Evaluation of Simultaneous Growth of Escherichia coli O157:H7, Salmonella spp., and Listeria monocytogenes in Ground Beef Samples in Different Growth Media
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Enrichment Media
2.3. Evaluation of Growth Kinetics in Single Cultures
2.4. Evaluation of Growth Kinetics in Co-Cultures
2.5. Kinetic Parameters Determination
2.6. Performance on Ground Beef Samples
2.7. Statistical Analysis
3. Results and Discussion
3.1. Evaluation of Growth Kinetics in Single Cultures
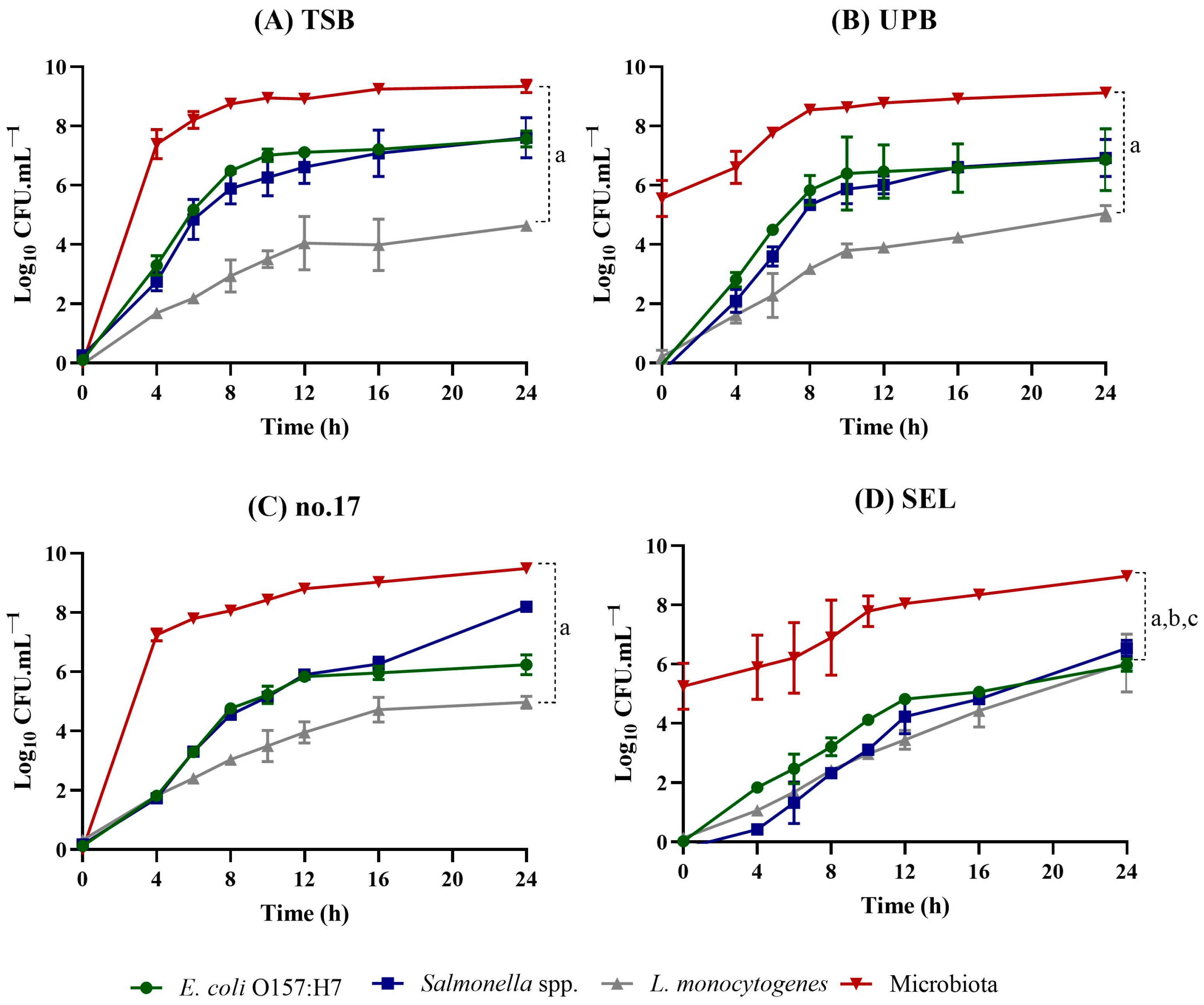
3.2. Evaluation of Growth Kinetics in Co-Cultures
3.3. Performance on Ground Beef Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fung, F.; Wang, H.S.; Menon, S. Food Safety in the 21st Century. Biomed. J. 2018, 41, 88–95. [Google Scholar] [CrossRef]
- Bintsis, T. Foodborne Pathogens. AIMS Microbiol. 2017, 3, 529–563. [Google Scholar] [CrossRef]
- Vieira, K.C.d.O.; Silva, H.R.A.d.; Rocha, I.P.M.; Barboza, E.; Eller, L.K.W. Foodborne Pathogens in the Omics Era. Crit. Rev. Food Sci. Nutr. 2022, 62, 6726–6741. [Google Scholar] [CrossRef]
- Jeon, B.; Luangtongkum, T.; Shen, Z.; Logue, C.M.; Lin, J. Editorial: Resistance and Tolerance in Food-Borne Pathogens: Mechanisms, Public Health Impact, and Control Measures. Front. Microbiol. 2021, 12, 769931. [Google Scholar] [CrossRef]
- Yeni, F.; Yavaş, S.; Alpas, H.; Soyer, Y. Most Common Foodborne Pathogens and Mycotoxins on Fresh Produce: A Review of Recent Outbreaks. Crit. Rev. Food Sci. Nutr. 2016, 56, 1532–1544. [Google Scholar] [CrossRef] [PubMed]
- Abdelhaseib, M.U.; Singh, A.K.; Bhunia, A.K. Simultaneous Detection of Salmonella enterica, Escherichia coli and Listeria monocytogenes in Food Using a Light Scattering Sensor. J. Appl. Microbiol. 2019, 126, 1496–1507. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.A.; Altemimi, A.B.; Alhelfi, N.; Ibrahim, S.A. Application of Biosensors for Detection of Pathogenic Food Bacteria: A Review. Biosensors 2020, 10, 58. [Google Scholar] [CrossRef]
- Bundidamorn, D.; Supawasit, W.; Trevanich, S. A New Single-Tube Platform of Melting Temperature Curve Analysis Based on Multiplex Real-Time PCR Using EvaGreen for Simultaneous Screening Detection of Shiga Toxin-Producing Escherichia coli, Salmonella spp. and Listeria monocytogenes in Food. Food Control 2018, 94, 195–204. [Google Scholar] [CrossRef]
- Jasson, V.; Jacxsens, L.; Luning, P.; Rajkovic, A.; Uyttendaele, M. Alternative Microbial Methods: An Overview and Selection Criteria. Food Microbiol. 2010, 27, 710–730. [Google Scholar] [CrossRef]
- Azinheiro, S.; Carvalho, J.; Prado, M.; Garrido-Maestu, A. Multiplex Detection of Salmonella spp., E. coli O157 and L. monocytogenes by qPCR Melt Curve Analysis in Spiked Infant Formula. Microorganisms 2020, 8, 1359. [Google Scholar] [CrossRef]
- Bhunia, A.K. One Day to One Hour: How Quickly Can Foodborne Pathogens Be Detected? Future Microbiol. 2014, 9, 935–946. [Google Scholar] [CrossRef] [PubMed]
- McMeekin, T.A.; Hill, C.; Wagner, M.; Dahl, A.; Ross, T. Ecophysiology of Food-Borne Pathogens: Essential Knowledge to Improve Food Safety. Int. J. Food Microbiol. 2010, 139, S64–S78. [Google Scholar] [CrossRef] [PubMed]
- Almeida, C.; Azevedo, N.F.; Iversen, C.; Fanning, S.; Keevil, C.W.; Vieira, M.J. Development and Application of a Novel Peptide Nucleic Acid Probe for the Specific Detection of Cronobacter Genomospecies (Enterobacter sakazakii) in Powdered Infant Formula. Appl. Environ. Microbiol. 2009, 75, 2925–2930. [Google Scholar] [CrossRef] [PubMed]
- Rohde, A.; Hammerl, J.A.; Appel, B.; Dieckmann, R.; Al Dahouk, S. FISHing for Bacteria in Food—A Promising Tool for the Reliable Detection of Pathogenic Bacteria? Food Microbiol. 2015, 46, 395–407. [Google Scholar] [CrossRef] [PubMed]
- Ding, T.; Suo, Y.; Zhang, Z.; Liu, D.; Ye, X.; Chen, S.; Zhao, Y. A Multiplex RT-PCR Assay for S. aureus, L. monocytogenes, and Salmonella spp. Detection in Raw Milk with Pre-Enrichment. Front. Microbiol. 2017, 8, 989. [Google Scholar] [CrossRef] [PubMed]
- Suo, B.; Wang, Y. Evaluation of a Multiplex Selective Enrichment Broth SEL for Simultaneous Detection of Injured Salmonella, Escherichia coli O157:H7 and Listeria monocytogenes. Braz. J. Microbiol. 2014, 44, 737–742. [Google Scholar] [CrossRef] [PubMed]
- Wu, V.C.H. A Review of Microbial Injury and Recovery Methods in Food. Food Microbiol. 2008, 25, 735–744. [Google Scholar] [CrossRef] [PubMed]
- Garrido, A.; Chapela, M.J.; Román, B.; Fajardo, P.; Vieites, J.M.; Cabado, A.G. In-House Validation of a Multiplex Real-Time PCR Method for Simultaneous Detection of Salmonella spp., Escherichia coli O157 and Listeria monocytogenes. Int. J. Food Microbiol. 2013, 164, 92–98. [Google Scholar] [CrossRef]
- Hahm, B.K.; Kim, H.; Singh, A.K.; Bhunia, A.K. Pathogen Enrichment Device (PED) Enables One-Step Growth, Enrichment and Separation of Pathogen from Food Matrices for Detection Using Bioanalytical Platforms. J. Microbiol. Methods 2015, 117, 64–73. [Google Scholar] [CrossRef]
- Germini, A.; Masola, A.; Carnevali, P.; Marchelli, R. Simultaneous Detection of Escherichia coli O175:H7, Salmonella spp., and Listeria monocytogenes by Multiplex PCR. Food Control 2009, 20, 733–738. [Google Scholar] [CrossRef]
- Costa-Ribeiro, A.; Lamas, A.; Prado, M.; Garrido-Maestu, A. Evaluation of the Novel MTA10 Selective Broth, MSB, for the Co-Enrichment and Detection of Salmonella spp., Escherichia coli O157 and Listeria monocytogenes in Ready-to-Eat Salad Samples. Foods 2023, 13, 63. [Google Scholar] [CrossRef]
- Wang, Y.; Suo, B. A New 7-Plex PCR Assay for Simultaneous Detection of Shiga Toxin-Producing Escherichia coli O157 and Salmonella Enteritidis in Meat Products. J. Verbraucherschutz Leb. 2011, 6, 441–447. [Google Scholar] [CrossRef]
- Boukharouba, A.; González, A.; García-Ferrús, M.; Ferrús, M.A.; Botella, S. Simultaneous Detection of Four Main Foodborne Pathogens in Ready-to-Eat Food by Using a Simple and Rapid Multiplex PCR (MPCR) Assay. Int. J. Environ. Res. Public Health 2022, 19, 1031. [Google Scholar] [CrossRef] [PubMed]
- Nam, H.M.; Murinda, S.E.; Nguyen, L.T.; Oliver, S.P. Evaluation of Universal Pre-Enrichment Broth for Isolation of Salmonella spp., Escherichia coli O157:H7, and Listeria monocytogenes from Dairy Farm Environmental Samples. Foodborne Pathog. Dis. 2004, 1, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, S.; Horikoshi, N.; Okada, Y.; Takeshita, K.; Sameshima, T.; Kawamoto, S. Multiplex PCR for Simultaneous Detection of Salmonella spp., Listeria monocytogenes, and Escherichia coli O157:H7 in Meat Samples. J. Food Prot. 2005, 68, 551–556. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Bhunia, A.K. SEL, a Selective Enrichment Broth for Simultaneous Growth of Salmonella Enterica, Escherichia coli O157:H7, and Listeria monocytogenes. Appl. Environ. Microbiol. 2008, 74, 4853–4866. [Google Scholar] [CrossRef] [PubMed]
- Omiccioli, E.; Amagliani, G.; Brandi, G.; Magnani, M. A New Platform for Real-Time PCR Detection of Salmonella Spp., Listeria monocytogenes and Escherichia coli O157 in Milk. Food Microbiol. 2009, 26, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Gehring, A.G.; Albin, D.M.; Bhunia, A.K.; Kim, H.; Reed, S.A.; Tu, S.I. Mixed Culture Enrichment of Escherichia coli O157:H7, Listeria monocytogenes, Salmonella Enterica, and Yersinia Enterocolitica. Food Control 2012, 26, 269–273. [Google Scholar] [CrossRef]
- Kim, J.; Demeke, T.; Clear, R.M.; Patrick, S.K. Simultaneous Detection by PCR of Escherichia coli, Listeria monocytogenes and Salmonella Typhimurium in Artificially Inoculated Wheat Grain. Int. J. Food Microbiol. 2006, 111, 21–25. [Google Scholar] [CrossRef]
- Bhaduri, S.; Cottrell, B. Sample Preparation Methods for PCR Detection of Escherichia coli O157:H7, Salmonella Typhimurium, and Listeria monocytogenes on Beef Chuck Shoulder Using a Single Enrichment Medium. Mol. Cell Probes 2001, 15, 267–274. [Google Scholar] [CrossRef]
- Sousa, M.; Rocha, R.; Araújo, D.; Castro, J.; Barbosa, A.; Azevedo, N.F.; Cerqueira, L.; Almeida, C. A New Peptide Nucleic Acid Fluorescence In Situ Hybridization Probe for the Specific Detection of Salmonella Species in Food Matrices. Foodborne Pathog. Dis. 2024, 21, 298–305. [Google Scholar] [CrossRef] [PubMed]
- Ramaswamy, V.; Cresence, V.M.; Rejitha, J.S.; Lekshmi, U.; Dharsana, K.S.; Prasad, P.; Vijila, M. Listeria-Review of Epidemiology and Pathogenesis. J. Microbiol. Immunol. Infect. 2007, 40, 4–13. [Google Scholar] [PubMed]
- Jacobsen, C.N. The Influence of Commonly Used Selective Agents on the Growth of Listeria monocytogenes. Int. J. Food Microbiol. 1999, 50, 221–226. [Google Scholar] [CrossRef]
- Mellefont, L.A.; McMeekin, T.A.; Ross, T. Effect of Relative Inoculum Concentration on Listeria monocytogenes Growth in Co-Culture. Int. J. Food Microbiol. 2008, 121, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Lianou, A.; Koutsoumanis, K.P. Evaluation of the Effect of Defrosting Practices of Ground Beef on the Heat Tolerance of Listeria monocytogenes and Salmonella Enteritidis. Meat Sci. 2009, 82, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Schlisselberg, D.B.; Kler, E.; Kalily, E.; Kisluk, G.; Karniel, O.; Yaron, S. Inactivation of Foodborne Pathogens in Ground Beef by Cooking with Highly Controlled Radio Frequency Energy. Int. J. Food Microbiol. 2013, 160, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Al-Zeyara, S.A.; Jarvis, B.; Mackey, B.M. The Inhibitory Effect of Natural Microflora of Food on Growth of Listeria monocytogenes in Enrichment Broths. Int. J. Food Microbiol. 2011, 145, 98–105. [Google Scholar] [CrossRef]
- Wang, B.; Wang, H.; Lu, X.; Zheng, X.; Yang, Z. Recent Advances in Electrochemical Biosensors for the Detection of Foodborne Pathogens: Current Perspective and Challenges. Foods 2023, 12, 2795. [Google Scholar] [CrossRef]
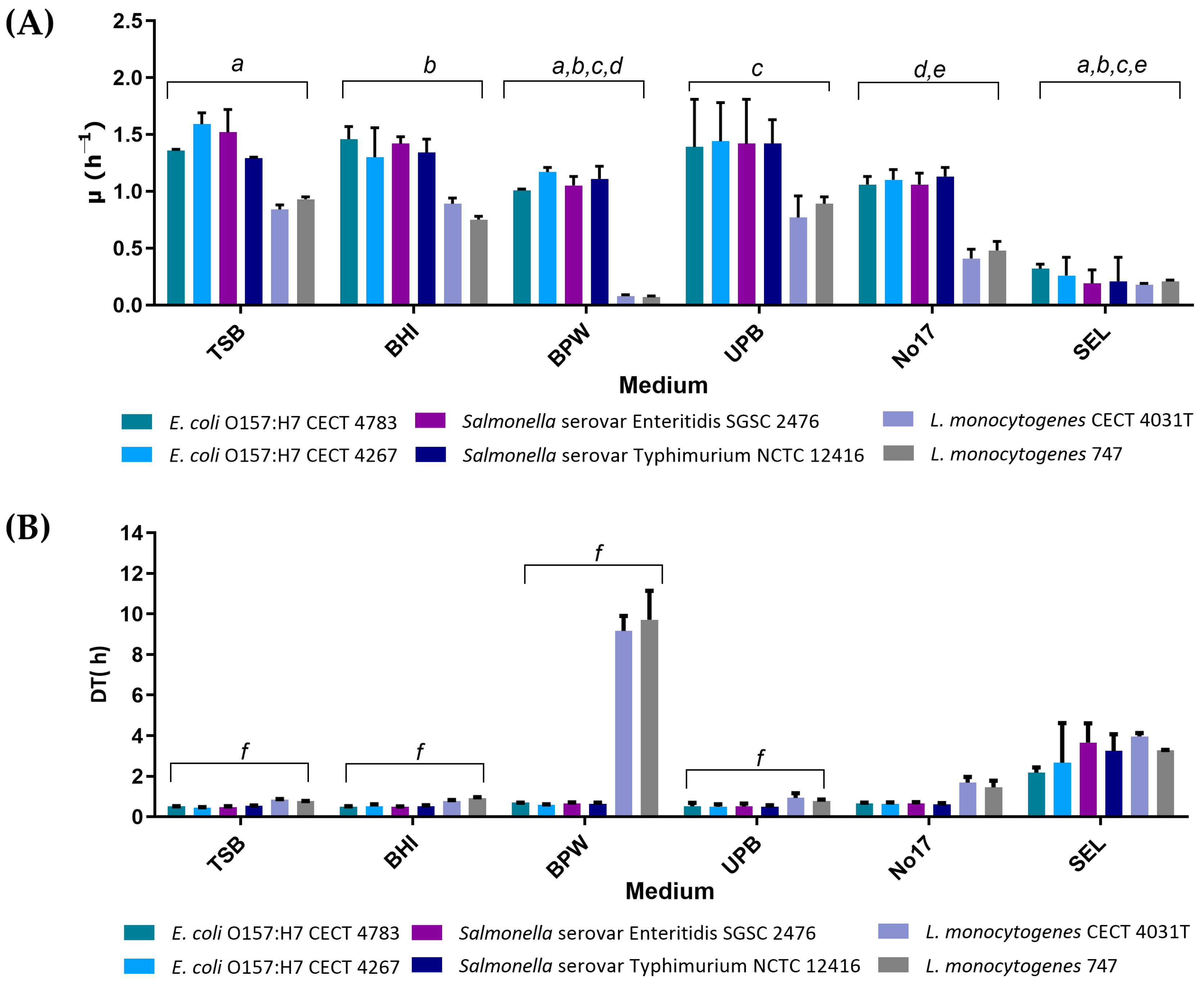
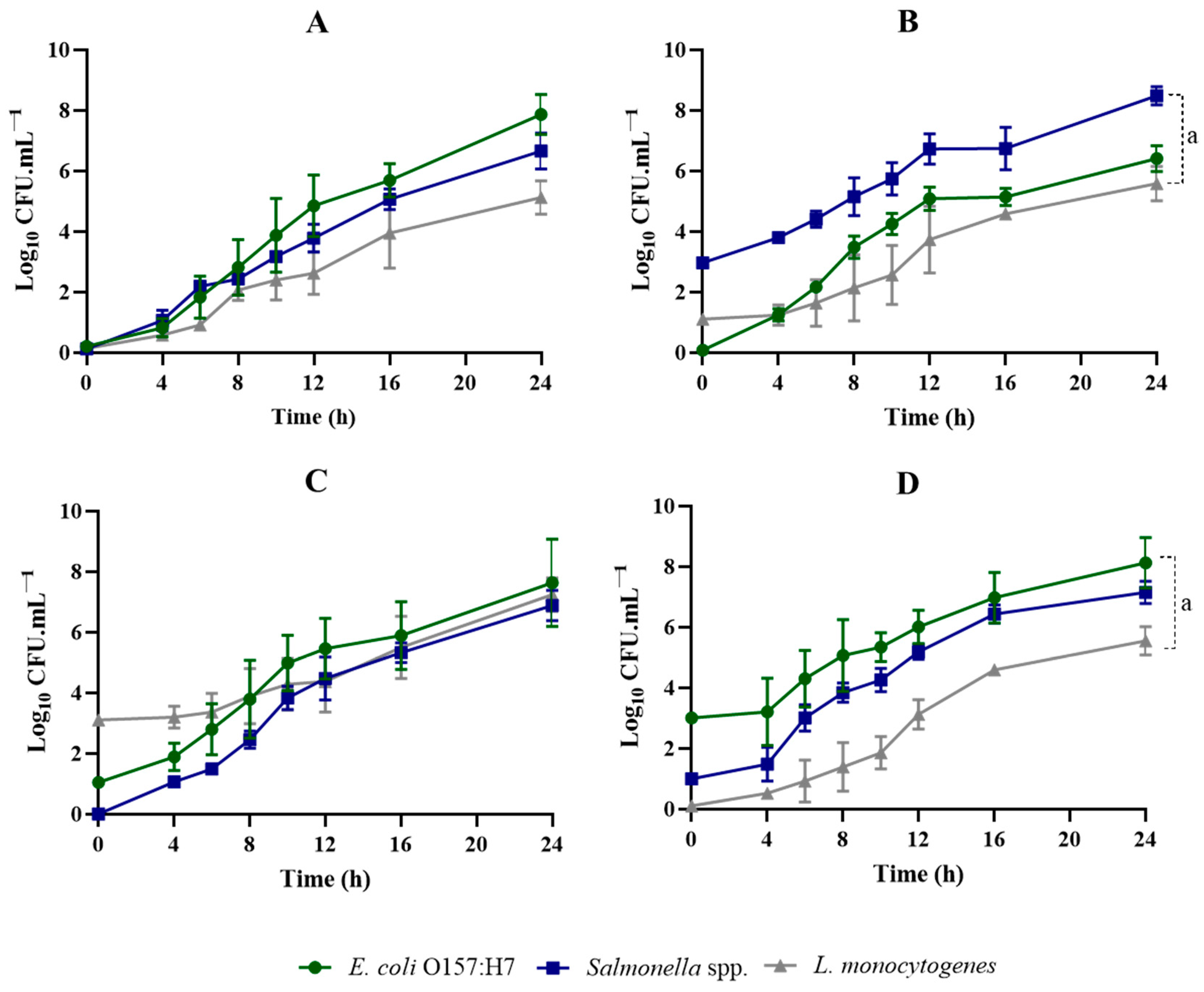
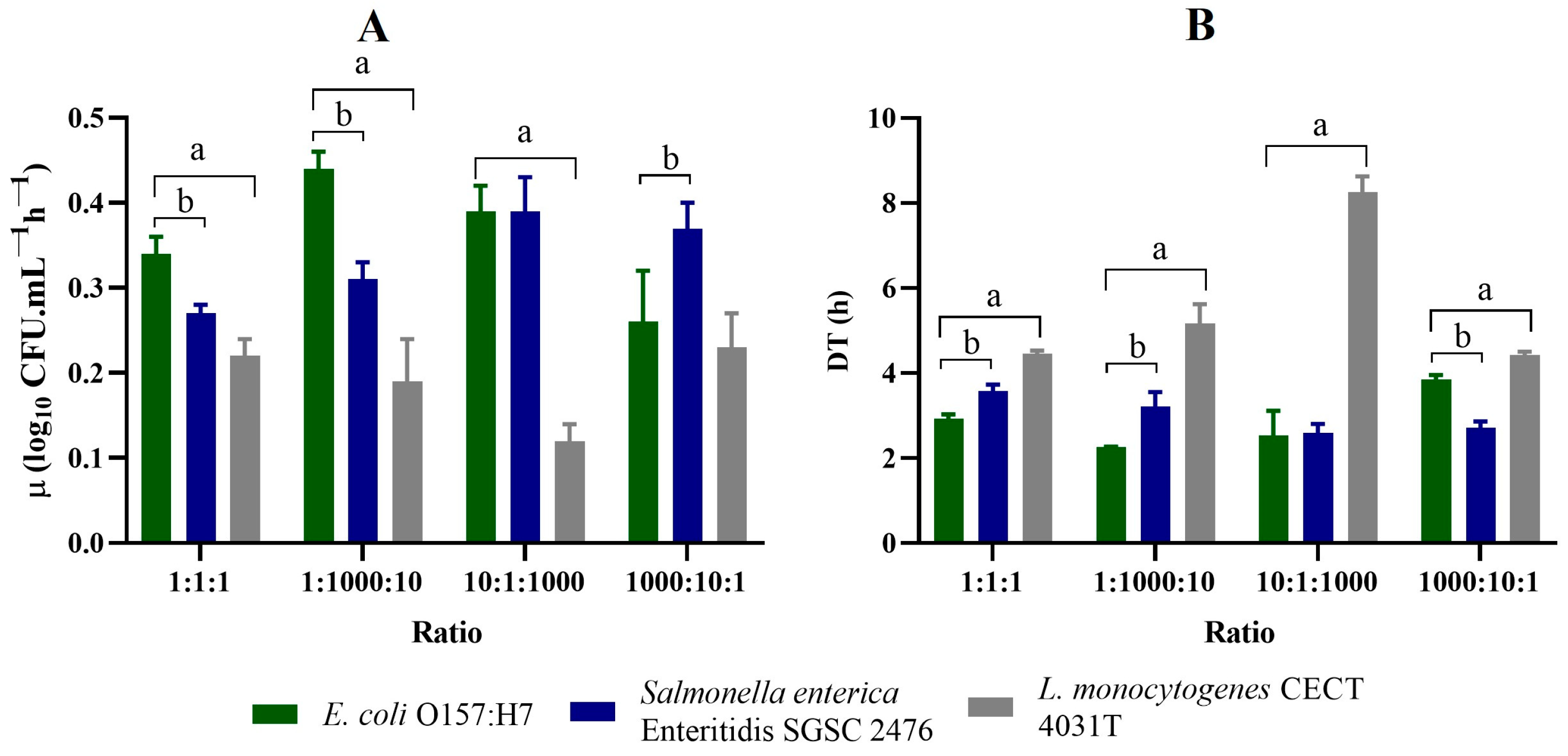
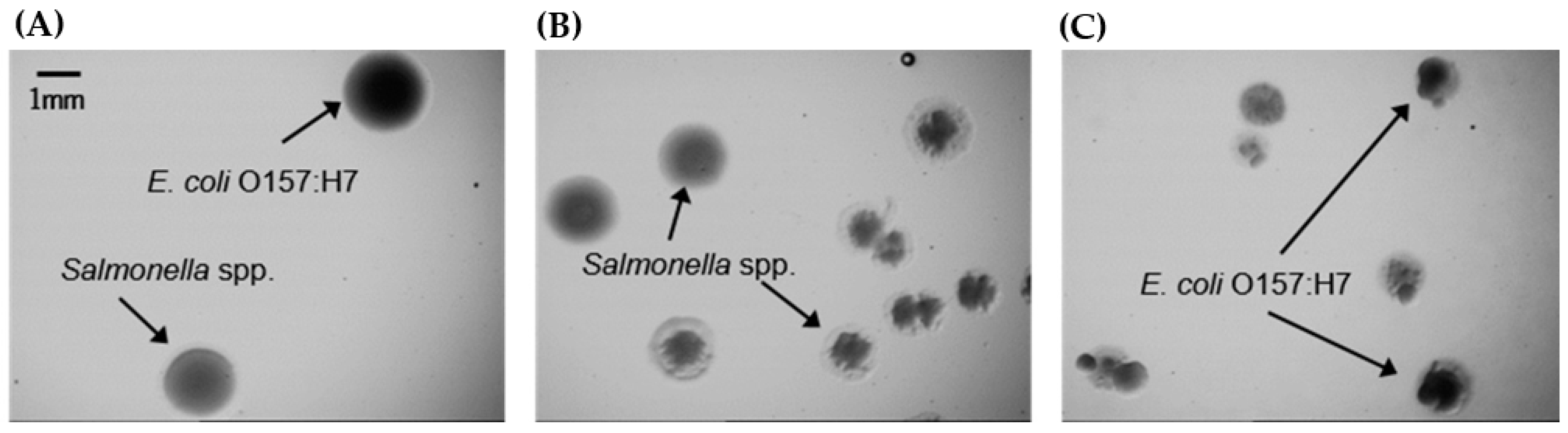
| Medium | Nutrient (s) | Buffer (s) | Salt (s) | Supplement (s) | pH | g/L H2O | Reference |
|---|---|---|---|---|---|---|---|
| BPW | Peptone | Phosphate | NaCl | - | 7.0 | 20 | [29] |
| TSB | Casein digest, soybean digest, dextrose | Phosphate | NaCl | - | 7.3 | 30 | [20] |
| UPB | Casein digest, soybean digest, dextrose, protease peptone, sodium pyruvate | Phosphate | NaCl, Mg2SO4, Ferric ammonium citrate | - | 6.3 | 38 | [30] |
| SEL | Pancreatic digest of casein, yeast extract, dextrose, soytone, sodium pyruvate | Phosphate | NaCl | Acriflavine, Cycloheximide, Fosfomycin, Nalidixic Acid | 7.0 | 48 | [26] |
| BHI | Brain heart infusion, casein digest, dextrose | Phosphate | NaCl | - | 7.4 | 37 | [27] |
| no. 17 | Tryptose, beef extract, yeast extract | Phosphate | NaCl | - | 7.2 | 48 | [18] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sousa, J.M.; Barbosa, A.; Araújo, D.; Castro, J.; Azevedo, N.F.; Cerqueira, L.; Almeida, C. Evaluation of Simultaneous Growth of Escherichia coli O157:H7, Salmonella spp., and Listeria monocytogenes in Ground Beef Samples in Different Growth Media. Foods 2024, 13, 2095. https://doi.org/10.3390/foods13132095
Sousa JM, Barbosa A, Araújo D, Castro J, Azevedo NF, Cerqueira L, Almeida C. Evaluation of Simultaneous Growth of Escherichia coli O157:H7, Salmonella spp., and Listeria monocytogenes in Ground Beef Samples in Different Growth Media. Foods. 2024; 13(13):2095. https://doi.org/10.3390/foods13132095
Chicago/Turabian StyleSousa, José Mário, Ana Barbosa, Daniela Araújo, Joana Castro, Nuno Filipe Azevedo, Laura Cerqueira, and Carina Almeida. 2024. "Evaluation of Simultaneous Growth of Escherichia coli O157:H7, Salmonella spp., and Listeria monocytogenes in Ground Beef Samples in Different Growth Media" Foods 13, no. 13: 2095. https://doi.org/10.3390/foods13132095
APA StyleSousa, J. M., Barbosa, A., Araújo, D., Castro, J., Azevedo, N. F., Cerqueira, L., & Almeida, C. (2024). Evaluation of Simultaneous Growth of Escherichia coli O157:H7, Salmonella spp., and Listeria monocytogenes in Ground Beef Samples in Different Growth Media. Foods, 13(13), 2095. https://doi.org/10.3390/foods13132095











