Lactiplantibacillus plantarum A72, a Strain with Antioxidant Properties, Obtained through ARTP Mutagenesis, Affects Caenorhabditis elegans Anti-Aging
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Preparation of Cell-Free Supernatant (CFS), Intact Cells (IC), and Cell-Free Extracts (CFE) from Lpb. plantarum
2.3. Free Radical Scavenging Capacity Assay of Lpb. plantarum
2.4. C. elegans Culture Conditions
2.5. Effects of Lpb. plantarum A72 Addition on the Physiological Conditions of C. elegans
2.5.1. Effect of Lpb. plantarum A72 Addition on the Lifespan of C. elegans
2.5.2. Effect of Lpb. plantarum A72 Addition on the Reproductive Capacity of C. elegans
2.5.3. Effect of Lpb. plantarum A72 Addition on the Head Swing of C. elegans
2.6. Resistance of C. elegans to Environmental Factors with Lpb. plantarum Supplement
2.6.1. Effect of Lpb. plantarum on the Survival Rate of C. elegans under Thermal Shock
2.6.2. Effect of Lpb. plantarum on the Survival Rate of C. elegans Stimulated by H2O2
2.7. Determination of the Antioxidant Enzyme Activities, MDA Levels and ROS Accumulation of C. elegans with Lpb. plantarum Supplement
2.8. Transcriptome Sequencing of C. elegans
2.9. Data Analysis
3. Results and Discussion
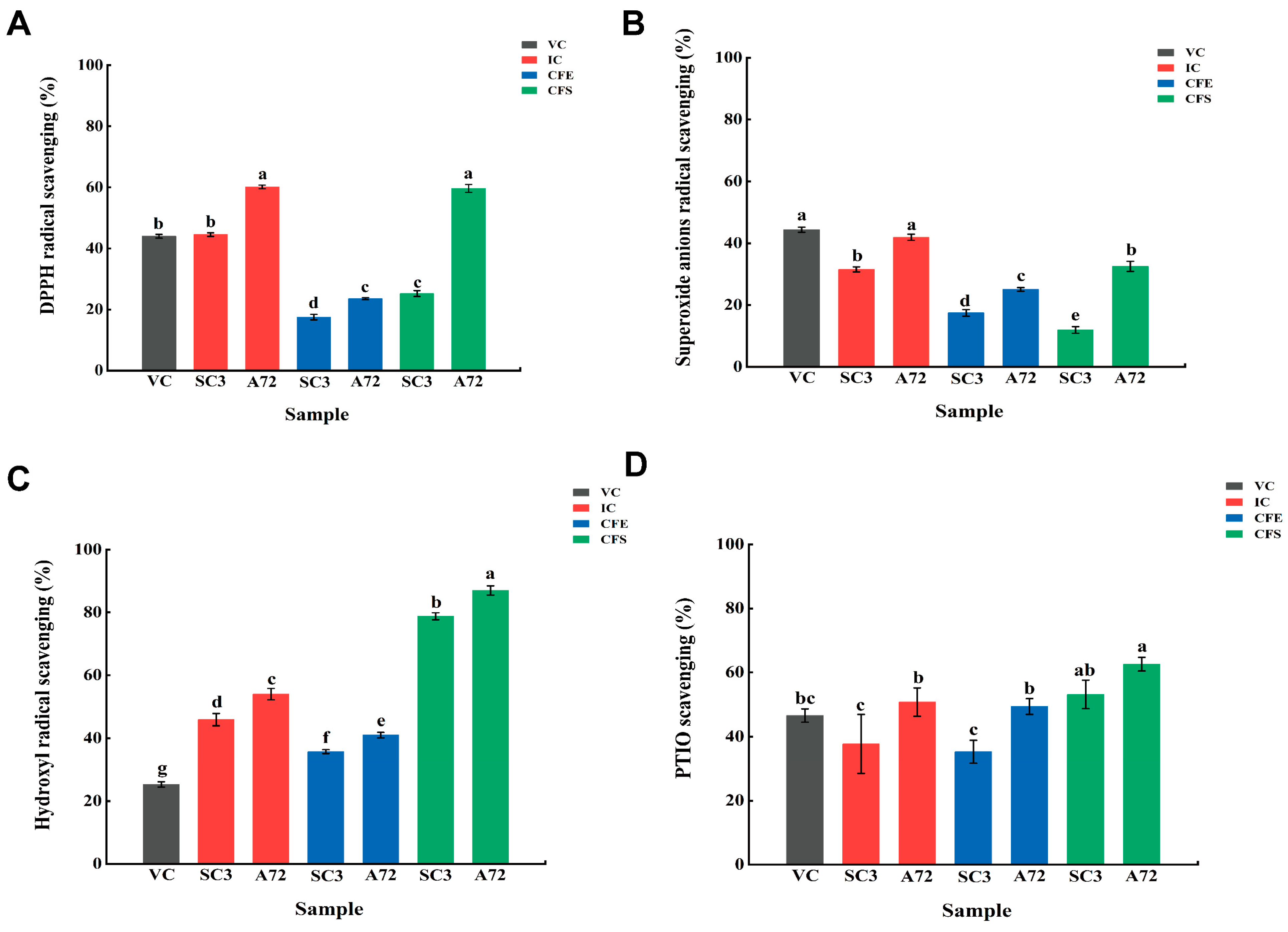
3.1. Antioxidant Activity In Vitro of Lpb. plantarum A72
3.2. Effects of Lpb. plantarum A72 Intact Cells Supplementation on C. elegans Physiological Conditions
3.3. Effect of Lpb. plantarum A72 Intact Cells on C. elegans Resistance to Environmentally Induced Oxidative Damage
3.4. Determination of the ROS Accumulation, Antioxidant Enzyme Activities and MDA Levels in C. elegans
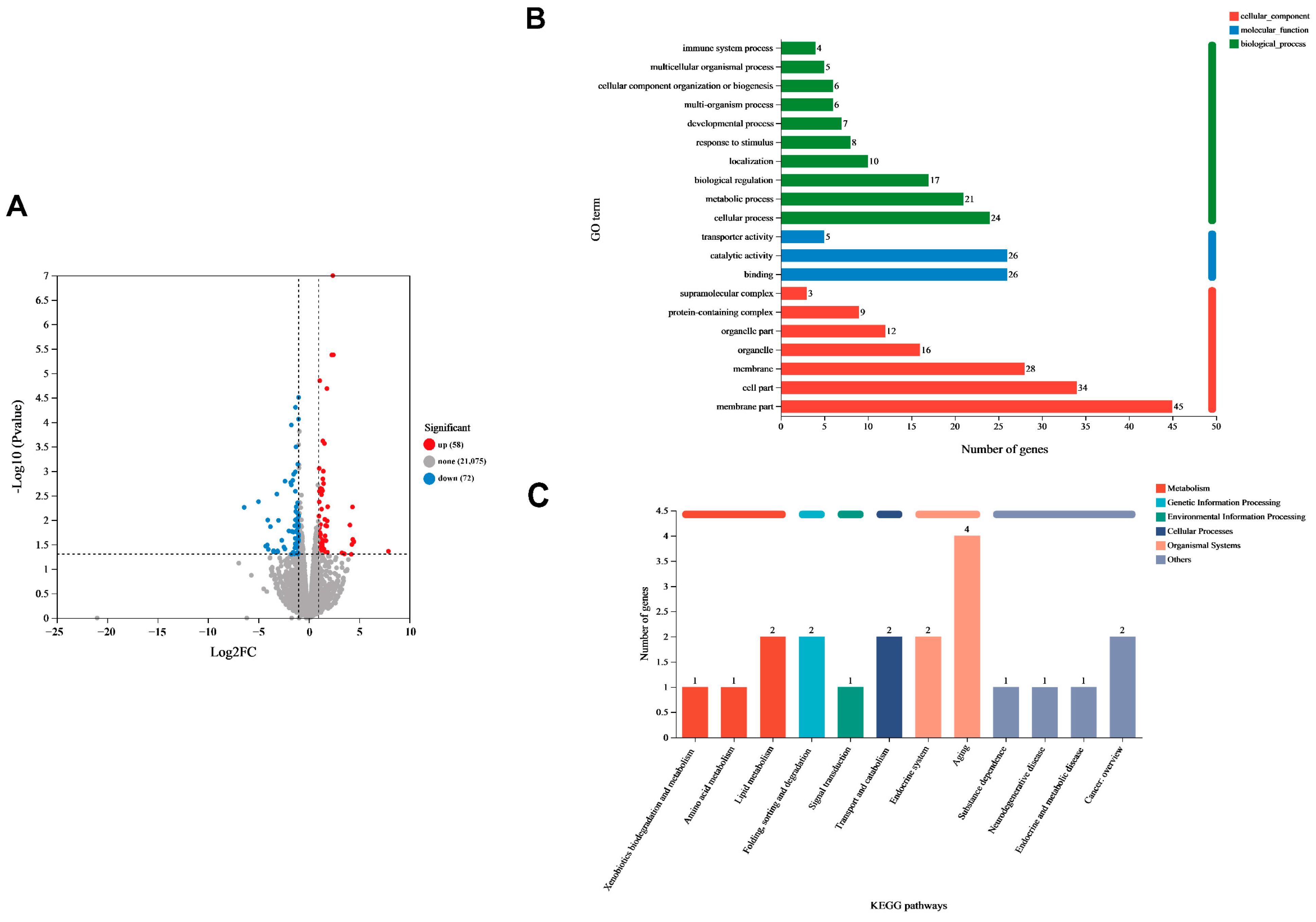
3.5. Differential Gene Expression of C. elegans Induced by Lpb. plantarum A72 Addition
3.6. Mechanistic Investigation of C. elegans Lifespan Extension by Lpb. plantarum A72 Supplement
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guedj, A.; Volman, Y.; Geiger-Maor, A.; Bolik, J.; Schumacher, N.; Künzel, S.; Baines, J.F.; Nevo, Y.; Elgavish, S.; Galun, E.; et al. Gut microbiota shape ‘inflamm-ageing’ cytokines and account for age-dependent decline in DNA damage repair. Gut 2020, 69, 1064–1075. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Chiang, H.H.; Luo, H.; Zheng, Z.; Qiao, Q.; Wang, L.; Tan, M.; Ohkubo, R.; Mu, W.C.; Zhao, S. An acetylation switch of the NLRP3 inflammasome regulates aging-associated chronic inflammation and insulin resistance. Cell Metab. 2020, 31, 580–591. [Google Scholar] [CrossRef] [PubMed]
- Ge, Q.; Yang, B.; Liu, R.; Jiang, D.; Yu, H.; Wu, M.; Zhang, W. Antioxidant activity of Lactobacillus plantarum NJAU-01 in an animal model of aging. BMC Microbiol. 2021, 21, 182. [Google Scholar] [CrossRef]
- Feng, T.; Wang, J. Oxidative stress tolerance and antioxidant capacity of lactic acid bacteria as probiotic: A systematic review. Gut Microbes 2020, 12, 1801944. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, W.; Wei, Z.; Yin, B.; Man, C.; Jiang, Y. Enhancement of functional characteristics of blueberry juice fermented by Lactobacillus plantarum. LWT-Food Sci. Technol. 2021, 139, 110590. [Google Scholar] [CrossRef]
- Pan, Y.; Wang, H.; Tan, F.; Yi, R.; Li, W.; Long, X.; Mu, J.; Zhao, X. Lactobacillus plantarum KFY02 enhances the prevention of CCl4-induced liver injury by transforming geniposide into genipin to increase the antioxidant capacity of mice. J. Funct. Foods 2020, 73, 104128. [Google Scholar] [CrossRef]
- Evans, K.S.; van Wijk, M.H.; McGrath, P.T.; Andersen, E.C.; Sterken, M.G. From QTL to gene: C. elegans facilitates discoveries of the genetic mechanisms underlying natural variation. Trends Genet. 2021, 37, 933–947. [Google Scholar] [CrossRef]
- Li, N.; Li, Q.; He, X.; Gao, X.; Wu, L.; Xiao, M.; Cai, W.; Liu, B.; Zeng, F. Antioxidant and anti-aging activities of Laminaria japonica polysaccharide in Caenorhabditis elegans based on metabonomic analysis. Int. J. Biol. Macromol. 2022, 221, 346–354. [Google Scholar] [CrossRef]
- Sun, J.; Zhong, X.; Sun, D.; Xu, L.; Shi, L.; Sui, J.; Liu, Y. Anti-aging effects of polysaccharides from ginseng extract residues in Caenorhabditis elegans. Int. J. Biol. Macromol. 2023, 225, 1072–1084. [Google Scholar] [CrossRef]
- Zou, S.B.; Zhao, M.W.; Ji, C.F.; Lin, S.F.; Sufang, Z.; Liang, H. Screening of high antioxidant activity lactic acid bacteria in traditional fermented Suancai of northeast China and its prebiotic studies. J. Food Saf. Qual. 2023, 14, 42–50. [Google Scholar]
- Wang, H.; Li, L. Comprehensive evaluation of probiotic property, hypoglycemic ability and antioxidant activity of lactic acid bacteria. Foods 2022, 11, 1363. [Google Scholar] [CrossRef]
- Li, H.; Huang, J.; Wang, Y.; Wang, X.; Ren, Y.; Yue, T.; Wang, Z.; Gao, Z. Study on the nutritional characteristics and antioxidant activity of dealcoholized sequentially fermented apple juice with Saccharomyces cerevisiae and Lactobacillus plantarum fermentation. Food Chem. 2021, 363, 130351. [Google Scholar] [CrossRef]
- Li, W.; Wang, T. Effect of solid-state fermentation with Bacillus subtilis lwo on the proteolysis and the antioxidative properties of chickpeas. Int. J. Food Microbiol. 2021, 338, 108988. [Google Scholar] [CrossRef] [PubMed]
- Li, X. 2-Phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl 3-oxide (PTIO•) radical scavenging: A new and simple antioxidant assay in vitro. J. Agric. Food Chem. 2017, 65, 6288–6297. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Fu, K.; Xiang, K.P.; Wang, L.Y.; Zhang, Y.F.; Luo, Y.P. Comparison of the chronic and multigenerational toxicity of racemic glufosinate and L-glufosinate to Caenorhabditis elegans at environmental concentrations. Chemosphere 2023, 316, 137863. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, Y.; Sun, Z.; Sun, T. Lacticaseibacillus rhamnosus Probio-M9 extends the lifespan of Caenorhabditis elegans. Commun. Biol. 2022, 5, 1139. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shi, J.; Liu, K.; Wang, Y.; Xu, Y.; Liu, Y. Metabolomics and gene expression levels reveal the positive effects of teaseed oil on lifespan and aging process in Caenorhabditis elegans. Food Sci. Hum. Wellness 2023, 12, 1391–1401. [Google Scholar] [CrossRef]
- Wang, L.; Xiao, M.F.; Chen, H.P.; Liu, B.; Zeng, F. Antioxidant activity of oyster peptide fraction OE-I and its anti-aging effect on Caenorhabditis elegans. Food Sci. 2022, 43, 152–160. [Google Scholar]
- Xue, Z.; Yinan, W.; Fei, M. Advances in the application of ARTP mutagenesis for biocatalyst modification in food and feed processing. Biotechnol. Bus. 2019, 3, 13–23. [Google Scholar]
- Luan, X.; Feng, M.; Sun, J. Effect of Lactobacillus plantarum on antioxidant activity in fermented sausage. Food Res. Int. 2021, 144, 110351. [Google Scholar] [CrossRef]
- Noureen, S.; Riaz, A.; Saif, A.; Arshad, M.; Qamar, M.; Arshad, N. Antioxidant properties of Lactobacillus brevis of Horse Origin and Commercial Lactic Acid Bacterial Strains: A Comparison. Pak. Vet. J. 2018, 38, 306–310. [Google Scholar] [CrossRef]
- Khubber, S.; Marti-Quijal, F.J.; Tomasevic, I.; Remize, F.; Barba, F.J. Lactic acid fermentation as a useful strategy to recover antimicrobial and antioxidant compounds from food and by-products. Curr. Opin. Food Sci. 2022, 43, 189–198. [Google Scholar] [CrossRef]
- Wu, S.; Chen, Y.; Chen, Z.; Zhou, Q.; Wei, F.; Li, P.; Gu, Q. Antioxidant properties and molecular mechanisms of Lactiplantibacillus plantarum ZJ316: A potential probiotic resource. LWT-Food Sci. Technol. 2023, 187, 115269. [Google Scholar] [CrossRef]
- Li, W.; Huang, W.; Ma, Y.; Muhammad, I.; Hanif, A.; Ding, Z.; Guo, X. Antioxidant properties of lactic acid bacteria isolated from traditional fermented yak milk and their probiotic effects on the oxidative senescence of Caenorhabditis elegans. Food Funct. 2022, 13, 3690–3703. [Google Scholar] [CrossRef]
- Sharma, K.; Pooranachithra, M.; Balamurugan, K.; Goel, G. Multivariate Analysis of Increase in Life Span of Caenorhabditis elegans Through Intestinal Colonization by Indigenous Probiotic Strains. Probiotics Antimicrob. Proteins 2018, 11, 865–873. [Google Scholar] [CrossRef]
- Gazzola, S.; Fontana, C.; Bassi, D.; Cocconcelli, P. Assessment of tetracycline and erythromycin resistance transfer during sausage fermentation by culture-dependent and-independent methods. Food Microbiol. 2012, 30, 348–354. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Chen, Z.; Wu, Y.; Chen, Y.; Wang, J.; Lin, Q.; Liang, Y. The rice bran peptide KF-8 extends the lifespan and improves the healthspan of Caenorhabditis elegans via skn-1 and daf-16. Food Funct. 2022, 13, 2427–2440. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Deng, N.; Zheng, B.; Li, T.; Liu, R.H. Rhodiola extract promotes longevity and stress resistance of Caenorhabditis elegans via DAF-16 and SKN-1. Food Funct. 2021, 12, 4471–4483. [Google Scholar] [CrossRef]
- Liu, H.; Wang, Y.; Zhang, W.; Sun, W.; Ji, X.; Zhang, S.; Qiao, K. Lentinan extends lifespan and increases oxidative stress resistance through DAF-16 and SKN-1 pathways in Caenorhabditis elegans. Int. J. Biol. Macromol. 2022, 202, 286–295. [Google Scholar] [CrossRef]
- Ritchie, D.J.; Friesen, C.R. Invited review: Thermal effects on oxidative stress in vertebrate ectotherms. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2022, 263, 111082. [Google Scholar] [CrossRef]
- Jin, S.Y.; Li, D.Q.; Lu, S.; Han, L.T.; Liu, D.H.; Huang, Z.; Huang, B.S.; Cao, Y. Ethanol extracts of Panax notoginseng increase lifespan and protect against oxidative stress in Caenorhabditis elegans via the insulin/IGF-1 signaling pathway. J. Funct. Foods 2019, 58, 218–226. [Google Scholar] [CrossRef]
- Kumar, H.; Dhalaria, R.; Guleria, S.; Cimler, R.; Sharma, R.; Siddiqui, S.A.; Valko, M.; Nepovimova, E.; Dhanjal, D.S.; Singh, R. Anti-oxidant potential of plants and probiotic spp. in alleviating oxidative stress induced by H2O2. Biomed. Pharmacother. 2023, 165, 115022. [Google Scholar] [CrossRef]
- Lu, H.; Sun, L.; Tong, S.; Jiang, F.; Chen, L.; Wang, Y. Latilactobacillus curvatus FFZZH5L isolated from pickled cowpea enhanced antioxidant activity in Caenorhabditis elegans by upregulating the level of glutathione S-transferase. Food Funct. 2023, 14, 8646–8660. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; He, Y.; Liu, Z.; Zhou, Y.; Chen, X.; Wang, G.; Sun, Z.; Zhao, J.; Zhang, H.; Chen, W. Lactic acid bacteria exhibit similar antioxidant capacities in Caenorhabditis elegans and Campylobacter jejuni-infected mice. RSC Adv. 2020, 10, 3329–3342. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Chen, R.; Qian, Y.; Ye, K.; Long, X.; Park, K.-Y.; Zhao, X. Antioxidant effect of Lactobacillus fermentum HFY02-fermented soy milk on D-galactose-induced aging mouse model. Food Sci. Hum. Wellness 2022, 11, 1362–1372. [Google Scholar] [CrossRef]
- Jiang, W.; Zhang, Y.; Zeng, J.; Yao, J.; Lu, A.; Fang, Z.; Wang, G.; Wang, W.; Zhang, Y. Composition analysis of acid hydrolysates from Cucurbita moschata Duch. polysaccharides and their effect on oxidative stress resistance of Caenorhabditis elegans. Food Sci. Hum. Wellness 2023, 12, 795–800. [Google Scholar] [CrossRef]
- Lin, C.; Xiao, J.; Xi, Y.; Zhang, X.; Zhong, Q.; Zheng, H.; Cao, Y.; Chen, Y. Rosmarinic acid improved antioxidant properties and healthspan via the IIS and MAPK pathways in Caenorhabditis elegans. BioFactors 2019, 45, 774–787. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Liu, Y.; Zhang, G.; Yang, Z.; Xu, W.; Chen, Q. The applications and mechanisms of superoxide dismutase in medicine, food, and cosmetics. Antioxidants 2023, 12, 1675. [Google Scholar] [CrossRef] [PubMed]
- Cuanalo-Contreras, K.; Schulz, J.; Mukherjee, A.; Park, K.-W.; Armijo, E.; Soto, C. Extensive accumulation of misfolded protein aggregates during natural aging and senescence. Front. Aging Neurosci. 2023, 14, 1090109. [Google Scholar] [CrossRef] [PubMed]
- Reinle, K.; Mogk, A.; Bukau, B. The diverse functions of small heat shock proteins in the proteostasis network. J. Mol. Biol. 2022, 434, 167157. [Google Scholar] [CrossRef]
- Qin, Y.; Chen, F.; Tang, Z.; Ren, H.; Wang, Q.; Shen, N.; Lin, W.; Xiao, Y.; Yuan, M.; Chen, H. Ligusticum chuanxiong Hort as a medicinal and edible plant foods: Antioxidant, anti-aging and neuroprotective properties in Caenorhabditis elegans. Front. Pharmacol. 2022, 13, 1049890. [Google Scholar] [CrossRef] [PubMed]
- Kumsta, C.; Chang, J.T.; Schmalz, J.; Hansen, M. Hormetic heat stress and HSF-1 induce autophagy to improve survival and proteostasis in C. elegans. Nat. Commun. 2017, 8, 14337. [Google Scholar] [CrossRef] [PubMed]
- Zečić, A.; Braeckman, B.P. DAF-16/FoxO in Caenorhabditis elegans and its role in metabolic remodeling. Cells 2020, 9, 109. [Google Scholar] [CrossRef]
- Xie, J.; Hou, X.; He, W.; Xiao, J.; Cao, Y.; Liu, X. Astaxanthin reduces fat storage in a fat-6/fat-7 dependent manner determined using high fat Caenorhabditis elegans. Food Funct. 2023, 14, 7347–7360. [Google Scholar] [CrossRef]
- Ackerman, D.; Gems, D. The mystery of C. elegans aging: An emerging role for fat: Distant parallels between C. elegans aging and metabolic syndrome? BioEssays 2012, 34, 466–471. [Google Scholar] [CrossRef] [PubMed]




| Group | Average Lifespan (Days) | Percentage Increase (%) | Maximum Lifespan (Days) |
|---|---|---|---|
| Control | 25.33 ± 0.47 d | 26 | |
| VC | 27.67 ± 0.47 c | 9.28 ± 3.84 b | 28 |
| 105 CFU/mL | 29.00 ± 0.82 bc | 14.56 ± 5.13 ab | 30 |
| 107 CFU/mL | 30.33 ± 1.25 ab | 19.85 ± 6.72 ab | 32 |
| 109 CFU/mL | 31.67 ± 1.25 a | 25.13 ± 7.08 a | 33 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, S.; Wu, Q.; Li, Z.; Zhang, S.; Dong, L.; Chen, Y.; Dai, Y.; Ji, C.; Liang, H.; Lin, X. Lactiplantibacillus plantarum A72, a Strain with Antioxidant Properties, Obtained through ARTP Mutagenesis, Affects Caenorhabditis elegans Anti-Aging. Foods 2024, 13, 924. https://doi.org/10.3390/foods13060924
Zou S, Wu Q, Li Z, Zhang S, Dong L, Chen Y, Dai Y, Ji C, Liang H, Lin X. Lactiplantibacillus plantarum A72, a Strain with Antioxidant Properties, Obtained through ARTP Mutagenesis, Affects Caenorhabditis elegans Anti-Aging. Foods. 2024; 13(6):924. https://doi.org/10.3390/foods13060924
Chicago/Turabian StyleZou, Sibo, Qi Wu, Zhigao Li, Sufang Zhang, Liang Dong, Yingxi Chen, Yiwei Dai, Chaofan Ji, Huipeng Liang, and Xinping Lin. 2024. "Lactiplantibacillus plantarum A72, a Strain with Antioxidant Properties, Obtained through ARTP Mutagenesis, Affects Caenorhabditis elegans Anti-Aging" Foods 13, no. 6: 924. https://doi.org/10.3390/foods13060924
APA StyleZou, S., Wu, Q., Li, Z., Zhang, S., Dong, L., Chen, Y., Dai, Y., Ji, C., Liang, H., & Lin, X. (2024). Lactiplantibacillus plantarum A72, a Strain with Antioxidant Properties, Obtained through ARTP Mutagenesis, Affects Caenorhabditis elegans Anti-Aging. Foods, 13(6), 924. https://doi.org/10.3390/foods13060924







