Identification, Safety Assessment, and Antimicrobial Characteristics of Cocci Lactic Acid Bacteria Isolated from Traditional Egyptian Dairy Products
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Isolation of Lactic Acid Bacteria
2.3. Identification and Cluster Analysis
2.3.1. DNA Extraction
2.3.2. PCR Amplification and Cluster Analysis
2.3.3. PCR and Sequencing
2.4. Hemolytic Activity
2.5. Antibiotic Susceptibility
2.6. Screening of Bacteriocin-like Inhibitory Substance (BLIS)-Producing LAB Strains
2.6.1. Preparation of Cell-Free Supernatants (CFSs)
2.6.2. Antibacterial Activity
2.6.3. Statistical Analysis
3. Results and Discussion
3.1. Lactic Acid Bacterial Content from Different Dairy Products
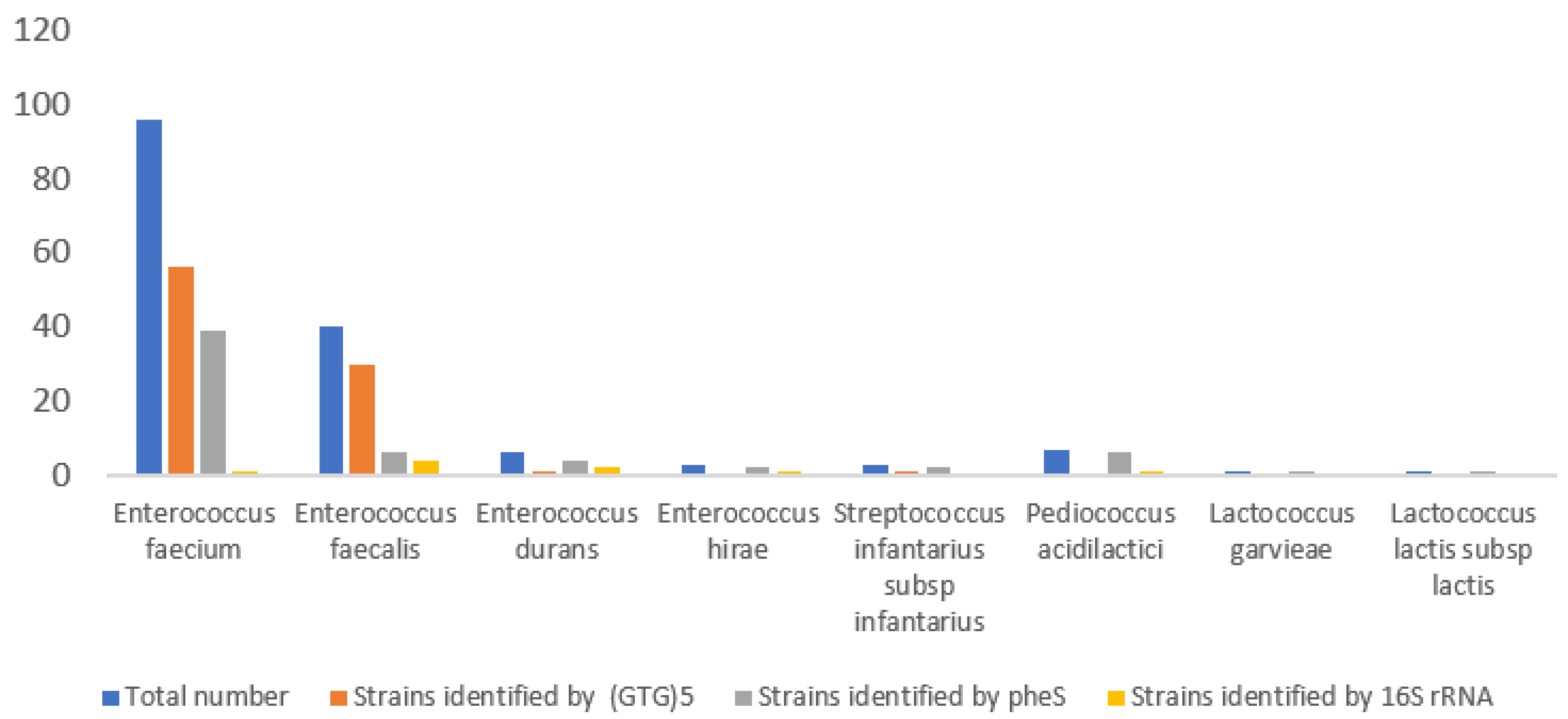
Strain Typing via Genotypic Methods
3.2. Hemolytic Activity
3.3. Antibiotic Resistance
3.4. Antibacterial Effect of LAB
Bacteriocin Production and Antagonistic Activity
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Awad, S.; Ahmed, N.; El Soda, M. Influence of microfiltration and adjunct culture on quality of Domiati cheese. J. Dairy Sci. 2010, 1, 1807–1814. [Google Scholar] [CrossRef] [PubMed]
- Aguirre-Garcia, Y.L.; Nery-Flores, S.D.; Campos-Muzquiz, L.G.; Flores-Gallegos, A.C.; Palomo-Ligas, L.; Ascacio-Valdés, J.A.; Sepúlveda-Torres, L.; Rodríguez-Herrera, R. Lactic Acid Fermentation in the Food Industry and Bio-Preservation of Food. Fermentation 2024, 15, 168. [Google Scholar] [CrossRef]
- Cuevas-González, P.F.; Reyes-Díaz, R.; Santiago-López, L.; Vallejo-Cordoba, B.; Hernández-Mendoza, A.; Beltrán-Barrientos, L.M.; González-Córdova, A.F. Microbiological quality and native lactic acid bacteria diversity of artisanal Mexican cheeses: A review. Food Res. Int. 2024, 6, 114876. [Google Scholar] [CrossRef]
- El-Soda, M.; El-Ziney, M.; Awad, S.; Osman, G.; Omran, N.; Gamal, G.; Ezzat, N.; El-Shafei, H. A culture collection of lactic acid bacteria isolated from raw milk and traditional Egyptian dairy products. Egypt. J. Dairy Sci. 2003, 31, 23–41. [Google Scholar]
- García-Díez, J.; Saraiva, C. Use of starter cultures in foods from animal origin to improve their safety. Int. J. Environ. Res. Public Health 2021, 4, 2544. [Google Scholar] [CrossRef]
- Maiouet, I.; Mahi, K.E.; Abouloifa, H.; Rhallabi, N. Technological characterization of lactic acid bacteria isolated from raw milk. Interactions 2024, 245, 218. [Google Scholar] [CrossRef]
- Fernandes, N.; Faria, A.S.; Carvalho, L.; Choupina, A.; Rodrigues, C.; Gonzales-Barron, U.; Cadavez, V. Genetic Identification and Technological Potential of Indigenous Lactic Acid Bacteria Isolated from Alheira, a Traditional Portuguese Sausage. Foods 2024, 16, 598. [Google Scholar] [CrossRef]
- Kagkli, D.M.; Vancanneyt, M.; Hill, C.; Vandamme, P.; Cogan, T.M. Enterococcus and Lactobacillus contamination of raw milk in a farm dairy environment. Int. J. Food Microbiol. 2007, 10, 243–251. [Google Scholar] [CrossRef]
- Vandamme, P.; Pot, B.; Gillis, M.; De Vos, P.; Kersters, K.; Swings, J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Micro Rev. 1996, 60, 407–438. [Google Scholar] [CrossRef]
- Svec, P.; Vancanneyt, M.; Seman, M.; Snauwaert, C.; Lefebvre, K.; Sedlacek, I.; Swings, J. Evaluation of (GTG)5-PCR for identification of Enterococcus spp. FEMS Microbiol. Lett. 2005, 247, 59–63. [Google Scholar]
- Naser, S.; Thompson, F.L.; Hoste, B.; Gevers, D.; Vandemeulebroecke, K.; Cleenwerck, I.; Thompson, C.C.; Vancanneyt, M.; Swings, J. Phylogeny and identification of enterococci using atpA gene sequence analysis. J. Clin. Microbiol. 2005, 43, 2224–2230. [Google Scholar] [CrossRef] [PubMed]
- Naser, S.M.; Thompson, F.L.; Hoste, B.; Gevers, D.; Dawyndt, P.; Vancanneyt, M.; Swings, J. Application of multilocus sequence analysis (MLSA) for rapid identification of Enterococcus species based on rpoA and pheS genes. Microbiology 2005, 151, 2141–2150. [Google Scholar] [CrossRef] [PubMed]
- Malek, R.; El-Attar, A.; Mohamed, M.; Anwar, S.; El-Soda, M.; Béal, C. Technological and safety properties display biodiversity among enterococci isolated from two Egyptian cheeses, “Ras” and “Domiati”. Int. J. Food Microbiol. 2012, 153, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Lee, D.H.; Seo, H.S.; Eun, S.H.; Lee, D.S.; Choi, Y.K.; Kim, T.Y. Genome-based taxonomic identification and safety assessment of an Enterococcus strain isolated from a homemade dairy product. Int. Microbiol. 2024, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Dapkevicius, M.D.L.E.; Sgardioli, B.; Câmara, S.P.; Poeta, P.; Malcata, F.X. Current trends of enterococci in dairy products: A comprehensive review of their multiple roles. Foods 2021, 10, 821. [Google Scholar] [CrossRef]
- Smout, E.; Palanisamy, N.; Valappil, S.P. Prevalence of vancomycin-resistant Enterococci in India between 2000 and 2022: A systematic review and meta-analysis. Antimicrob. Resist. Infect. Control 2023, 12, 79. [Google Scholar] [CrossRef]
- Teshome, E.; Forsido, S.F.; Rupasinghe, H.V.; Olika Keyata, E. Potentials of natural preservatives to enhance food safety and shelf life: A review. Sci. World J. 2022, 1, 990–1018. [Google Scholar] [CrossRef]
- Todorov, S.D.; Dioso, C.M.; Liong, M.T.; Nero, L.A.; Khosravi-Darani, K.; Ivanova, I.V. Beneficial features of pediococcus: From starter cultures and inhibitory activities to probiotic benefits. World J. Microbiol. Biotech. 2023, 39, 4. [Google Scholar] [CrossRef]
- Xiao, H.; Molina, G.E.S.; Tovar, M.; Quoc, H.M.; Hansen, E.B.; Bang-Berthelsen, C.H. Isolation and charac terization of plant-based lactic acid bacteria from spontaneously fermented foods using a new modified medium. LWT Food Sci. Technol. 2024, 192, 115695. [Google Scholar] [CrossRef]
- Adugna, M.; Andualem, B. Isolation, characterization and safety assessment of probiotic lactic acid bacteria from metata ayib (Traditional spiced cottage cheese). Food Hum. 2023, 1, 85–91. [Google Scholar] [CrossRef]
- Dosuky, A.S.; Elsayed, T.R.; Yousef, E.T.; Olfat Sayed Barakat, O.S.; Nasr Fawzy Nasr, N.F. Isolation, identification, and application of lactic acid-producing bacteria using salted cheese whey substrate and immobilized cells technology. J. Genet. Eng. Biotechnol. 2022, 20, 26. [Google Scholar] [CrossRef] [PubMed]
- Gevers, D.; Huys, G.; Swings, J. Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiol. Lett. 2001, 205, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Maragkoudakis, P.A.; Zoumpopoulou, G.; Miaris, C.; Kalantzopoulos, G.; Pot, B.; Tsakalidou, E. Probiotic potential of Lactobacillus strains isolated from dairy products. Int. Dairy J. 2006, 16, 189–199. [Google Scholar] [CrossRef]
- Fortina, M.G.; Ricci, G.; Borgo, F.; Manachini, P.L.; Arends, K.; Schiwon, K.; Grohmann, E. A survey on biotechnological potential and safety of the novel Enterococcus species of dairy origin, E. italicus. Int. J. Food Microbiol. 2008, 123, 204–211. [Google Scholar] [CrossRef]
- Ren, S.; Yuan, X.; Liu, F.; Fang, F.; Iqbal, H.M.; Zahran, S.A.; Bilal, M. Bacteriocin from Lacticaseibacillus rhamnosus sp. A5: Isolation, purification, characterization, and antibacterial evaluation for sustainable food processing. Sustainability 2022, 4, 9571. [Google Scholar] [CrossRef]
- Tejero-Sariñena, S.; Barlow, J.; Costabile, A.; Gibson, G.R.; Rowland, I. In vitro evaluation of the antimicrobial activity of a range of probiotics against pathogens: Evidence for the effects of organic acids. Anaerobe 2012, 18, 530–538. [Google Scholar] [CrossRef]
- Awad, S. Texture and flavour development in Ras cheese made from raw and pasteurised milk. Food Chem. 2006, 97, 394–400. [Google Scholar] [CrossRef]
- Awad, S.; Ahmed, N.; El Soda, M. Evaluation of isolated starter lactic acid bacteria in Ras cheese ripening and flavour development. Food Chem. 2007, 104, 1192–1199. [Google Scholar] [CrossRef]
- Bintsis, T.; Papademas, P. The Application of Protective Cultures in Cheese: A Review. Fermentation 2004, 10, 117. [Google Scholar] [CrossRef]
- Ayad, E.H.E.; Nashat, S.; El-Sadek, N.; Metwaly, H.; El-Soda, M. Selection of wild lactic acid bacteria isolated from traditional Egyptian dairy products according to production and technological criteria. Food Microbiol. 2004, 21, 715–725. [Google Scholar] [CrossRef]
- El-Baradei, G.; Delacroix-Buchet, A.; Ogier, J.C. Biodiversity of bacterial ecosystems in traditional Egyptian Domiati cheese. Appl. Environ. Microbiol. 2007, 73, 1248–1255. [Google Scholar] [CrossRef] [PubMed]
- Ouadghiri, M.; Vancanneyt, M.; Vandamme, P.; Naser, S.; Gevers, D.; Lefebvre, K.; Amar, M. Identification of lactic acid bacteria in Moroccan raw milk and traditionally fermented skimmed milk ‘lben’. J. Appl. Microbiol. 2009, 106, 486–495. [Google Scholar] [CrossRef] [PubMed]
- Abdelgadir, W.; Nielsen, D.S.; Hamad, S.; Jakobsen, M. A traditional Sudanese fermented camel’s milk product, Gariss, as a habitat of Streptococcus infantarius subsp. infantarius. Int. J. Food Microbiol. 2008, 127, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Gboko, K.D.; Traoré, S.G.; Sanhoun, A.R.; Kirioua, J.; Otaru, N.; Kurt, F.; Bonfoh, B. Risk factors for the carriage of Streptococcus infantarius subspecies infantarius isolated from African fermented dairy products. PLoS ONE 2019, 14, e0225452. [Google Scholar] [CrossRef]
- Dos Santos, K.M.O.; de Matos, C.R.; Salles, H.O.; de Melo Franco, B.D.G.; Arellano, K.; Holzapfel, W.H.; Todorov, S.D. Exploring beneficial/virulence properties of two dairy-related strains of Streptococcus infantarius subsp. infantarius. Probiotics Antimicrob. Proteins 2020, 12, 1524–1541. [Google Scholar] [CrossRef]
- De Vuyst, L.; Leroy, F. Bacteriocins from lactic acid bacteria: Production, purification, and food applications. J. Microbiol. Biotechnol. 2007, 13, 194–199. [Google Scholar] [CrossRef]
- El-Ghaish, S.; El-Baz, A.; Hwanhlem, N.; Zommara, M.; Ayad, E.; Choiset, Y.; Chobert, J.M. Bacteriocin production and safety evaluation of non-starter Enterococcus faecium IM1 and Enterococcus hirae IM1 strains isolated from homemade Egyptian dairy products. Eur. Food Res. Technol. 2015, 240, 1211–1223. [Google Scholar] [CrossRef]
- Peters, J.; Mac, K.; Wichmann-Schauer, H.; Klein, G.; Ellerbroek, L. Species distribution and antibiotic resistance patterns of enterococci isolated from food of animal origin in Germany. Int. J. Food Microbiol. 2003, 88, 311–314. [Google Scholar] [CrossRef]
- Russo, N.; Caggia, C.; Pino, A.; Coque, T.M.; Arioli, S.; Randazzo, C.L. Enterococcus spp. in ragusano PDO and pecorino siciliano cheese types: A snapshot of their antibiotic resistance distribution. Food Chem. Toxicol. 2018, 120, 277–286. [Google Scholar] [CrossRef]
- Mannu, L.; Paba, A.; Daga, E.; Comunian, R.; Zanetti, S.; Duprè, I.; Sechi, L.A. Comparison of the incidence of virulence determinants and antibiotic resistance between Enterococcus faecium strains of dairy, animal and clinical origin. Int. J. Food Microbiol. 2003, 88, 291–304. [Google Scholar] [CrossRef]
- Franciosi, E.; Settanni, L.; Cavazza, A.; Poznanski, E. Biodiversity and technological potential of wild lactic acid bacteria from raw cows’ milk. Int. Dairy J. 2009, 19, 3–11. [Google Scholar] [CrossRef]
- Valenzuela, A.S.; Omar, N.B.; Abriouel, H.; López, R.L.; Ortega, E.; Cañamero, M.M.; Gálvez, A. Risk factors in enterococci isolated from foods in Morocco: Determination of antimicrobial resistance and incidence of virulence traits. Food Chem. Toxicol. 2008, 46, 2648–2652. [Google Scholar] [CrossRef]
- Favaro, L.; Basaglia, M.; Casella, S.; Hue, I.; Dousset, X.; de Melo Franco, B.D.G.; Todorov, S.D. Bacteriocinogenic potential and safety evaluation of non-starter Enterococcus faecium strains isolated from home made white brine cheese. Food Microbiol. 2014, 38, 228–239. [Google Scholar] [CrossRef]
- Zuo, F.L.; Feng, X.J.; Chen, L.L.; Chen, S.W. Identification and partial characterization of lactic acid bacteria isolated from traditional dairy products produced by herders in the western Tianshan Mountains of China. Lett. Appl. Microbiol. 2014, 59, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Sireswar, S.; Dey, G.; Sreesoundarya, T.K.; Sarkar, D. Design of probiotic-fortified food matrices influence their antipathogenic potential. Food Biosci. 2017, 20, 28–35. [Google Scholar] [CrossRef]
- Alnakip, M.E.; Quintela-Baluja, M.; Böhme, K.; Caamaño-Antelo, S.; Bayoumi, M.A.; Kamal, R.M.; Barros-Velázquez, J. Molecular characterisation and typing the methicillin resistance of Staphylococcus spp. isolated from raw milk and cheeses in northwest Spain: A mini survey. Int. Dairy J. 2019, 89, 68–76. [Google Scholar] [CrossRef]
- Choi, G.H.; Fugaban, J.I.; Dioso, C.M.; Bucheli, J.E.; Holzapfel, W.H.; Todorov, S.D. Antimicrobial peptides (bacteriocins) produced by Lactococcus lactis and Pediococcus pentosaceus strains with activity against clinical and food-borne pathogens. Probiotics Antimicrob. Proteins 2023, 1, 1–22. [Google Scholar] [CrossRef]
- Zaghloul, E.H.; Halfawy, N.M. Marine Pediococcus pentosaceus E3 Probiotic Properties, Whole-Genome Sequence Analysis, and Safety Assessment. Probiotics Antimicrob. Proteins 2024, 15, 1–2. [Google Scholar] [CrossRef]
- Lima, J.M.; Carneiro, K.O.; Pinto, U.M.; Todorov, S.D. Bacteriocinogenic anti-listerial properties and safety assessment of Enterococcus faecium and Lactococcus garvieae strains isolated from Brazilian artisanal cheesemaking environment. J. Appl. Microbiol. 2024, 135, 159. [Google Scholar] [CrossRef]
- dos Santos Nascimento, J.; Fagundes, P.C.; de Paiva Brito, M.A.V.; Dos Santos, K.R.N.; de Freire Bastos, M.D.C. Production of bacteriocins by coagulase-negative staphylococci involved in bovine mastitis. Vet. Microbiol. 2005, 106, 61–71. [Google Scholar] [CrossRef]
- Wang, J.; Li, L.; Zhao, X.; Zhou, Z. Partial characteristics and antimicrobial mode of pediocin produced by Pediococcus acidilactici PA003. Ann. Microbiol. 2015, 65, 1753–1762. [Google Scholar] [CrossRef]
- Choeisoongnern, T.; Sirilun, S.; Waditee-Sirisattha, R.; Pintha, K.; Peerajan, S.; Chaiyasut, C. Potential probiotic Enterococcus faecium OV3-6 and its bioactive peptide as alternative bio-preservation. Foods 2021, 24, 2264. [Google Scholar] [CrossRef] [PubMed]
- Lengkey, H.A.; Balia, R.L.; Siwi, J.A.; Tasbac, B.A.; Togoe, I. Isolation and identification of Lactobacillus bacteria from culled hens meat for meat biopreservator. KnE Life Sci. 2017, 26, 518–528. [Google Scholar] [CrossRef]
| Primer Name | Sequence (5′–3′) | Position |
|---|---|---|
| pheS-21-F | CAYCCNGCHCGYGAYATGC | 557 |
| pheS-22-R | CCWARVCCRAARGCAAARCC | 1031 |
| pheS-23-R | GGRTGRACCATVCCNGCHCC | 968 |
| Dairy Products | Number of Cocci Isolates | Number of Identified Strains | Identified Strains | Method of Confirmation | ||
|---|---|---|---|---|---|---|
| GTG5 | pheS | 16S | ||||
| Buffalo milk | 4 | 4 | Enterococcus faecium | 1 | 3 | |
| Raw cow milk | 20 | 1 | St. infantarius subsp. infantarius | 1 | ||
| 13 | Enterococcus faecium | 7 | 6 | |||
| 5 | Enterococcus faecalis | 5 | ||||
| 1 | Enterococcus durans | 1 | ||||
| Domiati cheese | 19 | 12 | Enterococcus faecium | 6 | 5 | 1 |
| 4 | Enterococcus faecalis | 4 | ||||
| 1 | Lact garvieae | 1 | ||||
| 1 | Lc. Lactis subsp. lactis | 1 | ||||
| 1 | St. infantarius subsp. infantarius | 1 | ||||
| Karish cheese | 9 | 4 | Enterococcus faecium | 3 | 1 | |
| 4 | Enterococcus faecalis | 4 | ||||
| 1 | Enterococcus durans | 1 | ||||
| Kishk | 8 | 4 | Enterococcus faecium | 1 | 3 | |
| 1 | Enterococcus faecalis | 1 | ||||
| 1 | Enterococcus durans | 1 | ||||
| 2 | Pediococcus acidilactici | 1 | 1 | |||
| Ras cheese | 58 | 39 | Enterococcus faecium | 25 | 14 | |
| 14 | Enterococcus faecalis | 6 | 5 | 3 | ||
| 3 | Pediococcus acidilactici | 3 | ||||
| 2 | Enterococcus hirae | 1 | 1 | |||
| Rayab | 36 | 1 | St. infantarius subsp. infantarius | 1 | ||
| 18 | Enterococcus faecium | 12 | 6 | |||
| 12 | Enterococcus faecalis | 11 | 1 | |||
| 1 | Pediococcus acidilactici | 1 | ||||
| 3 | Enterococcus durans | 1 | 2 | |||
| 1 | Enterococcus hirae | 1 | ||||
| Ras whey | 2 | 1 | Enterococcus faecium | 1 | ||
| 1 | Pediococcus acidilactici | 1 | ||||
| Zabady | 1 | 1 | Enterococcus faecium | 1 | ||
| 157 | 157 | 89 | 60 | 8 | ||
| Total Number | β-Hemolysis | α-Hemolysis | Total | % β-Hemolysis | % α-Hemolysis | %Total | |
|---|---|---|---|---|---|---|---|
| Enterococcus faecium | 96 | 10 | 6 | 16 | 10.42 | 6.25 | 16.67 |
| Enterococcus faecalis | 40 | 2 | 1 | 3 | 5.00 | 2.50 | 7.50 |
| Enterococcus durans | 6 | 1 | 1 | 2 | 16.67 | 16.67 | 33.33 |
| Enterococcus hirae | 3 | 1 | 0 | 1 | 33.33 | 0.00 | 33.33 |
| Streptococcus infantarius subsp. infantarius | 3 | 0 | 1 | 1 | 0.00 | 33.33 | 33.33 |
| Pediococcus acidilactici | 7 | 0 | 0 | 0 | 0.00 | 0.00 | 0.00 |
| Lactococcus garvieae | 1 | 0 | 0 | 0 | 0.00 | 0.00 | 0.00 |
| Lactococcus lactis subsp. lactis | 1 | 0 | 0 | 0 | 0.00 | 0.00 | 0.00 |
| Total | 157 | 14 | 9 | 23 | 8.92 | 5.73 | 14.65 |
| Antibiotic Tests | En. faecium | En. faecalis | En. durans | En. hirae | St. infantarius Subsp infantarius | Ped. cidilactici | Lac. garvieae | L. lact. subsp. lact. | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R | S | R | S | R | S | R | S | R | S | R | S | R | S | R | S | |
| Number of tested strains | 80 | 37 | 4 | 2 | 2 | 7 | 1 | 1 | ||||||||
| Ampicillin | 2 | 78 | 4 | 33 | 0 | 4 | 0 | 2 | 0 | 2 | 0 | 7 | 0 | 1 | 0 | 1 |
| Chloramphenicol | 8 | 72 | 13 | 24 | 0 | 4 | 0 | 2 | 0 | 2 | 0 | 7 | 0 | 1 | 0 | 1 |
| Gentamicin | 7 | 73 | 25 | 12 | 0 | 4 | 0 | 2 | 0 | 2 | 0 | 7 | 0 | 1 | 0 | 1 |
| Norfloxacin | 22 | 58 | 15 | 22 | 0 | 4 | 0 | 2 | 0 | 2 | 0 | 7 | 0 | 1 | 0 | 1 |
| Nitrofurantoin | 6 | 74 | 3 | 34 | 0 | 4 | 0 | 2 | 0 | 2 | 0 | 7 | 0 | 1 | 0 | 1 |
| Fusidic acid | 2 | 78 | 3 | 34 | 0 | 4 | 0 | 2 | 0 | 2 | 0 | 7 | 0 | 1 | 0 | 1 |
| Lincomycin | 1 | 79 | 1 | 36 | 0 | 4 | 0 | 2 | 0 | 2 | 0 | 7 | 0 | 1 | 0 | 1 |
| Ofloxacin | 1 | 79 | 1 | 36 | 0 | 4 | 0 | 2 | 0 | 2 | 0 | 7 | 0 | 1 | 0 | 1 |
| Penicillin G | 0 | 80 | 0 | 37 | 0 | 4 | 0 | 2 | 0 | 2 | 0 | 7 | 0 | 1 | 0 | 1 |
| Pristinamycin | 0 | 80 | 0 | 37 | 0 | 4 | 0 | 2 | 0 | 2 | 0 | 7 | 0 | 1 | 0 | 1 |
| Rifampicin | 22 | 58 | 15 | 22 | 0 | 4 | 0 | 2 | 0 | 2 | 0 | 7 | 0 | 1 | 0 | 1 |
| Tetracycline | 9 | 71 | 27 | 10 | 0 | 4 | 0 | 2 | 0 | 2 | 0 | 7 | 0 | 1 | 0 | 1 |
| Vancomycin | 0 | 80 | 0 | 37 | 0 | 4 | 0 | 2 | 0 | 2 | 0 | 7 | 0 | 1 | 0 | 1 |
| Streptomycin | 0 | 80 | 0 | 37 | 0 | 4 | 0 | 2 | 0 | 2 | 0 | 7 | 0 | 1 | 0 | 1 |
| Nalidixic acid | 0 | 80 | 0 | 37 | 0 | 4 | 0 | 2 | 0 | 2 | 0 | 7 | 0 | 1 | 0 | 1 |
| Cefotaxime | 1 | 79 | 1 | 36 | 0 | 4 | 0 | 2 | 0 | 2 | 0 | 7 | 0 | 1 | 0 | 1 |
| Clindamycin | 8 | 72 | 2 | 35 | 0 | 4 | 0 | 2 | 0 | 2 | 0 | 7 | 0 | 1 | 0 | 1 |
| Erythromycin | 4 | 76 | 1 | 36 | 0 | 4 | 0 | 2 | 0 | 2 | 0 | 7 | 0 | 1 | 0 | 1 |
| Oxacillin | 4 | 76 | 3 | 34 | 0 | 4 | 0 | 2 | 0 | 2 | 0 | 7 | 0 | 1 | 0 | 1 |
| Tobramycin | 7 | 73 | 8 | 29 | 0 | 4 | 0 | 2 | 0 | 2 | 0 | 7 | 0 | 1 | 0 | 1 |
| Kanamycin | 16 | 64 | 7 | 30 | 0 | 4 | 0 | 2 | 0 | 2 | 0 | 7 | 0 | 1 | 0 | 1 |
| Ciprofloxacin | 24 | 56 | 12 | 25 | 0 | 4 | 0 | 2 | 0 | 2 | 0 | 7 | 0 | 1 | 0 | 1 |
| No | Source and Code of Isolate | E. coli ATTC 25922 | Salmonella typhimurium ATTC 14028 | Staphylococcus aureus ATTC 25923 | Listeria monocytogenes ATTC 7644 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crude | pH 7 | Catalase | Crude | pH 7 | Catalase | Crude | pH 7 | Catalase | Crude | pH 7 | Catalase | |||
| 1 | Pediococcus acidilactici | Ras-03 | 22.3 a | 18.9 b | 17.9 b | 17.5 b | 13.8 d | 12.5 d | 9.1 g | 5.6 n | 7.5 l | 8.9 h | 7.8 l | 4.5 m |
| 2 | Enterococcus faecium | Raw milk-62 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | Enterococcus faecium | Domiati-04 | 8.5 h | 0 | 0 | 0 | 0 | 0 | 6.5 l | 0 | 0 | 0.0 | 0 | 0 |
| 4 | Enterococcus faecium | Ras-31 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0 | 0 | 0 |
| 5 | Enterococcus faecalis | Ras-32 | 0 | 0 | 0 | 12.5 d | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 6 | Enterococcus faecium | Raw milk-60 | 11.2 e | 10.5 f | 10.1 fg | 4.6 n | 0 | 0 | 4.5 m | 0 | 0 | 0 | 0 | 0 |
| 7 | Enterococcus faecium | Rayeb-28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | Pediococcus acidilactici | Rayeb-48 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | Enterococcus faecalis | Ras-34 | 22.1 a | 0 | 0 | 6.8 i | 2.3 o | 0 | 21.2 a | 0 | 0 | 0 | 0 | 0 |
| 10 | Enterococcus faecalis | Ras-13 | 0 | 0 | 0 | 0 | 0 | 0 | 18.5 b | 16.5 c | 14.5 d | 12.6 d | 11.6 e | 10.5 f |
| 11 | Lact garvieae | Domiati-81 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | Lc. Lactis subsp. lactis | Domiati-89 | 21.5 a | 18.6 b | 16.1 c | 14.5 d | 12.5 d | 11.9 e | 9.2 g | 8.9 h | 9 gh | 5.2 l | 4.6 l | 4.7 lm |
| 13 | Pediococcus acidilactici | Whey-20 | 8.5 h | 8.4 h | 8.2 h | 6.1 j | 5.8k | 4.9 l | 0 | 0 | 0 | 4.1 m | 3.8 n | 3.4 n |
| 14 | Enterococcus faecalis | Raw milk-76 | 14.2 d | 10 fg | 11.6 e | 8.5 h | 7.9 i | 6.8 j | 6.1k | 0 | 0 | 5.6 kl | 5.2 l | 4.9 l |
| 15 | Enterococcus faecalis | Raw milk-37 | 4.2 m | 0 | 0 | 5.1 l | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | Enterococcus faecium | Rayeb-33 | 0 | 0 | 0 | 4.6 m | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | Enterococcus faecium | Ras-33 | 12.3 e | 10.5 f | 11.6 e | 10.2 f | 7.8 h | 7.2 ij | 12.5 d | 10.5 f | 9.8 fg | 11.4 f | 10.8 f | 9.7 g |
| 18 | Enterococcus faecium | Ras-24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | St. infantarius subsp. infantarius | Rayeb-20 | 8.6 h | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.3 m | 0 | 0 |
| 20 | Enterococcus faecalis | Ras-07 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 21 | Enterococcus faecium | Rayeb-16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 22 | Enterococcus faecium | Rayeb-53 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 23 | Enterococcus faecium | Raw milk-52 | 9.8 g | 2.3 o | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 24 | Enterococcus faecalis | Rayeb-23 | 4.5 lm | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 25 | Enterococcus faecium | Rayeb-21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elsaadany, K.; EL-Sayed, A.I.M.; Awad, S. Identification, Safety Assessment, and Antimicrobial Characteristics of Cocci Lactic Acid Bacteria Isolated from Traditional Egyptian Dairy Products. Foods 2024, 13, 3059. https://doi.org/10.3390/foods13193059
Elsaadany K, EL-Sayed AIM, Awad S. Identification, Safety Assessment, and Antimicrobial Characteristics of Cocci Lactic Acid Bacteria Isolated from Traditional Egyptian Dairy Products. Foods. 2024; 13(19):3059. https://doi.org/10.3390/foods13193059
Chicago/Turabian StyleElsaadany, Khaled, Abeer I. M. EL-Sayed, and Sameh Awad. 2024. "Identification, Safety Assessment, and Antimicrobial Characteristics of Cocci Lactic Acid Bacteria Isolated from Traditional Egyptian Dairy Products" Foods 13, no. 19: 3059. https://doi.org/10.3390/foods13193059
APA StyleElsaadany, K., EL-Sayed, A. I. M., & Awad, S. (2024). Identification, Safety Assessment, and Antimicrobial Characteristics of Cocci Lactic Acid Bacteria Isolated from Traditional Egyptian Dairy Products. Foods, 13(19), 3059. https://doi.org/10.3390/foods13193059





