of Age: Advances in Analysis of Oxidized Phospholipids in Physiology and Pathology. Antioxid. Redox Signal. 2015, 22, 1646–1666. [Google Scholar] [CrossRef] [PubMed]
- Tessmer, M.A.; Antoniolli, L.R.; Appezzato-da-Glória, B. Cuticle of “Gala” and “Galaxy” apples cultivars under different environmental conditions. Braz. Arch. Biol. Technol. 2012, 55, 709–714. [Google Scholar] [CrossRef]
- Lommen, A.; Godejohann, M.; Venema, D.P.; Hollman, P.C.; Spraul, M. Application of directly coupled HPLC-NMR-MS to the identification and confirmation of quercetin glycosides and phloretin glycosides in apple peel. Anal. Chem. 2000, 72, 1793–1797. [Google Scholar] [CrossRef]
- Awad, M.A.; de Jager, A.; van Westing, L.M. Flavonoid and chlorogenic acid levels in apple fruit: Characterisation of variation. Sci. Hort. 2000, 83, 249–263. [Google Scholar] [CrossRef]
| Analytical Method | Description of Apple Samples | Classification Factor | Number of Samples | Number of Classes to Be Distinguished within the Sample Set | Classification Method | Performance of Classification | Reference |
|---|---|---|---|---|---|---|---|
| NIR | Surface of whole apple fruits | Cultivar | 300 | 3 (Fuji, Red Star, Gala) | NN, SVM, ELM | Calibration set 98% (ELM) Prediction set 97% (ELM) | [4] |
| Geographical origin | 2 (grown in different Chinese provinces) | ||||||
| Fluorescent spectroscopy | Apple juice (squeezed with a juice extractor) | Cultivar | 89 | 2 (grown in different Chinese provinces) | PLS | Calibration set 100% Prediction set 96% | [5] |
| SPME-GC-MS | Apple juice (squeezed with a juicer) | Cultivar | 50 | 6 (Starkrimson, Qinguan, Gala, Jonagold, Golden Delicioius, Fuji) | LDA, SLDA | Predicition set 100% (SLDA) | [6] |
| Geographical origin | 5 (grown in different counties within Chinese province) | Predicition set 90% (SLDA) | |||||
| SPME-GC-MS | Apple juice (squeezed with hand press) | Cultivar | 4 (3 kg of apples per sample) | 4 (Rijo, Verde, Ribeiro, Azedo) | PLS-DA, HCA | Vague description of model performance | [7] |
| Geographical origin | 2 (different civil parishes of Madeira) | ||||||
| IR-MS + conventional methods | Pulp, juice | Cultivar | 19 | 6 (Topaz, Idared, Golden Delicious, Goldrush, Gala, Gloster) | LDA | Insufficient description of models | [2] |
| Geographical origin | 4 (different regions of Slovenia) | ||||||
| Agricultural practice | 2 (way of farming organic, conventional) | ||||||
| IR-MS | Whole apples, peel, pulp, seed | Cultivar | 128 | 4 (Cripps Pink, Gala, Golden Delicious, Granny Smith) | LDA | 71% correctly classified samples | [8] |
| Geographical origin | 4 (grown in different districts of northerm Italy) | 99% (LOOCV) | |||||
| IR-MS | Peel, petiole, pulp, seed | Geographical origin | 48 | 2 (grown in different districts of northern Italy) | LDA | Limited information on classification models performance | [9] |
| IR-MS, ICP-MS | Apple juice (concentrated to sugar content 65.0°Brix) | Geographical origin | 135 | 6 (grown in different Chinese provinces) | LDA, PLS-DA | Only description of sample clustering in PLS-DA model without information about model validation | [10] |
| Electronic nose, electronic tongue | Apple juice (centrifugal juicer) | Cultivar | 126 | 10 (Fuji, Jonagold, Corolla, Gala, Red Delicous, Red Chief Delicious, Cattle Apple, Ralls Janet, Ourin, Tail, Golden Delicous) | LDA, PLS-DA, SVM | 100% (prediction ability of PLS-DA) 100% (accuracy testing rate of SVM) | [3] |
| Geographical origin | 7 (grown in different Chinese provinces) |
| Marker Ion (m/z) | Retention Time [min] | Adduct Type | Elemental Formula | Mass Error [ppm] | Tentative Identification | PLSDA VIP Score | Confidence Level |
|---|---|---|---|---|---|---|---|
| 701.7138 | 14.03 | [M+H]+ | C48H92O2 | −5.4 | Wax ester (30:1/18:1) | 3.1 | 2 |
| 317.064 | 2.06 | [M+H]+ | C16H12O7 | −6.7 | Isorhamnetine | 2.9 | 3 |
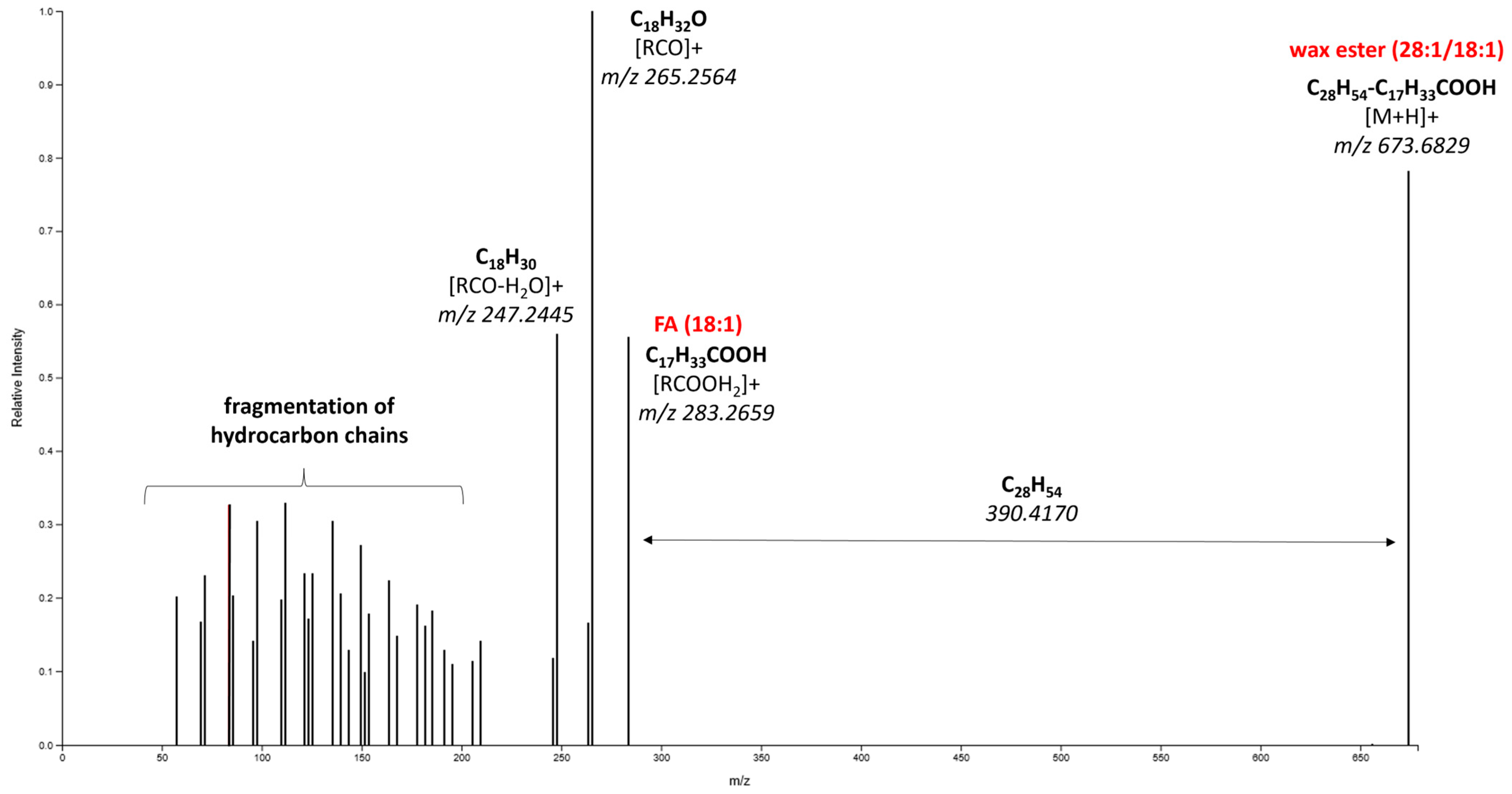
| 673.6829 | 13.69 | [M+H]+ | C46H88O2 | −5 | Wax ester (28:1/18:1) | 2.9 | 2 |
| 461.1111 | 2.14 | [M-H]− | C22H22O11 | 5.9 | Isorhamnetin rhamnoside | 2.8 | 3 |
| 671.6652 | 13.43 | [M+H]+ | C46H86O2 | -8 | Wax esters (46:3) | 2.8 | 3 |
| 699.691 | 14.64 | [M+Na]+ | C46H92O2 | −12.1 | Wax esters (46:0) | 2.7 | 3 |
| 979.8971 | 14.81 | [M+Na]+ | C63H120O5 | −6.4 | TAG (60:2) | 2.4 | 3 |
| 509.4234 | 12.24 | [M+H]+ | C31H56O5 | 5.6 | DAG (28:2) | 2.1 | 3 |
| 533.0917 | 1.33 | [M-H]− | C24H22O14 | 2.6 | Luteolin-O-malonyl glucoside | 2.1 | 3 |
| 663.3906 | 5.99 | [M+HCOO]− | C39H54O6 | 1.3 | Caffeoylbetulinic acid | 2 | 3 |
| 535.4747 | 12.87 | [M-H]− | C34H64O4 | 3.8 | FAHFA (18:1/16:0) | 1.9 | 2 |
| 549.3436 | 3.7 | [M+HCOO]− | C30H48O6 | 1.6 | Triterpenic acid | 1.6 | 3 |
| 749.6105 | 13.21 | [M-H]− | C49H82O5 | 2.8 | DAG (46:7) | 1.6 | 3 |
| OPLS-DA Model Parameters | ESI+ | ESI− | ||||||
|---|---|---|---|---|---|---|---|---|
| Gala | Golden Delicious | Idared | Jonagold | Gala | Golden Delicious | Idared | Jonagold | |
| number of features | 506 | 1048 | 156 | 11 | 13 | 44 | 24 | 9 |
| R2X | 0.783 | 0.596 | 0.570 | 0.946 | 0.667 | 0.567 | 0.850 | 0.921 |
| R2Y | 0.735 | 0.635 | 0.886 | 0.561 | 0.639 | 0.738 | 0.646 | 0.480 |
| Q2Y | 0.624 | 0.554 | 0.809 | 0.501 | 0.543 | 0.686 | 0.574 | 0.436 |
| RMSEE | 0.265 | 0.309 | 0.175 | 0.335 | 0.307 | 0.261 | 0.308 | 0.362 |
| p-value of permutation for R2Y | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| p-value of permutation for Q2Y | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| validity of the model over time | 82% | 65% | 85% | 88% | 78% | 77% | 78% | 88% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bechynska, K.; Sedlak, J.; Uttl, L.; Kosek, V.; Vackova, P.; Kocourek, V.; Hajslova, J. Metabolomics on Apple (Malus domestica) Cuticle—Search for Authenticity Markers. Foods 2024, 13, 1308. https://doi.org/10.3390/foods13091308
Bechynska K, Sedlak J, Uttl L, Kosek V, Vackova P, Kocourek V, Hajslova J. Metabolomics on Apple (Malus domestica) Cuticle—Search for Authenticity Markers. Foods. 2024; 13(9):1308. https://doi.org/10.3390/foods13091308
Chicago/Turabian StyleBechynska, Kamila, Jiri Sedlak, Leos Uttl, Vit Kosek, Petra Vackova, Vladimir Kocourek, and Jana Hajslova. 2024. "Metabolomics on Apple (Malus domestica) Cuticle—Search for Authenticity Markers" Foods 13, no. 9: 1308. https://doi.org/10.3390/foods13091308
APA StyleBechynska, K., Sedlak, J., Uttl, L., Kosek, V., Vackova, P., Kocourek, V., & Hajslova, J. (2024). Metabolomics on Apple (Malus domestica) Cuticle—Search for Authenticity Markers. Foods, 13(9), 1308. https://doi.org/10.3390/foods13091308







