Quality Characterization of Fava Bean-Fortified Bread Using Hyperspectral Imaging
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. Flour Preparation
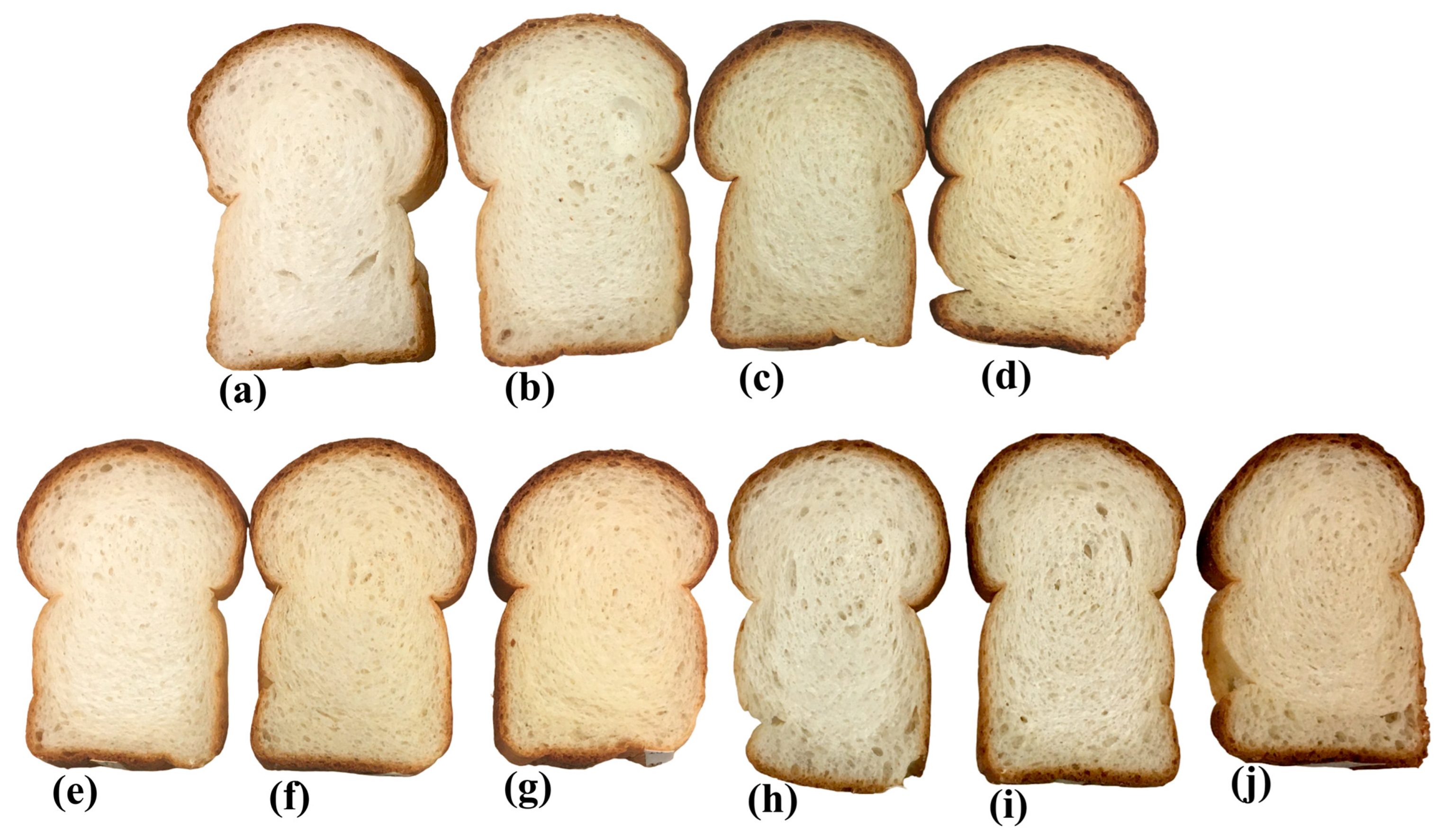
2.1.2. Bread Production
2.2. Methods
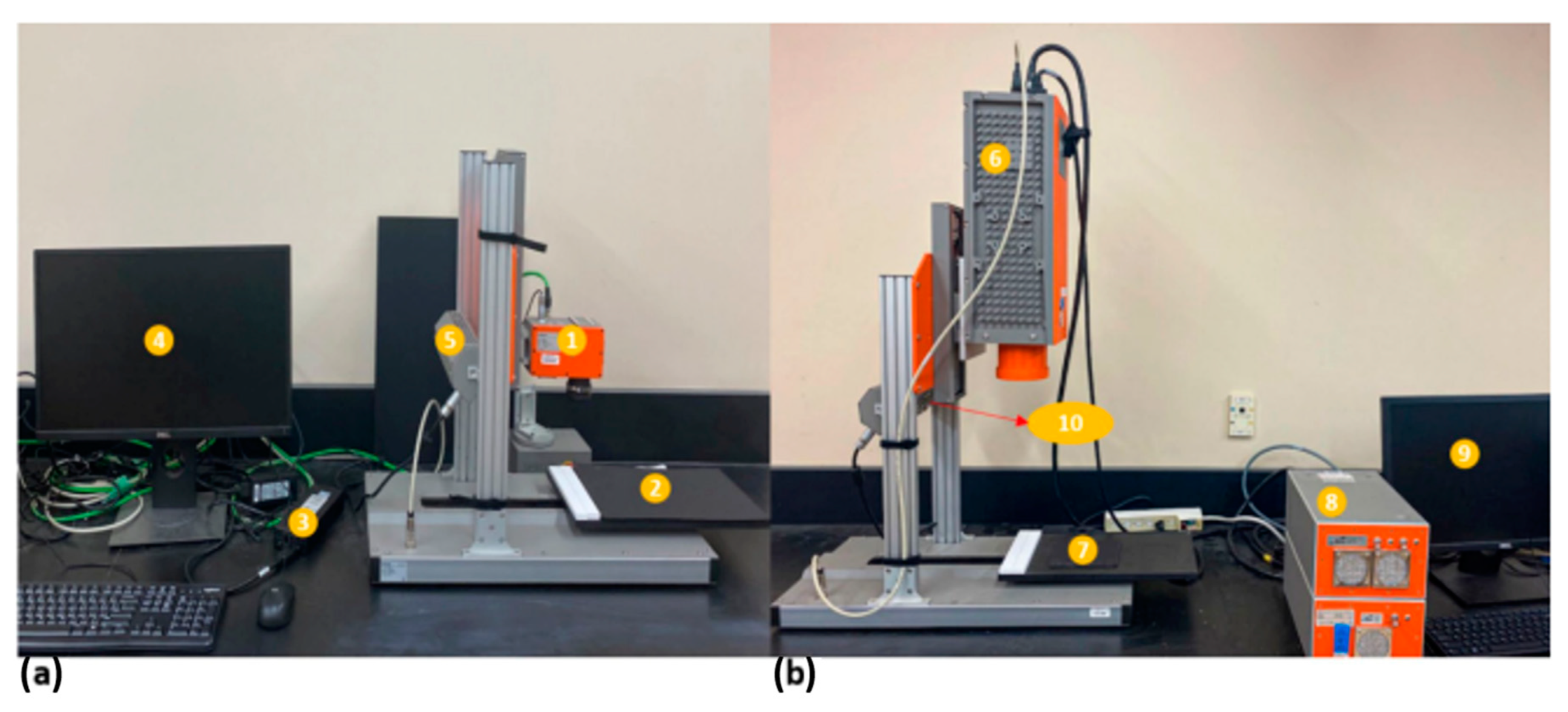
2.2.1. HSI Imaging System, Image Acquisition, and Correction
2.2.2. Image Processing and Spectral Extraction
2.2.3. Multivariate Data Analysis
2.2.4. Protein Content and Color Determination
3. Results and Discussion
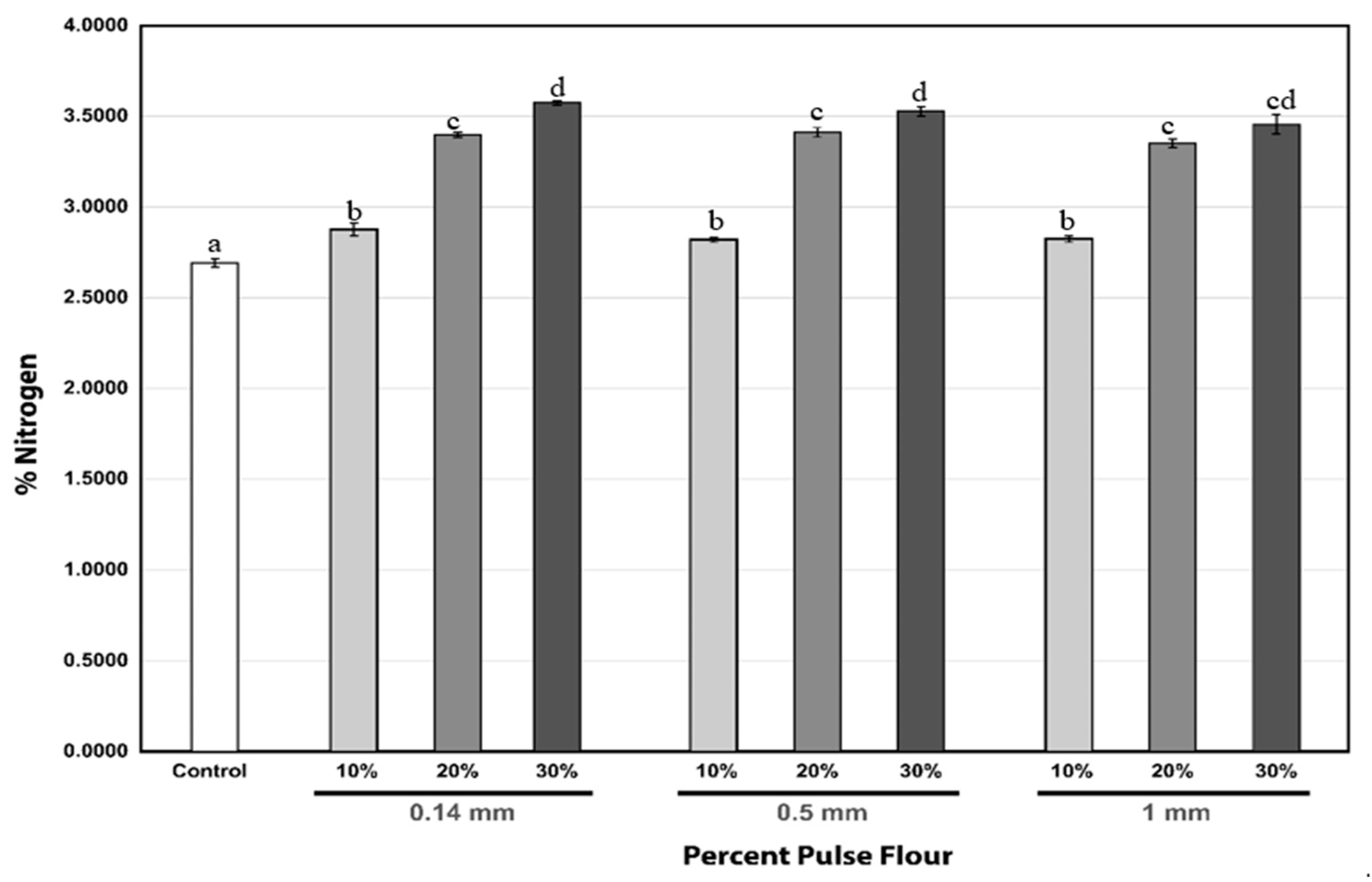
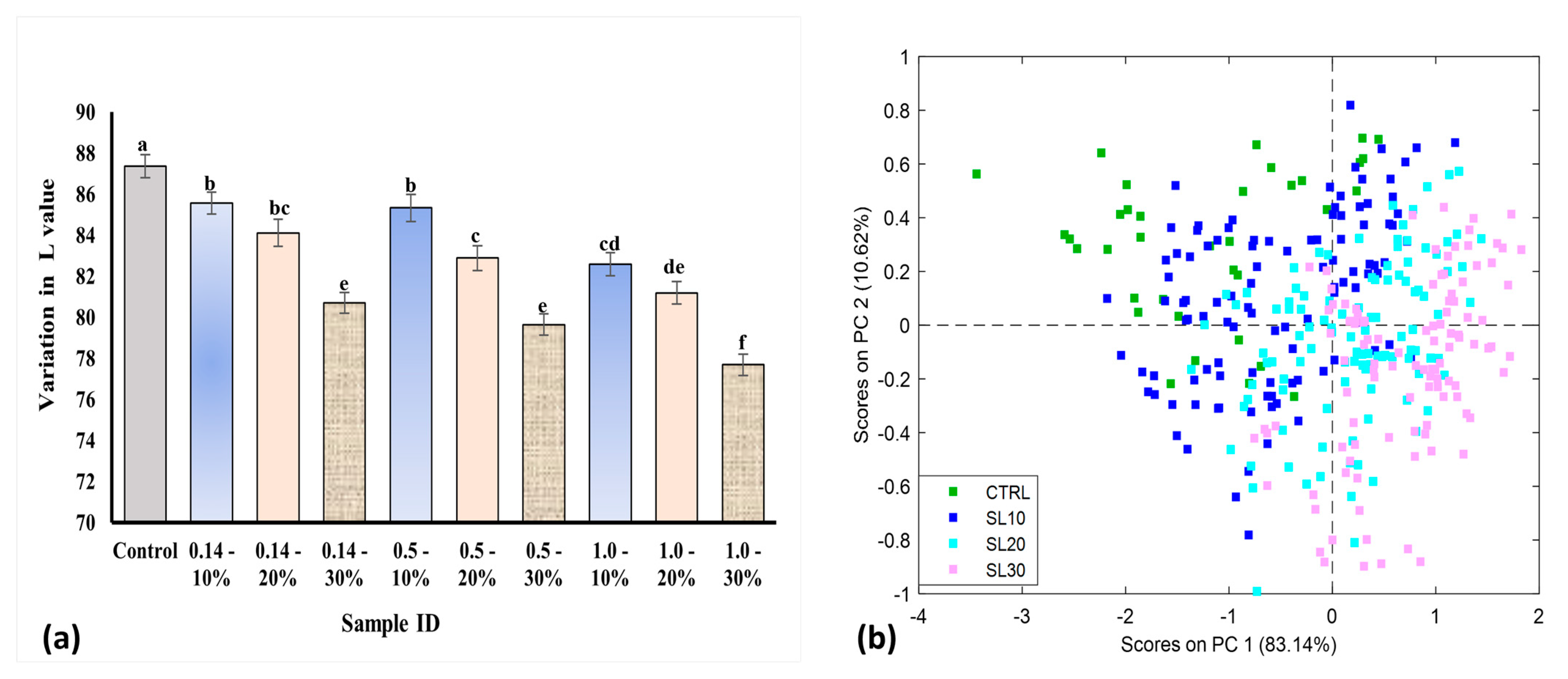
3.1. Analytical Techniques (Color and Protein Determination)
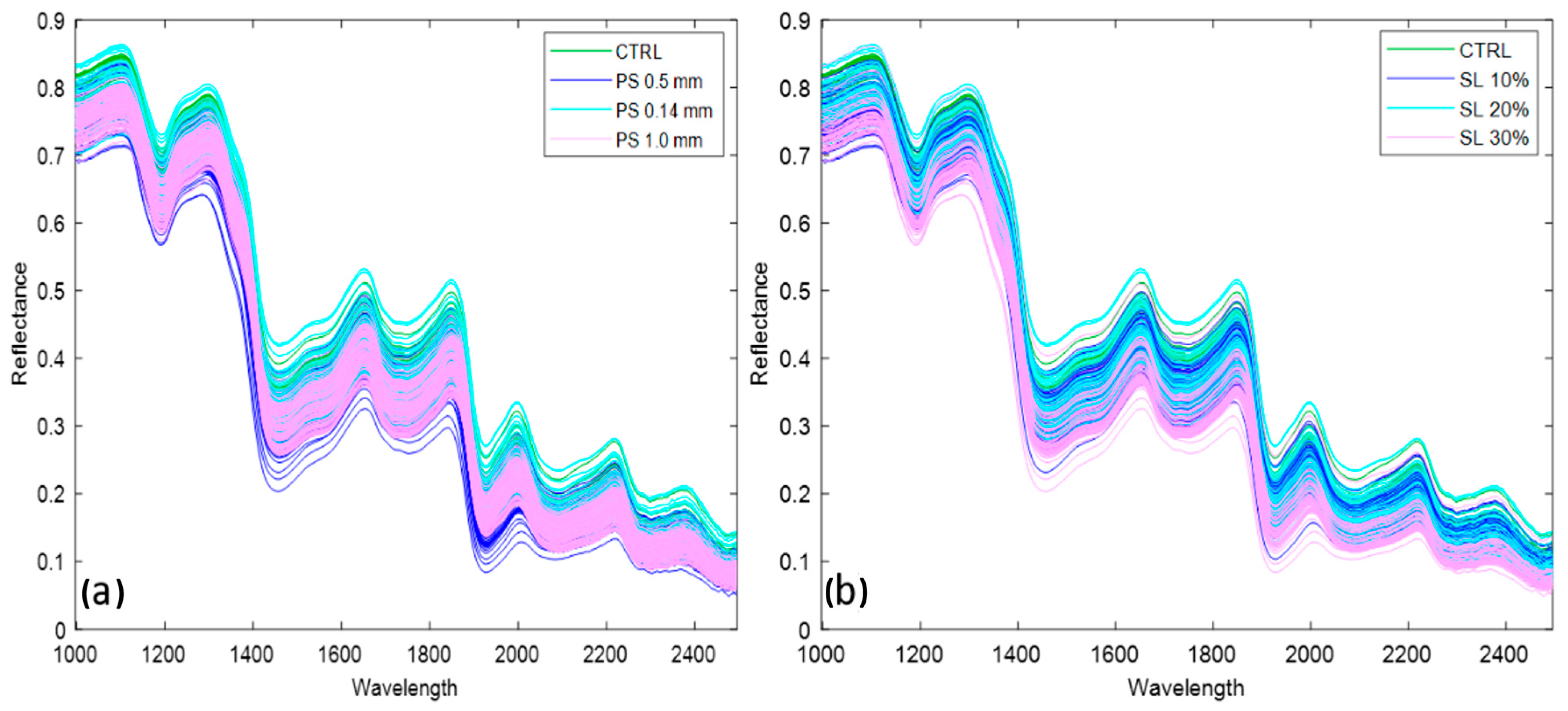
3.2. Spectral Profiles in the SWIR and Vis-NIR Ranges of the Dataset
3.3. Principal Component Analysis (PCA)
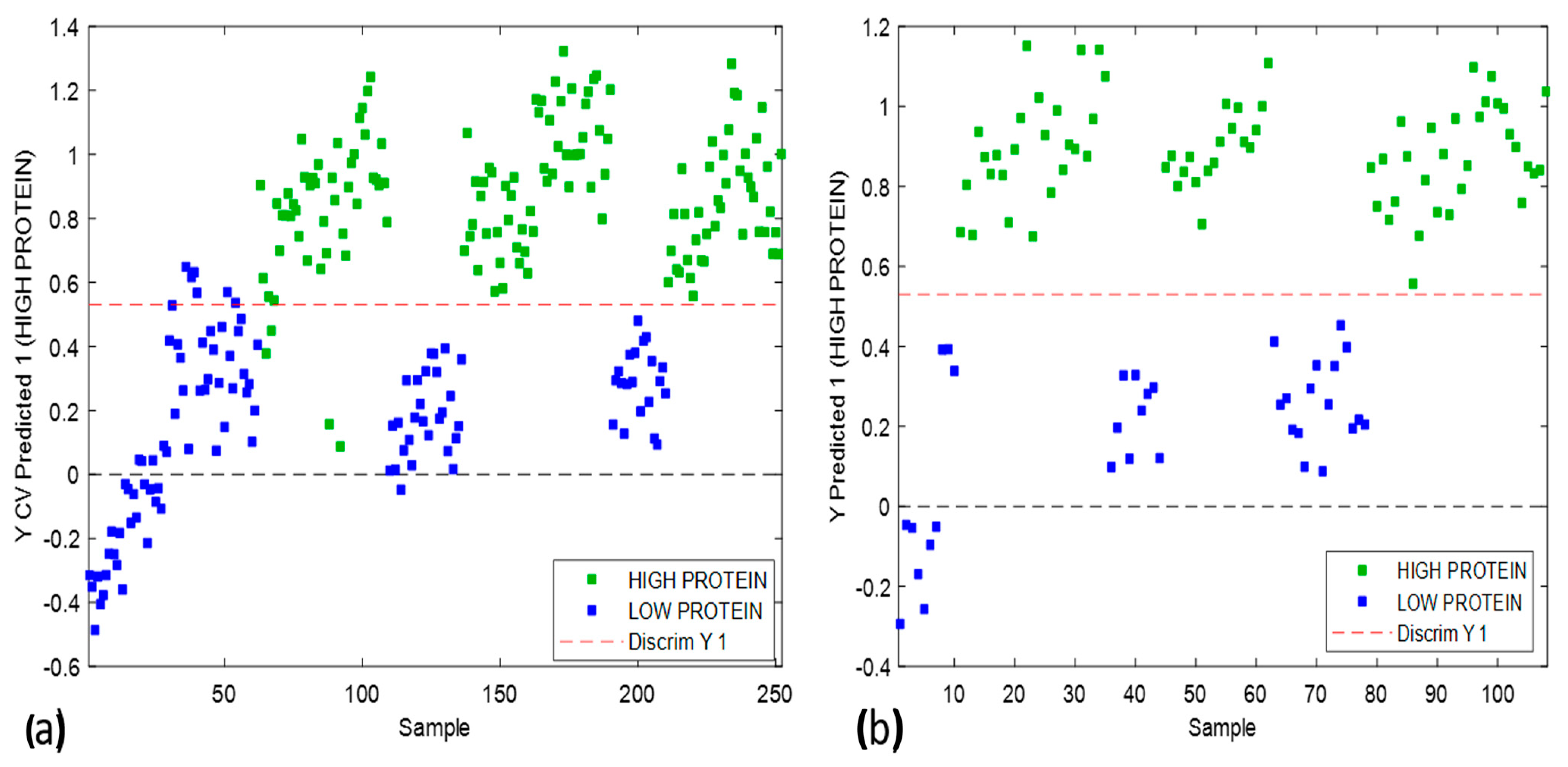
3.4. Supervised Classification Using Partial Least Squares Discriminant Analysis (PLS-DA)
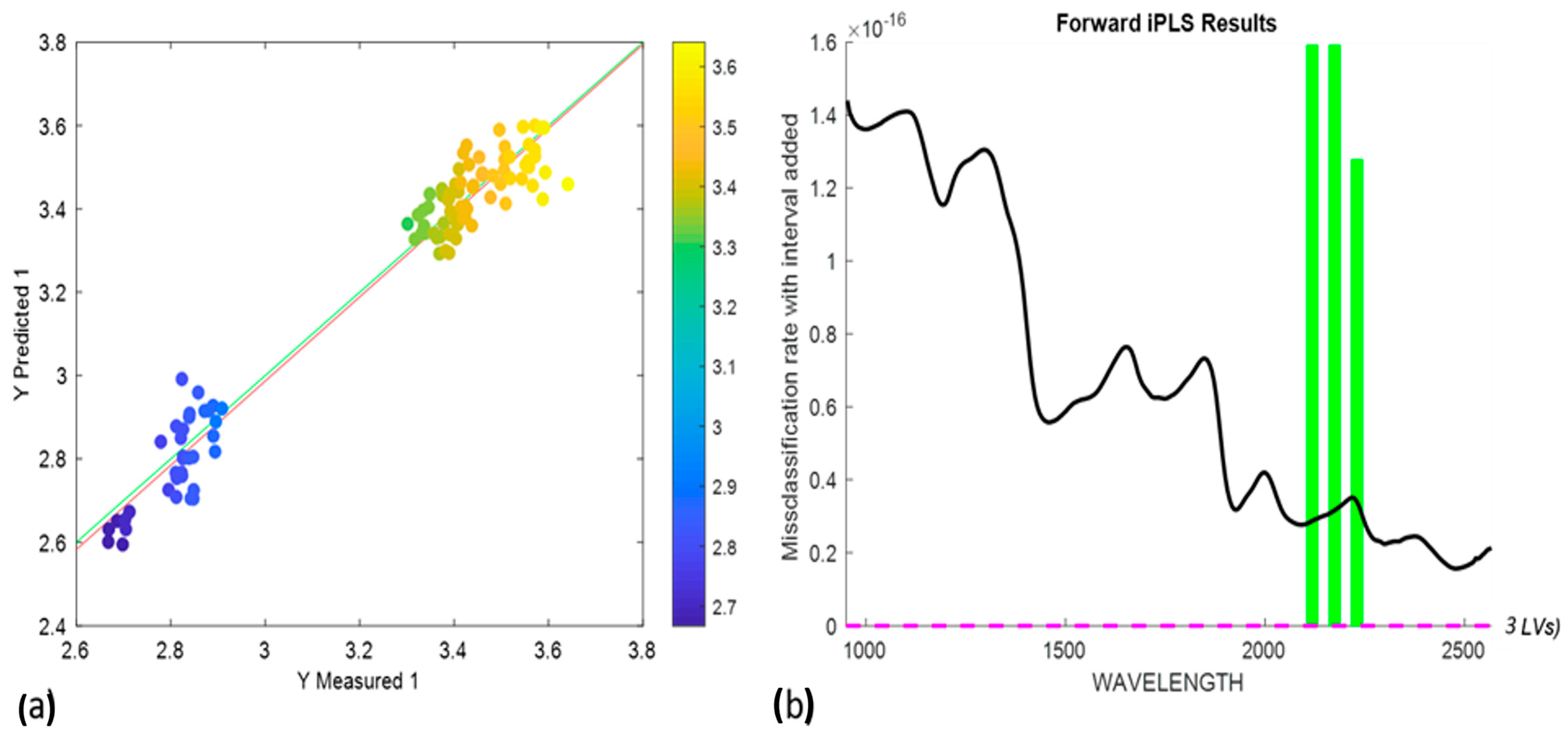
3.5. Protein Content Prediction Using Partial Least Squares Regression (PLSR)
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Olakanmi, S.J.; Jayas, D.S.; Paliwal, J. Implications of Blending Pulse and Wheat Flours on Rheology and Quality Characteristics of Baked Goods: A Review. Foods 2022, 11, 3287. [Google Scholar] [CrossRef]
- Carocho, M.; Morales, P.; Ciudad-Mulero, M.; Fernández-Ruiz, V.; Ferreira, E.; Heleno, S.; Rodrigues, P.; Barros, L.; Ferreira, I.C.F.R. Comparison of Different Bread Types: Chemical and Physical Parameters. Food Chem. 2020, 310, 125954. [Google Scholar] [CrossRef] [PubMed]
- Rossi, S.; Capobianco, F.; Sabatino, G.; Maurano, F.; Luongo, D.; Rossi, M. Pilot Scale Production of a Non-Immunogenic Soluble Gluten by Wheat Flour Transamidation with Applications in Food Processing for Celiac-Susceptible People. J. Cereal Sci. 2020, 96, 103117. [Google Scholar] [CrossRef]
- Demirkesen, I.; Kelkar, S.; Campanella, O.H.; Sumnu, G.; Sahin, S.; Okos, M. Characterization of Structure of Gluten-Free Breads by Using X-Ray Microtomography. Food Hydrocoll. 2014, 36, 37–44. [Google Scholar] [CrossRef]
- Giménez, M.A.; Drago, S.R.; De Greef, D.; Gonzalez, R.J.; Lobo, M.O.; Samman, N.C. Rheological, Functional and Nutritional Properties of Wheat/Broad Bean (Vicia faba) Flour Blends for Pasta Formulation. Food Chem. 2012, 134, 200–206. [Google Scholar] [CrossRef]
- Mohammed, I.; Ahmed, A.R.; Senge, B. Dough Rheology and Bread Quality of Wheat–Chickpea Flour Blends. Ind. Crops Prod. 2012, 36, 196–202. [Google Scholar] [CrossRef]
- Rahate, K.A.; Madhumita, M.; Prabhakar, P.K. Nutritional Composition, Anti-Nutritional Factors, Pretreatments-Cum-Processing Impact and Food Formulation Potential of Faba Bean (Vicia faba L.): A Comprehensive Review. LWT 2021, 138, 110796. [Google Scholar] [CrossRef]
- Millar, K.A.; Gallagher, E.; Burke, R.; McCarthy, S.; Barry-Ryan, C. Proximate Composition and Anti-Nutritional Factors of Fava-Bean (Vicia faba), Green-Pea and Yellow-Pea (Pisum sativum) Flour. J. Food Compos. Anal. 2019, 82, 103233. [Google Scholar] [CrossRef]
- Mayer Labba, I.C.; Frøkiær, H.; Sandberg, A.S. Nutritional and Antinutritional Composition of Fava Bean (Vicia faba L., Var. minor) Cultivars. Food Res. Int. 2021, 140, 110038. [Google Scholar] [CrossRef]
- Kukin, M.; Lavrenteva, N.; Nutchina, M.; Martirosyan, V. Adaptation of the Rapid Near-Infrared (NIR) Spectroscopy Technique to Determine the Mass Fraction of Protein and Moisture in Gluten-Free Bakery Products. BIO Web Conf. 2023, 64, 01001. [Google Scholar] [CrossRef]
- Cayuela-Sánchez, J.A.; Palarea-Albaladejo, J.; Zira, T.P.; Moriana-Correro, E. Compositional Method for Measuring the Nutritional Label Components of Industrial Pastries and Biscuits Based on Vis/NIR Spectroscopy. J. Food Compos. Anal. 2020, 92, 103572. [Google Scholar] [CrossRef]
- Xiao, Z.; Lai, K.; Du, R.; Shen, Y.; Sun, X.; Pan, Y.; Rasco, B.A.; Huang, Y. Fat and Moisture Content in Chinese Fried Bread Sticks: Assessment and Rapid near-Infrared Spectroscopic Method Development. J. Spectrosc. 2013, 1, 973623. [Google Scholar] [CrossRef]
- Nashat, S.; Abdullah, M.Z. Quality Evaluation of Bakery Products. In Computer Vision Technology for Food Quality Evaluation, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 525–589. [Google Scholar] [CrossRef]
- Olakanmi, S.J.; Jayas, D.S.; Paliwal, J. Applications of Imaging Systems for the Assessment of Quality Characteristics of Bread and Other Baked Goods: A Review. Compr. Rev. Food Sci. Food Saf. 2023, 22, 1817–1838. [Google Scholar] [CrossRef]
- Sivakumar, C.; Chaudhry, M.M.A.; Paliwal, J. Classification of Pulse Flours Using Near-Infrared Hyperspectral Imaging. LWT 2022, 154, 112799. [Google Scholar] [CrossRef]
- Kamal, M.; Karoui, R. Analytical Methods Coupled with Chemometric Tools for Determining the Authenticity and Detecting the Adulteration of Dairy Products: A Review. Trends Food Sci. Technol. 2015, 46, 27–48. [Google Scholar] [CrossRef]
- Nhouchi, Z.; Botosoa, E.P.; Karoui, R. Critical Assessment of Formulation, Processing and Storage Conditions on the Quality of Alveolar Baked Products Determined by Different Analytical Techniques: A Review. Trends Food Sci. Technol. 2018, 81, 159–171. [Google Scholar] [CrossRef]
- Feng, X.; Zhao, Y.; Zhang, C.; Cheng, P.; He, Y. Discrimination of Transgenic Maize Kernel Using NIR Hyperspectral Imaging and Multivariate Data Analysis. Sensors 2017, 17, 1894. [Google Scholar] [CrossRef]
- Kang, Z.; Zhao, Y.; Chen, L.; Guo, Y.; Mu, Q.; Wang, S. Advances in Machine Learning and Hyperspectral Imaging in the Food Supply Chain. Food Eng. Rev. 2022, 14, 596. [Google Scholar] [CrossRef]
- Ravikanth, L.; Jayas, D.S.; White, N.D.G.; Fields, P.G.; Sun, D.W. Extraction of Spectral Information from Hyperspectral Data and Application of Hyperspectral Imaging for Food and Agricultural Products. Food Bioprocess Technol. 2016, 10, 1–33. [Google Scholar] [CrossRef]
- Whitworth, M.B.; Millar, S.J.; Chau, A.; Whitworth, M.B.; Millar, S.J.; Chau, A. Food Quality Assessment by NIR Hyperspectral Imaging. SPIE 2010, 7676, 767605. [Google Scholar] [CrossRef]
- Liu, Z.; Møller, F. Bread Water Content Measurement Based on Hyperspectral Imaging. In Proceedings of the 2011 Scandinavian Workshop on Imaging Food Quality, Ystad, Sweden, 27 May 2011; pp. 93–98. [Google Scholar]
- Lancelot, E.; Courcoux, P.; Chevallier, S.; Le-Bail, A.; Jaillais, B. Prediction of Water Content in Biscuits Using near Infrared Hyperspectral Imaging Spectroscopy and Chemometrics. J. Near Infrared Spectrosc. 2020, 28, 140–147. [Google Scholar] [CrossRef]
- Lancelot, E.; Courcoux, P.; Chevallier, S.; Lebail, A.; Jaillais, B. Quantification of Water Content in Biscuit Using Near-Infrared Hyperspectral Imaging Spectroscopy and Chemometrics; AgroStat: LausanneLausanne, Switzerland, 2016. [Google Scholar]
- De Temmerman, A.; De Ryck, M.; Hellemans, T.; Verbeke, M. Infrared Hyperspectral Analysis for Non-Invasive, Inline Fat Content Determination in Bakery Products. In Proceedings of the 2023 IEEE 21st International Conference on Industrial Informatics (INDIN), Lemgo, Germany, 17–20 July 2023; pp. 1–7. [Google Scholar] [CrossRef]
- Polak, A.; Coutts, F.K.; Murray, P.; Marshall, S. Use of Hyperspectral Imaging for Cake Moisture and Hardness Prediction. IET Image Process. 2019, 13, 1152–1160. [Google Scholar] [CrossRef]
- Sricharoonratana, M.; Teerachaichayut, S. Prediction of Water Activity in Mamón (Filipino Sponge) Cakes by near Infrared Hyperspectral Imaging. Key Eng. Mater. 2020, 862, 7–11. [Google Scholar] [CrossRef]
- Morsey El, N.; Mokhtar, S.M.; Youssef, K.M. Accurate Quantification of Fungal Growth in Bread by Using Spectral Analysis. J. Food Dairy Sci. 2014, 5, 33–44. [Google Scholar] [CrossRef][Green Version]
- Amigo, J.M.; del Olmo, A.; Engelsen, M.M.; Lundkvist, H.; Engelsen, S.B. Staling of White Wheat Bread Crumb and Effect of Maltogenic α-Amylases. Part 3: Spatial Evolution of Bread Staling with Time by near Infrared Hyperspectral Imaging. Food Chem. 2021, 353, 129478. [Google Scholar] [CrossRef]
- Saleem, Z.; Hussain Khan, M.; Ahmad, M.; Sohaib, A.; Ayaz, H.; Mazzara, M. Prediction of Microbial Spoilage and Shelf-Life of Bakery Products through Hyperspectral Imaging. IEEE Access 2020, 8, 176986–176996. [Google Scholar] [CrossRef]
- Sricharoonratana, M.; Thompson, A.K.; Teerachaichayut, S. Use of near Infrared Hyperspectral Imaging as a Nondestructive Method of Determining and Classifying Shelf Life of Cakes. LWT 2021, 136, 110369. [Google Scholar] [CrossRef]
- Hasan, M.M.; Chaudhry, M.M.A.; Erkinbaev, C.; Paliwal, J.; Suman, S.P.; Rodas-Gonzalez, A. Application of Vis-NIR and SWIR Spectroscopy for the Segregation of Bison Muscles Based on Their Color Stability. Meat Sci. 2022, 188, 108774. [Google Scholar] [CrossRef]
- Achata, E.M.; Inguglia, E.S.; Esquerre, C.A.; Tiwari, B.K.; O’Donnell, C.P. Evaluation of Vis-NIR Hyperspectral Imaging as a Process Analytical Tool to Classify Brined Pork Samples and Predict Brining Salt Concentration. J. Food Eng. 2019, 246, 134–140. [Google Scholar] [CrossRef]
- Khan, A.; Munir, M.T.; Yu, W.; Young, B. Wavelength Selection FOR Rapid Identification of Different Particle Size Fractions of Milk Powder Using Hyperspectral Imaging. Sensors 2020, 20, 4645. [Google Scholar] [CrossRef]
- Erkinbaev, C.; Derksen, K.; Paliwal, J. Single Kernel Wheat Hardness Estimation Using near Infrared Hyperspectral Imaging. Infrared Phys. Technol. 2019, 98, 250–255. [Google Scholar] [CrossRef]
- Khalid, K.H.; Ohm, J.B.; Simsek, S. Influence of Bread-Making Method, Genotype, and Growing Location on Whole-Wheat Bread Quality in Hard Red Spring Wheat. Cereal Chem. 2022, 99, 467–481. [Google Scholar] [CrossRef]
- Erkinbaev, C.; Henderson, K.; Paliwal, J. Discrimination of Gluten-Free Oats from Contaminants Using near Infrared Hyperspectral Imaging Technique. Food Control 2017, 80, 197–203. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Horwitz, W. Agricultural Chemicals, Contaminants, Drugs. In Official Methods of Analysis of AOAC International; AOAC International: Gaithersburg, MD, USA, 1997; Volume I. [Google Scholar]
- Chaudhry, M.M.A.; Hasan, M.M.; Erkinbaev, C.; Paliwal, J.; Suman, S.; Rodas-Gonzalez, A. Bison Muscle Discrimination and Color Stability Prediction Using Near-Infrared Hyperspectral Imaging. Biosyst. Eng. 2021, 209, 1–13. [Google Scholar] [CrossRef]
- López-Maestresalas, A.; Keresztes, J.C.; Goodarzi, M.; Arazuri, S.; Jarén, C.; Saeys, W. Non-Destructive Detection of Blackspot in Potatoes by Vis-NIR and SWIR Hyperspectral Imaging. Food Control 2016, 70, 229–241. [Google Scholar] [CrossRef]
- Kandpal, L.M.; Lee, S.; Kim, M.S.; Bae, H.; Cho, B.K. Short Wave Infrared (SWIR) Hyperspectral Imaging Technique for Examination of Aflatoxin B1 (AFB1) on Corn Kernels. Food Control 2015, 51, 171–176. [Google Scholar] [CrossRef]
- Ambrose, A.; Cho, B.-K. A Review of Technologies for Detection and Measurement of Adulterants in Cereals and Cereal Products. J. Biosyst. Eng. 2014, 39, 357–365. [Google Scholar] [CrossRef]
- Do Nascimento, J.F.; Hein, P.R.G.; Davide, A.C.; De Melo, L.A.; Trugilho, P.F. Essential Oil Content in Eremanthus Erythropappus Wood Powder Can Be Estimated Using near Infrared Spectroscopy. J. Near Infrared Spectrosc. 2015, 23, 33–39. [Google Scholar] [CrossRef]
- Xing, Q.; Kyriakopoulou, K.; Zhang, L.; Boom, R.M.; Schutyser, M.A.I. Protein Fortification of Wheat Bread Using Dry Fractionated Chickpea Protein-Enriched Fraction or Its Sourdough. LWT 2021, 142, 110931. [Google Scholar] [CrossRef]
- Boukid, F.; Zannini, E.; Carini, E.; Vittadini, E. Pulses for Bread Fortification: A Necessity or a Choice? Trends Food Sci. Technol. 2019, 88, 416–428. [Google Scholar] [CrossRef]
- Mohammed, I.; Ahmed, A.R.; Senge, B. Effects of Chickpea Flour on Wheat Pasting Properties and Bread Making Quality. J. Food Sci. Technol. 2014, 51, 1902–1910. [Google Scholar] [CrossRef] [PubMed]
- Man, S.; Păucean, A.; Muste, S.; Pop, A. Effect of the Chickpea (Cicer arietinum L.) Flour Addition on Physicochemical Properties of Wheat Bread. Bull. Univ. Agric. Sci. Vet. Med. Cluj-Napoca. Food Sci. Technol. 2015, 72, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Chen, J.; Zhang, J.; Fu, F.; Wu, C. Effect of Penetration Depth and Particle Size on Detection of Wheat Flour Adulterant Using Hyperspectral Imaging. Biosyst. Eng. 2021, 204, 64–78. [Google Scholar] [CrossRef]
- Fu, X.; Kim, M.S.; Chao, K.; Qin, J.; Lim, J.; Lee, H.; Garrido-Varo, A.; Pérez-Marín, D.; Ying, Y. Detection of Melamine in Milk Powders Based on NIR Hyperspectral Imaging and Spectral Similarity Analyses. J. Food Eng. 2014, 124, 97–104. [Google Scholar] [CrossRef]
- Lin, S.; Gao, J.; Jin, X.; Wang, Y.; Dong, Z.; Ying, J.; Zhou, W. Whole-Wheat Flour Particle Size Influences Dough Properties, Bread Structure and in Vitro Starch Digestibility. Food Funct. 2020, 11, 3610–3620. [Google Scholar] [CrossRef]
- Esteller, M.S.; Zancanaro, O.; Nunes, C.; Palmeira, S.; Caetano, S.; Lannes, S. The Effect of Kefir Addition on Microstructure Parameters and Physical Properties of Porous White Bread. Eur. Food Res. Technol. 2005, 222, 26–31. [Google Scholar] [CrossRef]
- Hansen, P.M.; Jørgensen, J.R.; Thomsen, A. Predicting Grain Yield and Protein Content in Winter Wheat and Spring Barley Using Repeated Canopy Reflectance Measurements and Partial Least Squares Regression. J. Agric. Sci. 2002, 139, 307–318. [Google Scholar] [CrossRef]
- Navea, S.; Tauler, R.; De Juan, A. Application of the Local Regression Method Interval Partial Least-Squares to the Elucidation of Protein Secondary Structure. Anal. Biochem. 2005, 336, 231–242. [Google Scholar] [CrossRef]
- Sharabiani, V.R.; Nazarloo, A.S.; Taghinezhad, E. Prediction of Protein Content of Winter Wheat by Canopy of Near Infrared Spectroscopy (NIRS), Using Partial Least Squares Regression (PLSR) and Artificial Neural Network (ANN) Models. Yuz. Yıl Univ. J. Agric. Sci. 2019, 29, 43–51. [Google Scholar] [CrossRef]
- Macciotta, N.P.P.; Dimauro, C.; Bacciu, N.; Fresi, P.; Cappio-Borlino, A. Use of a Partial Least-Squares Regression Model to Predict Test Day of Milk, Fat and Protein Yields in Dairy Goats. Anim. Sci. 2006, 82, 463–468. [Google Scholar] [CrossRef][Green Version]
- Brestich, N.; Rüdt, M.; Büchler, D.; Hubbuch, J. Selective Protein Quantification for Preparative Chromatography Using Variable Pathlength UV/Vis Spectroscopy and Partial Least Squares Regression. Chem. Eng. Sci. 2018, 176, 157–164. [Google Scholar] [CrossRef]
- Vongsvivut, J.; Heraud, P.; Zhang, W.; Kralovec, J.A.; McNaughton, D.; Barrow, C.J. Rapid Determination of Protein Contents in Microencapsulated Fish Oil Supplements by ATR-FTIR Spectroscopy and Partial Least Square Regression (PLSR) Analysis. Food Bioprocess Technol. 2014, 7, 265–277. [Google Scholar] [CrossRef]
- Cheng, J.-H.; Sun, D.-W. Partial Least Squares Regression (PLSR) Applied to NIR and HSI Spectral Data Modeling to Predict Chemical Properties of Fish Muscle. Food Eng. Rev. 2016, 9, 36–49. [Google Scholar] [CrossRef]
- Flynn, C.C.K.; Frazier, A.E.; Admas, S. Nutrient Prediction for Tef (Eragrostis tef) Plant and Grain with Hyperspectral Data and Partial Least Squares Regression: Replicating Methods and Results across Environments. Remote Sens. 2020, 12, 2867. [Google Scholar] [CrossRef]
- Wilcox, K.E.; Blanch, E.W.; Doig, A.J. Determination of Protein Secondary Structure from Infrared Spectra Using Partial Least-Squares Regression. Biochemistry 2016, 55, 3794–3802. [Google Scholar] [CrossRef]
- Mahesh, S.; Jayas, D.S.; Paliwal, J.; White, N.D.G. Comparison of Partial Least Squares Regression (PLSR) and Principal Components Regression (PCR) Methods for Protein and Hardness Predictions Using the Near-Infrared (NIR) Hyperspectral Images of Bulk Samples of Canadian Wheat. Food Bioprocess Technol. 2015, 8, 31–40. [Google Scholar] [CrossRef]
- Bonfatti, V.; Tiezzi, F.; Miglior, F.; Carnier, P. Comparison of Bayesian Regression Models and Partial Least Squares Regression for the Development of Infrared Prediction Equations. J. Dairy Sci. 2017, 100, 7306–7319. [Google Scholar] [CrossRef]

| Ingredients | Mixing Proportions (%) |
|---|---|
| Flour blends (14%, db) | 100 |
| Water | Farinograph + 1% |
| Yeast, fresh | 4.0 |
| Sugar, refined | 4.0 |
| Salt, refined | 1.2 |
| Whey, powder | 4.0 |
| Shortening | 3.0 |
| Ammonium phosphate | 0.1 |
| Doh-Tone 2 | 0.03 |
| Ascorbic acid | 60 ppm |
| Gluten | 4.0 |
| Calibration | Actual Class | N | Global | |||
| High Protein | Low Protein | SENS | SPEC | |||
| Predicted class | High protein | 142 | 0 | 142 | 1.000 | 1.000 |
| Low protein | 0 | 109 | 109 | 1.000 | 1.000 | |
| Total | 251 | |||||
| Cross Validation | Actual class | N | Global | |||
| High protein | Low protein | SENS | SPEC | |||
| Predicted class | High protein | 142 | 1 | 142 | 1.000 | 0.991 |
| Low protein | 0 | 108 | 109 | 0.992 | 1.000 | |
| Total | 251 | |||||
| External Prediction | Actual class | N | Global | |||
| High protein | Low protein | SENS | SPEC | |||
| Predicted class | High protein | 72 | 0 | 73 | 0.986 | 1.000 |
| Low protein | 1 | 35 | 35 | 1.000 | 0.986 | |
| Total | 108 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olakanmi, S.J.; Jayas, D.S.; Paliwal, J.; Chaudhry, M.M.A.; Findlay, C.R.J. Quality Characterization of Fava Bean-Fortified Bread Using Hyperspectral Imaging. Foods 2024, 13, 231. https://doi.org/10.3390/foods13020231
Olakanmi SJ, Jayas DS, Paliwal J, Chaudhry MMA, Findlay CRJ. Quality Characterization of Fava Bean-Fortified Bread Using Hyperspectral Imaging. Foods. 2024; 13(2):231. https://doi.org/10.3390/foods13020231
Chicago/Turabian StyleOlakanmi, Sunday J., Digvir S. Jayas, Jitendra Paliwal, Muhammad Mudassir Arif Chaudhry, and Catherine Rui Jin Findlay. 2024. "Quality Characterization of Fava Bean-Fortified Bread Using Hyperspectral Imaging" Foods 13, no. 2: 231. https://doi.org/10.3390/foods13020231
APA StyleOlakanmi, S. J., Jayas, D. S., Paliwal, J., Chaudhry, M. M. A., & Findlay, C. R. J. (2024). Quality Characterization of Fava Bean-Fortified Bread Using Hyperspectral Imaging. Foods, 13(2), 231. https://doi.org/10.3390/foods13020231






