Genetic Identification and Technological Potential of Indigenous Lactic Acid Bacteria Isolated from Alheira, a Traditional Portuguese Sausage
Abstract
1. Introduction
2. Materials and Methods
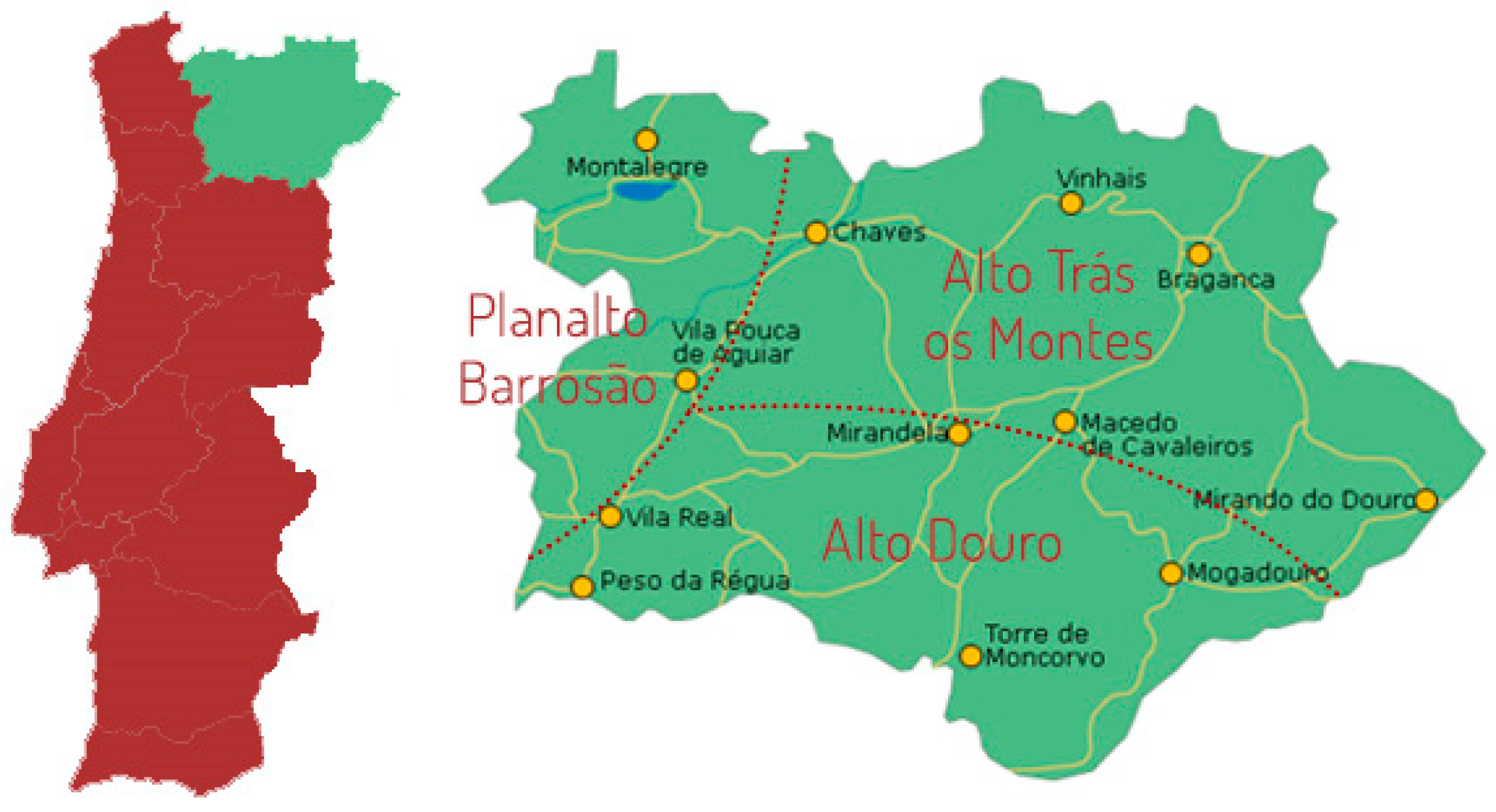
2.1. Sampling
2.2. Physicochemical and Microbiological Analysis of Sausages
2.3. LAB Isolation
2.4. Phenotypic Characterization
2.4.1. Antimicrobial Activity
2.4.2. Proteolytic Activity
2.4.3. Acidifying Capacity
2.4.4. L-Lactic Acid Production
2.5. Genotypic Characterization
2.5.1. DNA Extraction
2.5.2. 16S rRNA Amplification
2.5.3. 16S rRNA Sequencing
2.6. Data Analysis
2.6.1. Species Identification
2.6.2. Phylogenetic Tree
2.6.3. Phenotypic Characterization of LAB
3. Results and Discussion
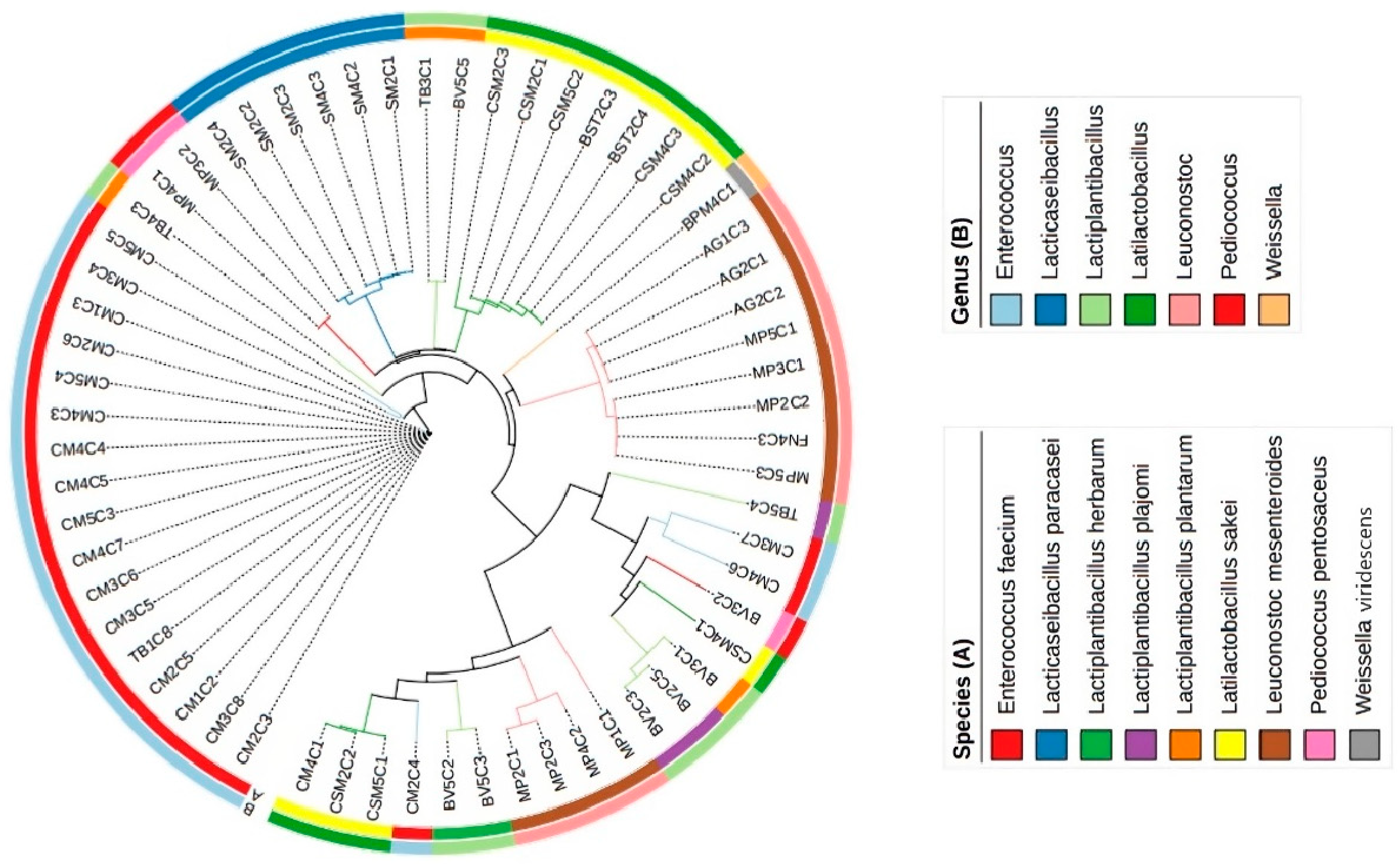
3.1. Genetic Identification of Lactic Acid Bacteria Isolates
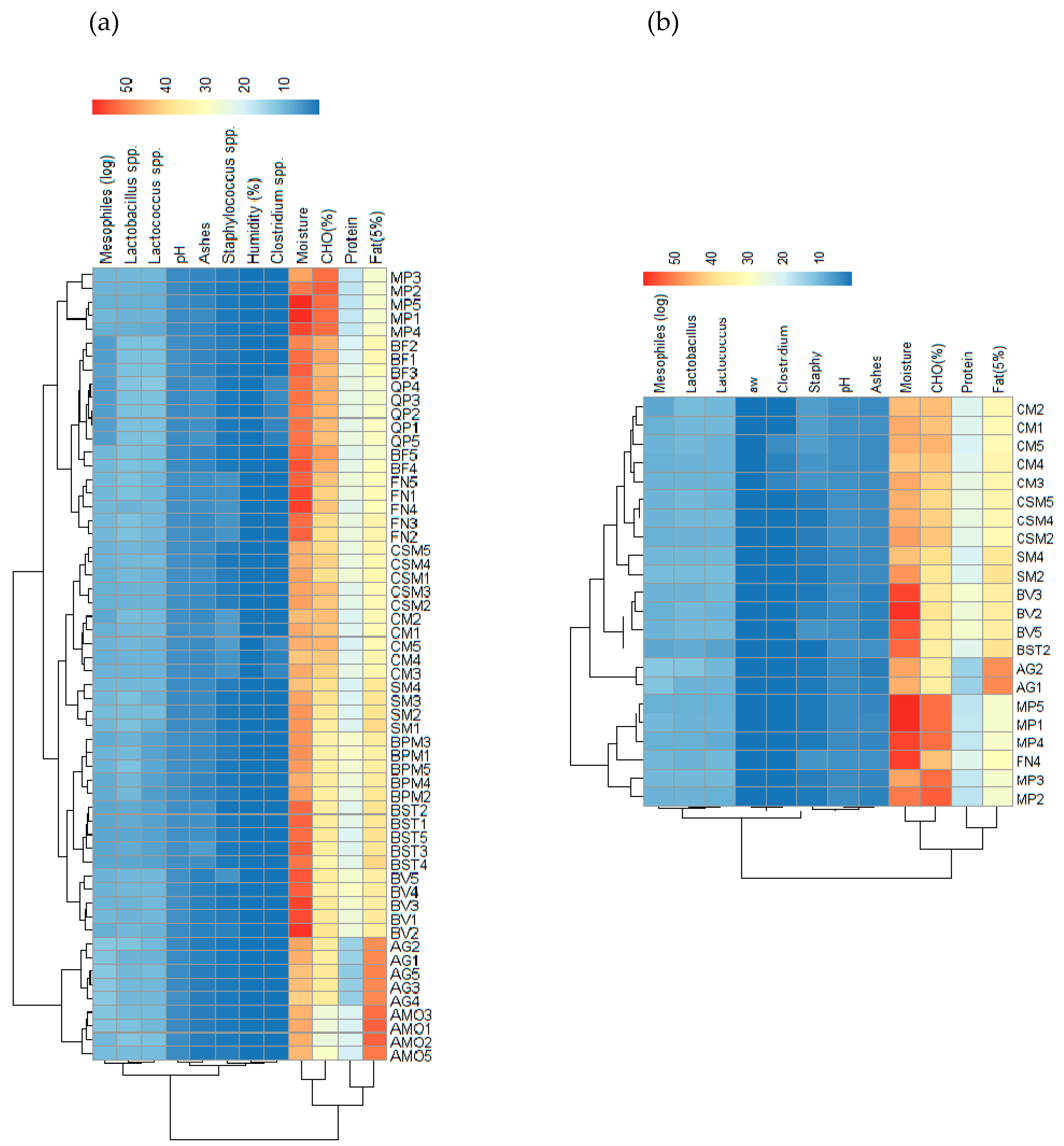
3.2. Alheira Physicochemical and Microbiological Analysis
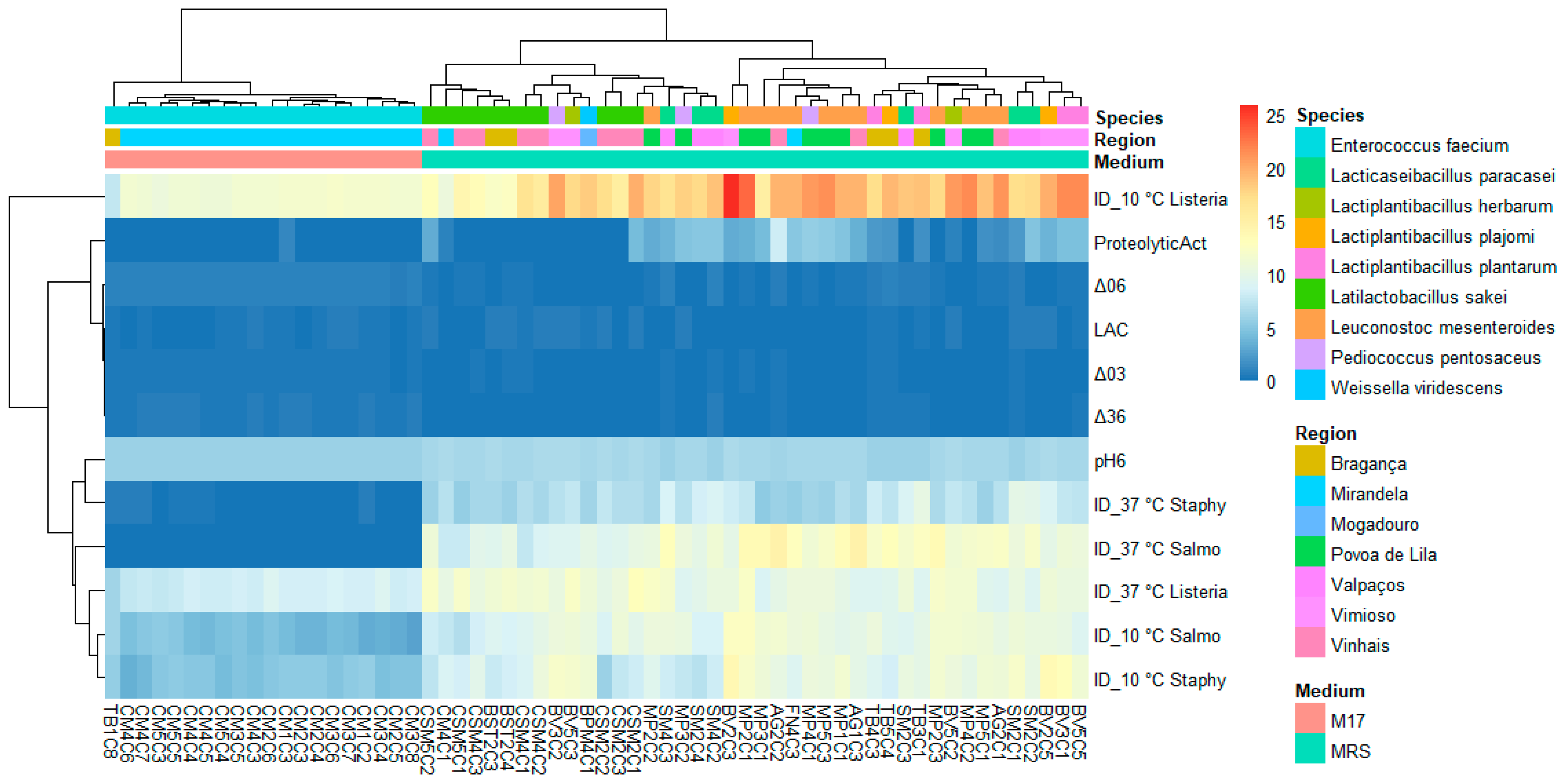
3.3. Lactic Acid Bacteria Phenotypic and Genetic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ferreira, V.; Barbosa, J.; Silva, J.; Felício, M.T.; Mena, C.; Hogg, T.; Gibbs, P.; Teixeira, P. Characterisation of Alheiras, Traditional Sausages Produced in the North of Portugal, with Respect to Their Microbiological Safety. Food Control 2007, 18, 436–440. [Google Scholar] [CrossRef]
- Patarata, L.; Judas, I.; Silva, J.A.; Esteves, A.; Martins, C. A Comparison of the Physicochemical and Sensory Characteristics of Alheira Samples from Different-Sized Producers. Meat Sci. 2008, 79, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Albano, H.; Oliveira, M.; Aroso, R.; Cubero, N.; Hogg, T.; Teixeira, P. Antilisterial Activity of Lactic Acid Bacteria Isolated from “Alheiras” (Traditional Portuguese Fermented Sausages): In Situ Assays. Meat Sci. 2007, 76, 796–800. [Google Scholar] [CrossRef] [PubMed]
- Campos, S.D.; Alves, R.C.; Mendes, E.; Costa, A.S.G.; Casal, S.; Oliveira, M.B.P.P. Nutritional Value and Influence of the Thermal Processing on a Traditional Portuguese Fermented Sausage (Alheira). Meat Sci. 2013, 93, 914–918. [Google Scholar] [CrossRef] [PubMed]
- Coelho-Fernandes, S.; Zefanias, O.; Rodrigues, G.; Faria, A.S.; Fernandes, Â.; Barros, L.; Cadavez, V.; Gonzales-Barron, U. Microbiological and Physicochemical Assessment of Artisanally Produced “Alheira” Fermented Sausages in Northern Portugal. Proceedings 2020, 70, 16. [Google Scholar] [CrossRef]
- Doyle, M.P.; Steenson, L.R.; Meng, J. Bacteria in Food and Beverage Production. In The Prokaryotes: Applied Bacteriology and Biotechnology; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 241–256. [Google Scholar] [CrossRef]
- Albano, H.; Henriques, I.; Correia, A.; Hogg, T.; Teixeira, P. Characterization of Microbial Population of ‘Alheira’ (a Traditional Portuguese Fermented Sausage) by PCR-DGGE and Traditional Cultural Microbiological Methods. J. Appl. Microbiol. 2008, 105, 2187–2194. [Google Scholar] [CrossRef]
- Moracanin, V.-M.; Turubatovic, L.; Skrinjar, M.; Obradovic, D. Antilisterial Activity of Bacteriocin Isolated from Leuconostoc Mesenteroides Ssp Mesenteroides IMAU:10231 in the Production of Sremska Sausages: Lactic Acid Bacteria Isolation, Bacteriocin Identification and Meat Application Experiments. Food Technol. Biotechnol. 2013, 51, 247–256. [Google Scholar]
- ISO 1442:1997; Meat and Meat Products—Determination of Moisture Content. ISO: Geneva, Switzerland, 1997.
- ISO 937:2023; Meat and Meat Products—Determination of Nitrogen Content. ISO: Geneva, Switzerland, 2023.
- ISO 936:1998; Meat and Meat Products—Determination of Total Ash. ISO: Geneva, Switzerland, 1998.
- Magalhães, A.L.; Ramalhosa, E.; Pereira, E. Microbiological Characterization of Alheira, a Typical Portuguese Fermented Sausage, and Its Relation with Hygienic Conditions of the Processing Environments. In Proceedings of the International Food Congress—Novel Approaches in Food Industry, NAFI 2011, İzmir, Türkiye, 26–29 May 2011; pp. 41–46. [Google Scholar]
- Esteves, A.; Patarata, L.; Saraiva, C.; Martins, C. Assessment of the Microbiological Characteristics of Industrially Produced Alheira, with Particular Reference to Foodborne Pathogens. J. Food Saf. 2008, 28, 88–102. [Google Scholar] [CrossRef]
- Zefanias, O.L.A. Caracterização Fisico-Química e Microbiológica de Alheiras Produzidas Artesanalmente na Terra Fria Transmontana. Master’s Thesis, Instituto Politécnico de Bragança, Bragança, Portugal, 2020. Available online: https://bibliotecadigital.ipb.pt/handle/10198/22671 (accessed on 26 January 2024).
- ISO 6888-1:2021; Microbiology of the Food Chain—Horizontal Method for the Enumeration of Coagulase-Positive Staphylococci (Staphylococcus aureus and Other Species). Part 1: Method Using Baird-Parker Agar Medium. ISO: Geneva, Switzerland, 2021.
- Dwivedi, H.P.; Smiley, R.D.; Pincus, D.H. 11. Rapid Methods for the Detection and Identification of Foodborne Pathogens. In Compendium of Methods for the Microbiological Examination of Foods; American Public Health Association: Washington, DC, USA, 2013. [Google Scholar] [CrossRef]
- Andrews, W.H.; Wang, H.; Jacobson, A.; Ge, B.; Zhang, G.; Hammack, T. BAM Chapter 5: Salmonella; FDA: White Oak, MD, USA, 2023. [Google Scholar]
- Faria, A.S.; Fernandes, N.; Cadavez, V.; Gonzales-Barron, U. Screening of Lactic Acid Bacteria Isolated from Artisanally Produced Alheira Fermented Sausages as Potential Starter Cultures. In Proceedings of the 2nd International Electronic Conference on Foods, Online, 15 October 2021. [Google Scholar]
- Franciosi, E.; Settanni, L.; Cavazza, A.; Poznanski, E. Biodiversity and Technological Potential of Wild Lactic Acid Bacteria from Raw Cows’ Milk. Int. Dairy. J. 2009, 19, 3–11. [Google Scholar] [CrossRef]
- Noll, F. L-(+)-Lactate. In Methods of Enzymatic Analysis, 3rd ed.; Bergmeyer, H.U., Ed.; VCH Publishers (UK) Ltd.: Cambridge, UK, 1988; Volume VI, pp. 582–588. [Google Scholar]
- Fernandes, N.; Faria, A.S.; Carvalho, L.; Choupina, A.; Rodrigues, C.; Cadavez, V.; Gonzales-Barron, U. Molecular Identification of Lactic Acid Producing Bacteria Isolated from Alheira, a Traditional Portuguese Fermented Sausage. Biol. Life Sci. Forum 2022, 18, 73. [Google Scholar] [CrossRef]
- Hou, Q.; Bai, X.; Li, W.; Gao, X.; Zhang, F.; Sun, Z.; Zhang, H. Design of Primers for Evaluation of Lactic Acid Bacteria Populations in Complex Biological Samples. Front. Microbiol. 2018, 9, 2045. [Google Scholar] [CrossRef] [PubMed]
- Mohania, D.; Nagpal, R.; Kumar, M.; Bhardwaj, A.; Yadav, M.; Jain, S.; Marotta, F.; Singh, V.; Parkash, O.; Yadav, H. Molecular Approaches for Identification and Characterization of Lactic Acid Bacteria. J. Dig. Dis. 2008, 9, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Holzapfel, W.H.; Wood, B.J. Lactic Acid Bacteria: Biodiversity and Taxonomy; Wiley-Blackwell: Chichester, UK, 2014. [Google Scholar]
- R: Core Team. R: A Language and Environment for Statistical Computing. 2021. Available online: https://www.R-project.org/ (accessed on 20 January 2024).
- Bonatesta, E.; Horejs-Kainrath, C.; Bodenhofer, U. Msa: Multiple Sequence Alignment, Version 1.32, Bioinformatics; Oxford University Press: Oxford, UK, 2015. [CrossRef]
- Charif, D.; Clerc, O.; Frank, C.; Lobry, J.R.; Necşulea, A.; Palmeira, L.; Penel, S.; Perrière, G. Seqinr: Biological Sequences Retrieval and Analysis. 2023. Available online: https://cran.r-project.org/web/packages/seqinr/index.html (accessed on 15 November 2023).
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Lam, T.T.-Y.; Xu, S.; Li, L.; Jones, B.; Silverman, J.; Iwasaki, W.M.; Xia, Y.; Huang, R. Ggtree: An R Package for Visualization of Tree and Annotation Data, Version 3.2.8. Methods Ecol Evol; British Ecological Society, Wiley-Blackwell: London, UK, 2022. [CrossRef]
- Dinov, I.D.; Rubin, D.; Lorensen, W.; Dugan, J.; Ma, J.; Murphy, S.; Kirschner, B.; Bug, W.; Sherman, M.; Floratos, A.; et al. iTools: A Framework for Classification, Categorization and Integration of Computational Biology Resources. PLoS ONE 2008, 3, e2265. [Google Scholar] [CrossRef] [PubMed]
- Revelle, W. Psych: Procedures for Psychological, Psychometric, and Personality Research. 2024. Available online: https://cran.r-project.org/web/packages/psych/index.html (accessed on 22 June 2023).
- Kolde, R. Pheatmap: Pretty Heatmaps. 2019. Available online: https://cran.r-project.org/web/packages/pheatmap/index.html (accessed on 25 June 2023).
- Fraqueza, M.J. Antibiotic Resistance of Lactic Acid Bacteria Isolated from Dry-Fermented Sausages. Int. J. Food Microbiol. 2015, 212, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Correia Santos, S.; Fraqueza, M.J.; Elias, M.; Salvador Barreto, A.; Semedo-Lemsaddek, T. Traditional Dry Smoked Fermented Meat Sausages: Characterization of Autochthonous Enterococci. LWT-Food Sci. Technol. 2017, 79, 410–415. [Google Scholar] [CrossRef]
- Albano, H.; van Reenen, C.A.; Todorov, S.D.; Cruz, D.; Fraga, L.; Hogg, T.; Dicks, L.M.T.; Teixeira, P. Phenotypic and Genetic Heterogeneity of Lactic Acid Bacteria Isolated from “Alheira”, a Traditional Fermented Sausage Produced in Portugal. Meat Sci. 2009, 82, 389–398. [Google Scholar] [CrossRef]
- Franciosa, I.; Ferrocino, I.; Giordano, M.; Mounier, J.; Rantsiou, K.; Cocolin, L. Specific Metagenomic Asset Drives the Spontaneous Fermentation of Italian Sausages. Food Res. Int. 2021, 144, 110379. [Google Scholar] [CrossRef]
- Ferreira, V.; Barbosa, J.; Vendeiro, S.; Mota, A.; Silva, F.; Monteiro, M.J.; Hogg, T.; Gibbs, P.; Teixeira, P. Chemical and Microbiological Characterization of Alheira: A Typical Portuguese Fermented Sausage with Particular Reference to Factors Relating to Food Safety. Meat Sci. 2006, 73, 570–575. [Google Scholar] [CrossRef]
- Borges, A.F.; Cózar, A.; Patarata, L.; Gama, L.T.; Alfaia, C.M.; Fernandes, M.J.; Fernandes, M.H.; Pérez, H.V.; Fraqueza, M.J. Effect of High Hydrostatic Pressure Challenge on Biogenic Amines, Microbiota, and Sensory Profile in Traditional Poultry- and Pork-Based Semidried Fermented Sausage. J. Food Sci. 2020, 85, 1256–1264. [Google Scholar] [CrossRef]
- Silva, J.; Barbosa, J.; Albano, H.; Sequeira, M.; Pinto, A.; Bonito, C.C.; Saraiva, M.; Teixeira, P. Microbiological Characterization of Different Formulations of Alheiras (Fermented Sausages). AIMS Agric. Food 2019, 4, 399–413. [Google Scholar] [CrossRef]
- Price, J.F.; Schweigert, B.S. The Science of Meat and Meat Products; W.H. Freeman: New York, NY, USA, 1971. [Google Scholar]
- Bintsis, T. Foodborne Pathogens. AIMS Microbiol. 2017, 3, 529–563. [Google Scholar] [CrossRef]
- Gilbert, R.J.; de Louvois, J.; Donovan, T.; Little, C.; Nye, K.; Ribeiro, C.D.; Richards, J.; Roberts, D.; Bolton, F.J. Guidelines for the Microbiological Quality of Some Ready-to-Eat Foods Sampled at the Point of Sale. PHLS Advisory Committee for Food and Dairy Products. Commun. Dis. Public Health 2000, 3, 163–167. [Google Scholar]
- Sarantinopoulos, P.; Andrighetto, C.; Georgalaki, M.D.; Rea, M.C.; Lombardi, A.; Cogan, T.M.; Kalantzopoulos, G.; Tsakalidou, E. Biochemical Properties of Enterococci Relevant to Their Technological Performance. Int. Dairy J. 2001, 11, 621–647. [Google Scholar] [CrossRef]
- Barbosa, J.; Ferreira, V.; Teixeira, P. Antibiotic Susceptibility of Enterococci Isolated from Traditional Fermented Meat Products. Food Microbiol. 2009, 26, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Albano, H.; Todorov, S.D.; van Reenen, C.A.; Hogg, T.; Dicks, L.M.T.; Teixeira, P. Characterization of Two Bacteriocins Produced by Pediococcus Acidilactici Isolated from “Alheira”, a Fermented Sausage Traditionally Produced in Portugal. Int. J. Food Microbiol. 2007, 116, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Albano, H.; Pinho, C.; Leite, D.; Barbosa, J.; Silva, J.; Carneiro, L.; Magalhães, R.; Hogg, T.; Teixeira, P. Evaluation of a Bacteriocin-Producing Strain of Pediococcus Acidilactici as a Biopreservative for “Alheira”, a Fermented Meat Sausage. Food Control 2009, 20, 764–770. [Google Scholar] [CrossRef]
- Azevedo, I.; Barbosa, J.; Albano, H.; Nogueira, T.; Teixeira, P. Lactic Acid Bacteria Isolated from Traditional and Innovative Alheiras as Potential Biocontrol Agents. Food Microbiol. 2024, 119, 104450. [Google Scholar] [CrossRef] [PubMed]
- Kieliszek, M.; Pobiega, K.; Piwowarek, K.; Kot, A.M. Characteristics of the Proteolytic Enzymes Produced by Lactic Acid Bacteria. Molecules 2021, 26, 1858. [Google Scholar] [CrossRef] [PubMed]
- Toldrá, F. Proteolysis and Lipolysis in Flavour Development of Dry-Cured Meat Products. Meat Sci. 1998, 49, S101–S110. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.-C.; Feng, M.-Q.; Sun, J.; Xu, X.-L.; Zhou, G.-H. Screening of Lactic Acid Bacteria with High Protease Activity from Fermented Sausages and Antioxidant Activity Assessment of Its Fermented Sausages. CyTA -J. Food 2019, 17, 347–354. [Google Scholar] [CrossRef]


| N. Isolates | Species | GenBank_ID | Identity (%) |
|---|---|---|---|
| 20 | Enterococcus faecium | NR_115764.1 | 99.2 [98.4;100.0] |
| 6 | Lacticaseibacillus paracasei | NR_025880.1 | 100.0 [100;100.0] |
| 2 | Lactiplantibacillus herbarum | NR_145899.1 | 99.5 [99.3;99.6] |
| 3 | Lactiplantibacillus plajomi | NR_136785.1 | 98.9 [98;99.8] |
| 4 | Lactiplantibacillus plantarum | NR_042394.1 | 98.5 [97;100.0] |
| 11 | Latilactobacillus sakei | NR_115172.1 | 99.8 [99.6;100.0] |
| 4 | Leuconostoc mesenteroides | NR_074957.1 | 98.5 [97;100.0] |
| 8 | Leuconostoc mesenteroides subsp. jonggajibkimchii | NR_157602.1 | 99.9 [99.8;100.0] |
| 3 | Pediococcus pentosaceus | NR_042058.1 | 99.5 [99.4;99.6] |
| 1 | Weissella viridescens | NR_040813.1 | 100.0 |
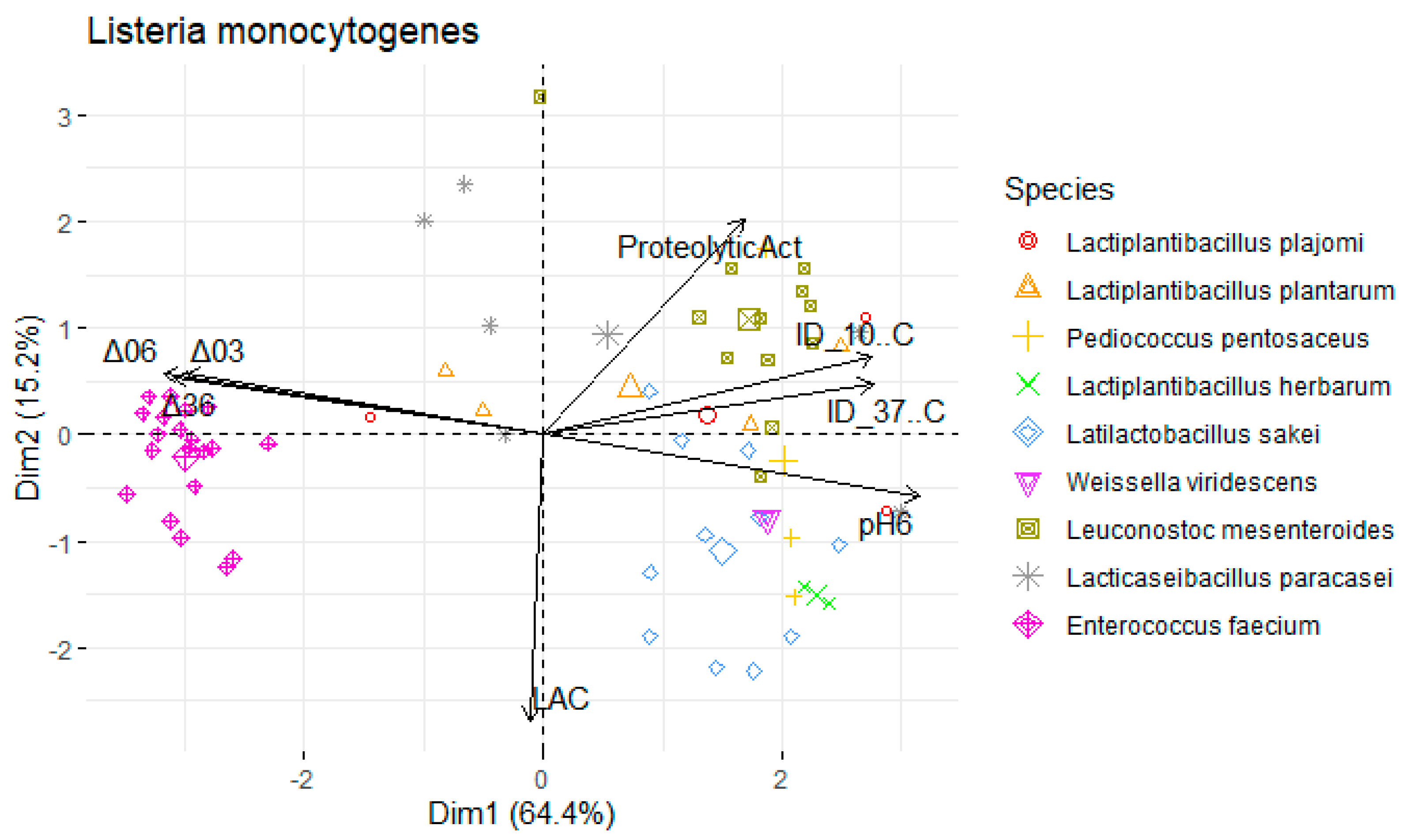
| Variables | S. Typhimurium | L. monocytogenes | S. aureus | |||
|---|---|---|---|---|---|---|
| PC1 | PC2 | PC1 | PC2 | PC1 | PC2 | |
| ProteolyticAct | 0.538 | 0.609 | 0.494 | 0.653 | 0.508 | 0.612 |
| pH6 | 0.942 | −0.232 | 0.960 | −0.159 | 0.950 | −0.175 |
| ΔpH03 | −0.896 | 0.212 | −0.903 | 0.137 | −0.901 | 0.173 |
| ΔpH06 | −0.956 | 0.225 | −0.968 | 0.152 | −0.961 | 0.173 |
| ΔpH36 | −0.927 | 0.218 | −0.940 | 0.149 | −0.931 | 0.162 |
| LAC | −0.068 | −0.771 | −0.032 | −0.783 | −0.032 | −0.812 |
| ID_10C | 0.884 | 0.209 | 0.758 | 0.387 | 0.834 | 0.218 |
| ID_37C | 0.872 | 0.299 | 0.784 | −0.099 | 0.834 | 0.143 |
| % of variance | 66.3 | 16.2 | 62.4 | 16.1 | 64.4 | 15.2 |
| Cumulative % of var. | 66.3 | 82.447 | 62.4 | 78.5 | 64.4 | 79.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernandes, N.; Faria, A.S.; Carvalho, L.; Choupina, A.; Rodrigues, C.; Gonzales-Barron, U.; Cadavez, V. Genetic Identification and Technological Potential of Indigenous Lactic Acid Bacteria Isolated from Alheira, a Traditional Portuguese Sausage. Foods 2024, 13, 598. https://doi.org/10.3390/foods13040598
Fernandes N, Faria AS, Carvalho L, Choupina A, Rodrigues C, Gonzales-Barron U, Cadavez V. Genetic Identification and Technological Potential of Indigenous Lactic Acid Bacteria Isolated from Alheira, a Traditional Portuguese Sausage. Foods. 2024; 13(4):598. https://doi.org/10.3390/foods13040598
Chicago/Turabian StyleFernandes, Nathália, Ana Sofia Faria, Laís Carvalho, Altino Choupina, Carina Rodrigues, Ursula Gonzales-Barron, and Vasco Cadavez. 2024. "Genetic Identification and Technological Potential of Indigenous Lactic Acid Bacteria Isolated from Alheira, a Traditional Portuguese Sausage" Foods 13, no. 4: 598. https://doi.org/10.3390/foods13040598
APA StyleFernandes, N., Faria, A. S., Carvalho, L., Choupina, A., Rodrigues, C., Gonzales-Barron, U., & Cadavez, V. (2024). Genetic Identification and Technological Potential of Indigenous Lactic Acid Bacteria Isolated from Alheira, a Traditional Portuguese Sausage. Foods, 13(4), 598. https://doi.org/10.3390/foods13040598







