Adaptation of a Commercial Qualitative BAX® Real-Time PCR Assay to Quantify Campylobacter spp. in Whole Bird Carcass Rinses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Campylobacter spp. Pure Cultures
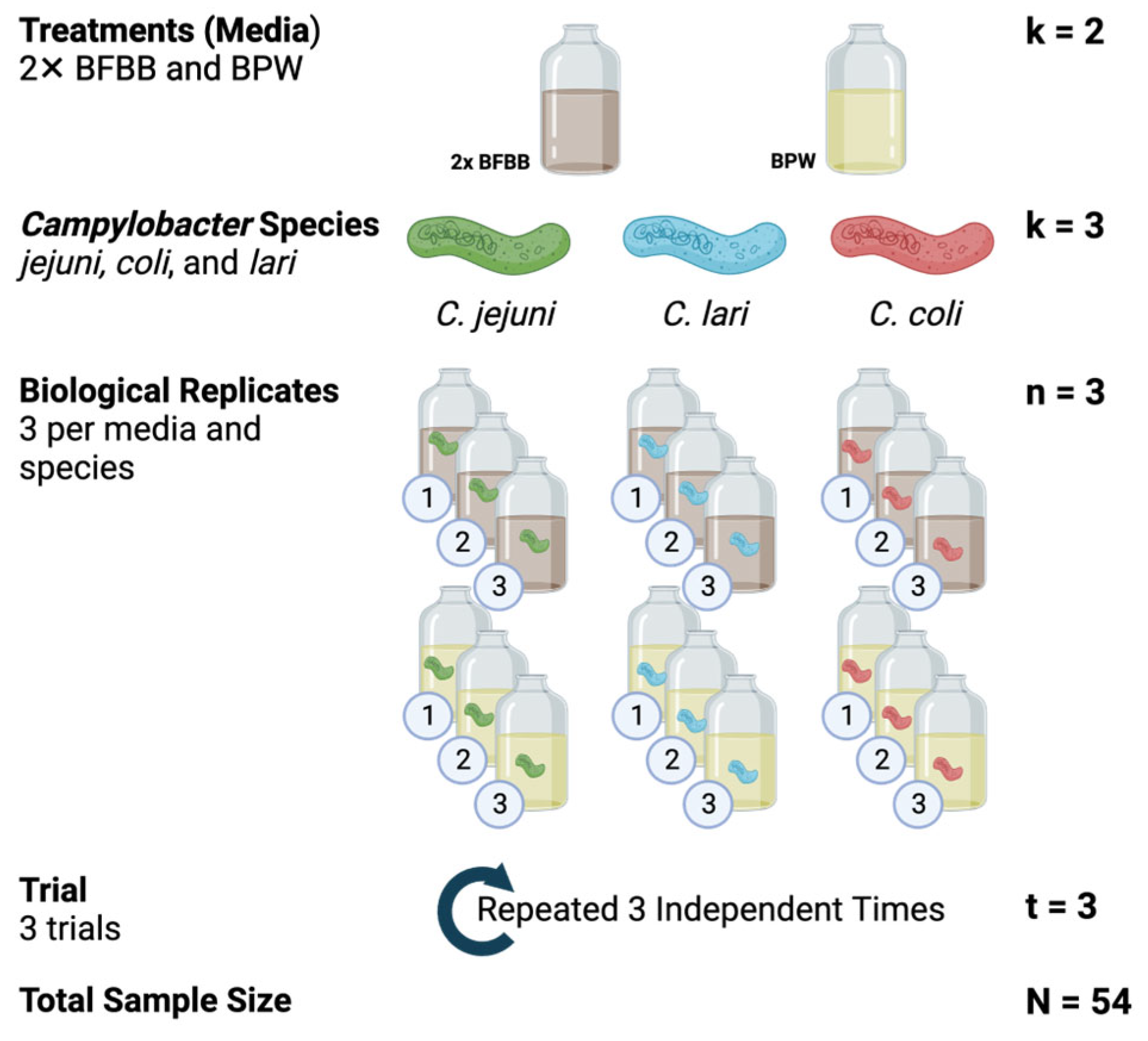
2.2. Objective 1: Determining Optimal Enrichment Media and Enumeration Range of BAX® System Real-Time PCR Assay
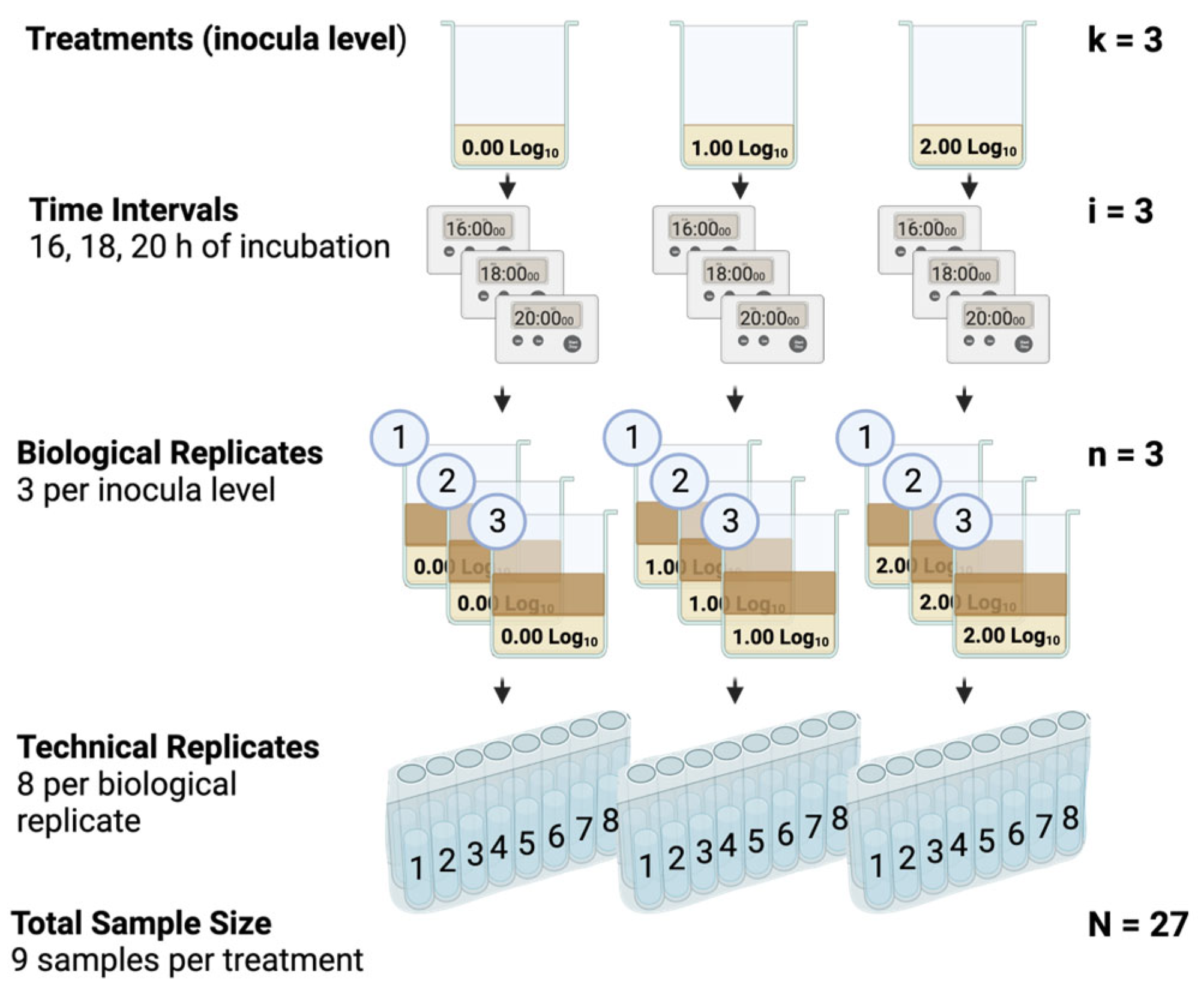
2.3. Objective 2: Time of Detection of Campylobacter spp. in Enriched Poultry Rinsates
2.4. Objective 3: Validation and Standard Curve Development of Quantification of BAX® System Real-Time PCR Assay
2.5. Statistical Analyses
2.5.1. Objective 1: Determining Optimal Enrichment Media and Enumeration Range of BAX® System Real-Time PCR Assay
2.5.2. Objective 2: Time of Detection of Campylobacter spp. in Enriched WBCR
2.5.3. Objective 3: Validation and Standard Curve Development of Quantification of BAX® System Real-Time PCR Assay
3. Results
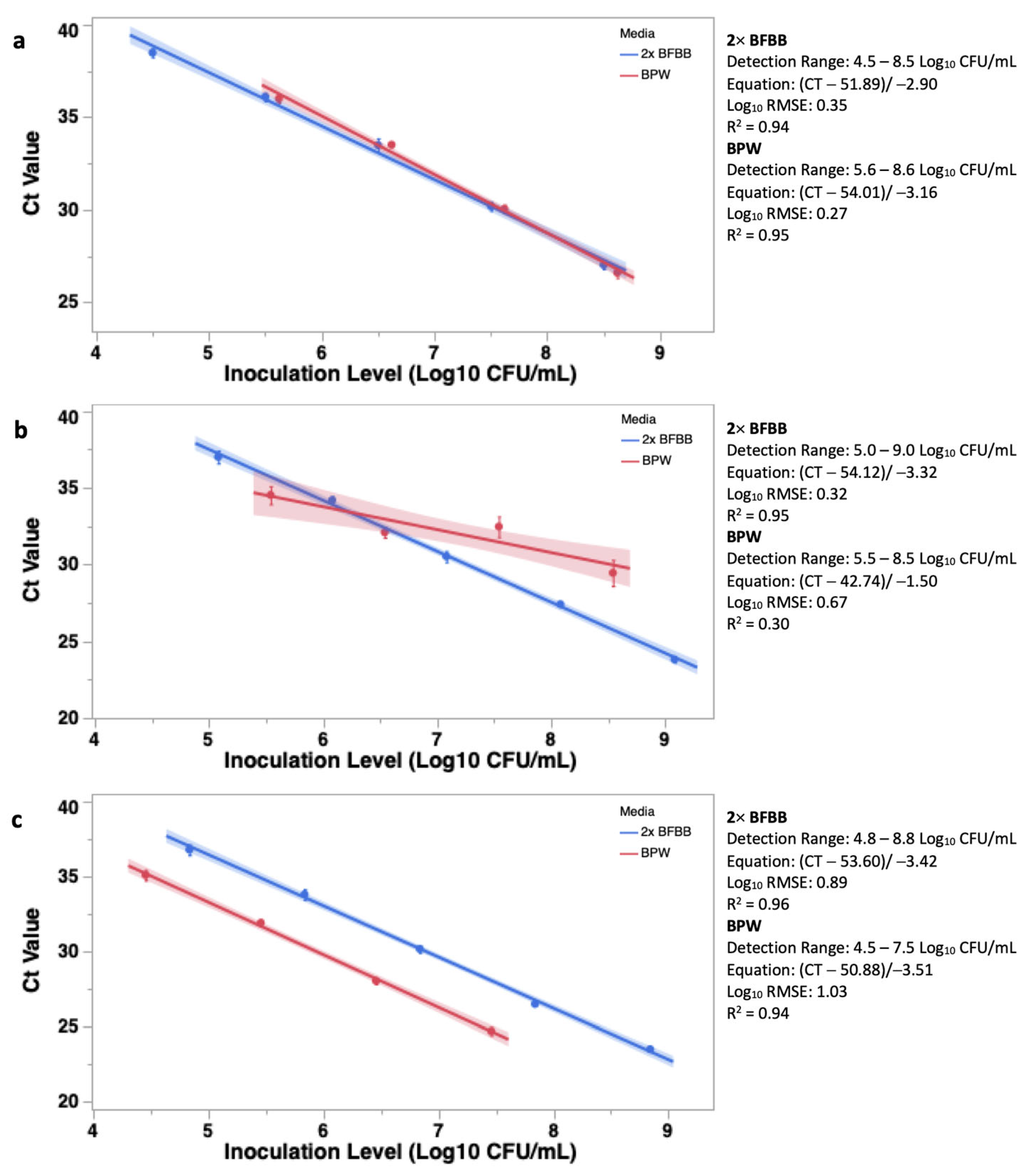
3.1. Objective 1: Determining Optimal Enrichment Media and Enumeration Range of BAX® System Real-Time PCR Assay
3.1.1. Optimal Enrichment Media
3.1.2. Enumeration Range and Linear Fit
3.2. Objective 2: Time of Detection of Campylobacter spp. in Enriched Poultry WBCR
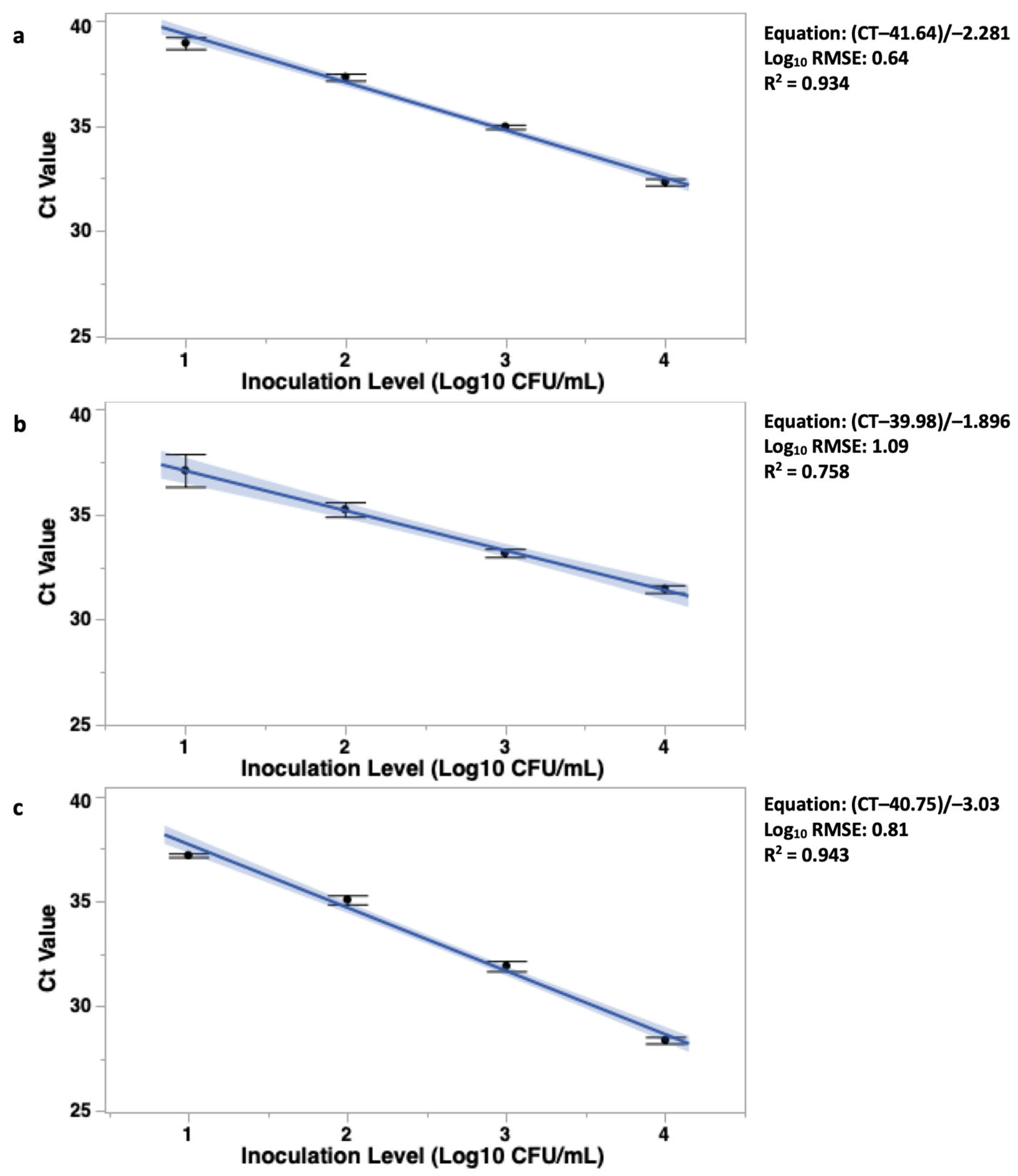
3.3. Objective 3: Validation and Standard Curve Development of Quantification of BAX® System Real-Time PCR Assay
3.3.1. CampyQuant™ Linear Fit and Subsequent Equation
3.3.2. Campylobacter spp. Levels from Each Quantification Method
3.3.3. Performance Parameters
4. Discussion
4.1. Objective 1: Determining Optimal Enrichment Media and Enumeration Range of BAX® System Real-Time PCR Assay
4.2. Objective 2: Time of Detection of Campylobacter spp. in Enriched Poultry WBCR
4.3. Objective 3: Validation and Standard Curve Development of Quantification of BAX® System Real-Time PCR Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Wagenaar, J.A.; French, N.P.; Havelaar, A.H. Preventing Campylobacter at the Source: Why is it so difficult? Clin. Infect. Dis. 2013, 57, 1600–1606. [Google Scholar] [CrossRef] [PubMed]
- Kaakoush, N.O.; Castaño-Rodríguez, N.; Mitchell, H.M.; Man, S.M. Global epidemiology of Campylobacter infection. Clin. Microbiol. Rev. 2015, 3, 687–720. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, C. Campylobacter. Clin. Lab. Med. 2015, 35, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Hazeleger, W.C.; Janse, J.D.; Koenraad, P.M.; Beumer, R.R.; Rombouts, F.M.; Abee, T. Temperature-dependent membrane fatty acid and cell physiology changes in coccoid forms of Campylobacter jejuni. Appl. Environ. Microbiol. 1995, 61, 2713–2719. [Google Scholar] [CrossRef] [PubMed]
- Hazeleger, W.C.; Jacobs-Reitsma, W.F.; Besten, H.M.W.D. Quantification of growth of Campylobacter and extended spectrum β-lactamase producing bacteria sheds light on black box of enrichment procedures. Front. Microbiol. 2016, 7, 1430. [Google Scholar] [CrossRef] [PubMed]
- Hill, G.N.; Bratcher, C.L.; Wang, L.; Singh, M.; Tapp, W.N.; McKee, S.R. Optimizing enrichment of Campylobacter on poultry. J. Appl. Poult. Res. 2017, 26, 307–315. [Google Scholar] [CrossRef]
- Facciolà, A.; Riso, R.; Avventuroso, E.; Visalli, G.; Delia, S.A.; Laganà, P. Campylobacter: From microbiology to prevention. J. Prev. Med. Hyg. 2017, 58, E79–E92. [Google Scholar]
- Mihaljevic, R.R.; Sikic, M.; Klancnik, A.; Brumini, G.; Mozina, S.S.; Abram, M. Environmental stress factors affecting survival and virulence of Campylobacter jejuni. Microb. Pathog. 2007, 43, 120–125. [Google Scholar] [CrossRef]
- Di Giannatale, E.; Calistri, P.; Di Donato, G.; Decastelli, L.; Goffredo, E.; Adriano, D.; Mancini, M.E.; Galleggiante, A.; Neri, D.; Antoci, S.; et al. Thermotolerant Campylobacter spp. in chicken and bovine meat in Italy: Prevalence, level of contamination and molecular characterization of isolates. PLoS ONE 2019, 14, e0225957. [Google Scholar] [CrossRef]
- Geissler, A.L.; Carrillo, F.B.; Swanson, K.; E Patrick, M.; E Fullerton, K.; Bennett, C.; Barrett, K.; E Mahon, B. Increasing Campylobacter infections, outbreaks, and antimicrobial resistance in the United States, 2004–2012. Clin. Infect. Dis. 2017, 65, 1624–1631. [Google Scholar] [CrossRef]
- Brena, M.C.; Mekonnen, Y.; Bettridge, J.M.; Williams, N.J.; Wigley, P.; Tessema, T.S.; Christley, R.M. Changing risk of environmental Campylobacter exposure with emerging poultry production systems in Ethiopia. Epidemiol. Infect. 2016, 144, 567–575. [Google Scholar] [CrossRef]
- Vizzini, P.; Braidot, M.; Vidic, J.; Manzano, M. Electrochemical and optical biosensors for the detection of Campylobacter and listeria: An update look. Micromachines 2019, 10, 500. [Google Scholar] [CrossRef] [PubMed]
- Tholozan, J.L.; Cappelier, J.M.; Tissier, J.P.; Delattre, G.; Federighi, M. Physiological characterization of viable-but-nonculturable Campylobacter jejuni cells. Appl. Environ. Microbiol. 1999, 65, 1110–1116. [Google Scholar] [CrossRef] [PubMed]
- Papić, B.; Pate, M.; Henigman, U.; Zajc, U.; Gruntar, I.; Biasizzo, M.; Ocepek, M.; Kušar, D. New approaches on quantification of Campylobacter jejuni in poultry samples: The use of digital PCR and real-time PCR against the ISO standard plate count method. Front. Microbiol. 2017, 8, 331. [Google Scholar] [CrossRef] [PubMed]
- Ricke, S.C.; Feye, K.M.; Chaney, W.E.; Shi, Z.; Pavlidis, H.; Yang, Y. Developments in rapid detection methods for foodborne Campylobacter in the United States. Front. Microbiol. 2019, 9, 3280. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Hughes, A.; Wilt, G.; Knabel, S.J. The BAX PCR Assay for screening Listeria monocytogenes targets a partial putative gene lmo 2234. J. Food Prot. 2004, 6, 1507–1511. [Google Scholar] [CrossRef] [PubMed]
- Hygiena. 2022. Available online: https://www.hygiena.com/patents/ (accessed on 30 October 2023).
- Silbernagel, K.; Jechorek, R.; Carver, C.; Barbour, W.M.; Mrozinski, P.; Albert, A.; Andaloro, B.; Anderson, G.; Beacorn, F.; Brooks, R.; et al. Evaluation of the BAX® system for detection of Salmonella in selected foods: Collaborative study. J. AOAC Int. 2003, 86, 1149–1159. [Google Scholar] [CrossRef] [PubMed]
- United States Department of Agriculture – Food Safety and Inspection Service. MLG 4.14. Isolation and Identification of Salmo-nella from Meat, Poultry, Pasteurized Egg, and Siluriformes (Fish) Products and Carcass and Environmental Sponges. 2023. Available online: https://www.fsis.usda.gov/sites/default/files/media_file/documents/MLG-4.14.pdf (accessed on 30 October 2023).
- United States Department of Agriculture—Food Safety and Inspection Service. MLG 41.05 Isolation and Identification of Campylobacter jejuni/coli/lari from Poultry Rinse, Sponge and Raw Product Samples. 2021. Available online: https://www.fsis.usda.gov/sites/default/files/media_file/2021-06/MLG-41.pdf (accessed on 30 October 2023).
- Wages, J.A.; Dittoe, D.K.; Feye, K.M.; Ricke, S.C. Consequences of implementing neutralizing buffered peptone water in commercial poultry processing on the microbiota of whole bird carcass rinses and the subsequent microbiological analyses. Front. Microbiol. 2022, 13, 813461. [Google Scholar] [CrossRef]
- United States Department of Agriculture – Food Safety and Inspection Service MLG 4.13 Isolation and Identification of Campylobacter jejuni/coli/lari from Poultry Rinse, Sponge and Raw Product Samples. 2023. Available online: https://www.fsis.usda.gov/sites/default/files/media_file/documents/MLG-4.13.pdf (accessed on 30 October 2023).
- Hygiena. 2021. Available online: https://cms.hygiena.com/sites/default/files/media/BAX-System-Q7-Users-Guide.pdf (accessed on 30 October 2023).
- Bodie, A.R.; Rothrock, M.J., Jr.; Ricke, S.C. Comparison of optical density-based growth kinetics for pure culture Campylobacter jejuni, coli and lari grown in blood-free Bolton broth. J. Environ. Sci. Health Part B 2023, 58, 671–678. [Google Scholar] [CrossRef]
- United States Department of Agriculture Food Safety and Inspection Service. 2016 FRN Docket No. FSIS-2014-0023. Available online: https://www.federalregister.gov/documents/2016/02/11/2016-02586/new-performance-standards-for-salmonella-and-campylobacter-in-not-ready-to-eat-comminuted-chicken (accessed on 30 October 2023).
- ISO/TS 10272-1:2017; Microbiology of Food and Animal Feed- Horizontal Method for the Detection and Enumeration of Campylobacter spp.—Part 1: Detection of Salmonella spp. International Organization for Standardization: Geneva, Switzerland, 2017.
- Chaney, W.E.; Englishbey, A.K.; Stephens, T.P.; Applegate, S.F.; Sanchez-Plata, M.X. Application of a commercial Salmonella real-time polymerase chain reaction (PCR) assay for the detection and quantitation of Salmonella enterica in poultry ceca. J. Food Prot. 2021, 85, 527–533. [Google Scholar] [CrossRef]
- O’Bryan, C.A.; Ricke, S.C.; Marcy, J.A. Public health impact of Salmonella spp. on raw poultry: Current concepts and future prospects in the United States. Food Control. 2022, 132, 108539. [Google Scholar] [CrossRef]
- Solís-Soto, L.Y.; García, S.; Wesley, I.; Heredia, N. A charcoal- and blood-free enrichment broth for isolation and PCR detection of Campylobacter jejuni and Campylobacter coli in chicken. J. Food Prot. 2011, 74, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Emanowicz, M.; Meade, J.; Bolton, D.; Golden, O.; Gutierrez, M.; Byrne, W.; Egan, J.; Lynch, H.; O’Connor, L.; Coffey, A.; et al. The impact of key processing stages and flock variables on the prevalence and levels of Campylobacter on broiler carcasses. Food Microbiol. 2021, 95, 103688. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.-J.; Qiao, B.; Xu, X.B.; Zhang, J.Z. Development and application of a real-time polymerase chain reaction method for Campylobacter jejuni detection. World J. Gastroenterol. 2013, 1, 3090–3095. [Google Scholar] [CrossRef]
- Keegstra, J.M.; Kamino, K.; Anquez, F.; Lazova, M.D.; Emonet, T.; Shimizu, T.S. Phenotypic diversity and temporal variability in a bacterial signaling network revealed by single cell FRET. eLife 2017, 6, e27455. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-H.; Chelliah, R.; Ramakrishnan, S.R.; Perumal, A.S.; Bang, W.-S.; Rubab, M.; Daliri, E.B.-M.; Barathikannan, K.; Elahi, F.; Park, E.; et al. Review on stress tolerance in Campylobacter jejuni. Front. Cell. Infect. Microbiol. 2021, 10, 596570. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.A.; Park, S.H.; Lee, S.I.; Owens, C.M.; Ricke, S.C. Assessment of chicken carcass microbiome responses during processing in the presence of commercial antimicrobials using a next generation sequencing approach. Sci. Rep. 2017, 7, srep43354. [Google Scholar] [CrossRef] [PubMed]
- Jung, G.H.; Lim, E.S.; Woo, M.-A.; Lee, J.Y.; Kim, J.-S.; Paik, H.-D. Inverse correlation between extracellular DNase activity and biofilm formation among chicken-derived Campylobacter strains. J. Microbiol. Biotechnol. 2017, 27, 1942–1951. [Google Scholar] [CrossRef]
- He, Z.; Gharaibeh, R.Z.; Newsome, R.C.; Pope, J.L.; Dougherty, M.W.; Tomkovich, S.; Pons, B.; Mirey, G.; Vignard, J.; Hendrixson, D.R.; et al. Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut 2019, 68, 289–300. [Google Scholar] [CrossRef]
- Yeow, M.; Liu, F.; Ma, R.; Williams, T.J.; Riordan, S.M.; Zhang, L. Analyses of energy metabolism and stress defense provide insights into Campylobacter concisus growth and pathogenicity. Gut Pathog. 2020, 12, 13. [Google Scholar] [CrossRef]
- Wagley, S.; Newcombe, J.; Laing, E.; Yusuf, E.; Sambles, C.M.; Studholme, D.J.; La Ragione, R.M.; Titball, R.W.; Champion, O.L. Differences in carbon source utilization distinguish Campylobacter jejuni from Campylobacter coli. BMC Microbiol. 2014, 14, 262. [Google Scholar] [CrossRef] [PubMed]
- Axelsson-Olsson, D.; Ellström, P.; Waldenström, J.; Haemig, P.D.; Brudin, L.; Olsen, B. Acanthamoeba—Campylobacter coculture as a novel method for enrichment of Campylobacter species. Appl. Environ. Microbiol. 2007, 73, 6864–6869. [Google Scholar] [CrossRef] [PubMed]
- Battersby, T.; Walsh, D.; Whyte, P.; Bolton, D.J. Campylobacter growth rates in four different matrices: Broiler caecal material, live birds, Bolton broth, and brain heart infusion broth. Infect. Ecol. Epidemiol. 2016, 6, 31217. [Google Scholar] [CrossRef] [PubMed]
- Ghssein, G.; Awada, R.; Salami, A.; Bahmad, H.F.; Awad, A.; Joumaa, W.H.; El Roz, A. Prevalence, laboratory findings and clinical characteristics of campylobacteriosis agents among hospitalized children with acute gastroenteritis in Lebanon. Pediatr. Gastroenterol. Hepatol. Nutr. 2021, 24, 346–356. [Google Scholar] [CrossRef]
- Ibrahim, J.N.; Eghnatios, E.; El Roz, A.; Fardoun, T.; Ghssein, G. Prevalence, antimicrobial resistance and risk factors for Campylobacteriosis in Lebanon. J. Infect. Dev. Ctries. 2019, 13, 11–20. [Google Scholar] [CrossRef]
- Paintsil, E.K.; Ofori, L.A.; Adobea, S.; Akenten, C.W.; Phillips, R.O.; Maiga-Ascofare, O.; Lamshöft, M.; May, J.; Danso, K.O.; Krumkamp, R.; et al. Prevalence and antibiotic resistance in Campylobacter spp. isolated from humans and food-producing animals in West Africa: A systematic review and meta-analysis. Pathogens 2022, 11, 140. [Google Scholar] [CrossRef]
| C. jejuni | C. coli | C. lari | Effects 3 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Media | 2× BFBB | BPW | 2× BFBB | BPW | 2× BFBB | BPW | Interaction 4 | Media | Species |
| Sensitivity (%) | 93.3 | 76.0 | 96.3 | 64.0 | 88.0 | 80.0 | 0.102 | <0.0001 | 0.277 |
| Accuracy (%) | 93.8 a | 81.9 bc | 96.3 a | 79.0 c | 89.8 ab | 84.0 bc | 0.024 | <0.0001 | 0.855 |
| Prevalence (%) | 90.0 | 73.1 | 87.2 | 61.5 | 84.6 | 74.9 | 0.273 | 0.0022 | 0.366 |
| NLR (%) | 6.7 | 24.0 | 4.0 | 36.0 | 12.0 | 20.0 | 0.063 | 0.003 | 0.57 |
| NPV (%) | 38.9 | 14.5 | 55.6 | 11.2 | 27.9 | 16.7 | 0.312 | 0.009 | 0.578 |
| C. jejuni | C. coli | C. lari | Effects | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time | 16 h | 18 h | 20 h | 16 h | 18 h | 20 h | 16 h | 18 h | 20 h | Interaction | Time | Species |
| Sensitivity | 66.7 cd | 86.1 ab | 94.4 a | 14.6 e | 25.7 e | 63.9 cd | 13.9 e | 53.0 d | 72.2 bc | 0.0040 | <0.0001 | <0.0001 |
| Accuracy | 42.7 bc | 71.4 a | 78.1 a | 3.0 d | 8.6 d | 39.2 bc | 1.2 d | 27.3 c | 50.4 b | 0.0100 | <0.0001 | <0.0001 |
| Prevalence | 64.0 | 82.7 | 90.7 | 17.3 | 33.3 | 61.3 | 21.3 | 49.3 | 69.3 | 0.1610 | <0.0001 | <0.0001 |
| NLR | 33.3 bc | 13.9 de | 5.6 e | 85.1 a | 74.3 a | 36.1 bc | 86.1 a | 47.0 b | 27.7 cd | 0.0004 | <0.0001 | <0.0001 |
| NPV | 10.7 bc | 17.0 b | 44.4 a | 6.8 bc | 6.0 bc | 10.4 bc | 5.1 c | 6.8 bc | 12.6 bc | <0.0001 | <0.0001 | <0.0001 |
| C. jejuni | C. coli | C. lari | Effects 1 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Log10 CFU/mL | CampyQuant™ | Campy-Cefex | CampyQuant™ | Campy-Cefex | CampyQuant™ | Campy-Cefex | Interaction 2 | Method | Species |
| 1.00 | 1.18 b ± 0.16 | 1.64 a ± 0.16 | 1.22 ab ± 0.10 | 1.260 ab ± 0.12 | 1.18 b ± 0.16 | 0.70 c ± 0.10 | 0.002 | 0.913 | 0.001 |
| 2.00 | 1.89 bc ± 0.05 | 2.18 a ± 0.05 | 2.09 ab ± 0.06 | 1.74 c ± 0.05 | 1.88 bc ± 0.05 | 1.50 d ± 0.05 | <0.001 | <0.001 | 0.002 |
| 3.00 | 2.93 ab ± 0.04 | 3.05 ab ± 0.10 | 2.74 b ± 0.09 | 3.23 a ± 0.09 | 2.92 b ± 0.09 | 2.68 b ± 0.09 | 0.001 | 0.100 | 0.639 |
| 4.00 | 4.08 ab ± 0.08 | 4.03 ab ± 0.05 | 3.91 b ± 0.07 | 4.13 a ± 0.05 | 4.09 ab ± 0.07 | 3.55 c ± 0.05 | <0.001 | 0.005 | <0.001 |
| C. jejuni | C. coli | C. lari | Effects 1 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Method | CampyQuant™ | Campy-Cefex | CampyQuant™ | Campy-Cefex | CampyQuant™ | Campy-Cefex | Interaction | Method | Species |
| Sensitivity (%) | 88.3 | 94.5 | 83.3 | 80.5 | 86.7 | 83.3 | 0.411 | 1.000 | 0.079 |
| Accuracy (%) | 74.7 | 82.5 | 66.5 | 60.0 | 71.7 | 64.5 | 0.415 | 0.7046 | 0.074 |
| Prevalence (%) | 84.1 | 87.2 | 79.4 | 74.3 | 82.5 | 76.9 | 0.433 | 0.405 | 0.083 |
| NLR (%) | 11.7 | 5.5 | 16.7 | 19.5 | 15.0 | 16.6 | 0.475 | 0.863 | 0.073 |
| NPV (%) | 34.4 | 30.5 | 20.6 | 15.9 | 26.1 | 27.8 | 0.733 | 0.527 | 0.020 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bodie, A.R.; Dittoe, D.K.; Applegate, S.F.; Stephens, T.P.; Ricke, S.C. Adaptation of a Commercial Qualitative BAX® Real-Time PCR Assay to Quantify Campylobacter spp. in Whole Bird Carcass Rinses. Foods 2024, 13, 56. https://doi.org/10.3390/foods13010056
Bodie AR, Dittoe DK, Applegate SF, Stephens TP, Ricke SC. Adaptation of a Commercial Qualitative BAX® Real-Time PCR Assay to Quantify Campylobacter spp. in Whole Bird Carcass Rinses. Foods. 2024; 13(1):56. https://doi.org/10.3390/foods13010056
Chicago/Turabian StyleBodie, Aaron R., Dana K. Dittoe, Savannah F. Applegate, Tyler P. Stephens, and Steven C. Ricke. 2024. "Adaptation of a Commercial Qualitative BAX® Real-Time PCR Assay to Quantify Campylobacter spp. in Whole Bird Carcass Rinses" Foods 13, no. 1: 56. https://doi.org/10.3390/foods13010056
APA StyleBodie, A. R., Dittoe, D. K., Applegate, S. F., Stephens, T. P., & Ricke, S. C. (2024). Adaptation of a Commercial Qualitative BAX® Real-Time PCR Assay to Quantify Campylobacter spp. in Whole Bird Carcass Rinses. Foods, 13(1), 56. https://doi.org/10.3390/foods13010056








