A Comparative Study of Chemical Profiling and Bioactivities between Thai and Foreign Hemp Seed Species (Cannabis sativa L.) Plus an In-Silico Investigation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
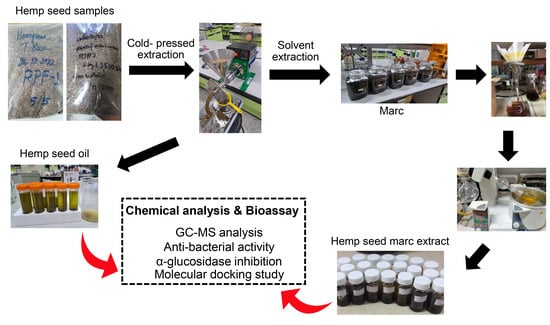
2.2. Hemp Seed Extraction
2.3. Gas Chromatography-Mass Spectrometry (GC–MS) Analysis
2.4. Antibacterial Activity
2.4.1. Test Microorganisms
2.4.2. Preliminary Antibacterial Test of Hemp Seed Oil and Extract
2.4.3. Determination of Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC)
2.5. α-Glucosidase Inhibition Activity
2.6. Combination Study
2.7. Molecular Docking Simulation
2.7.1. Docking Preparation
2.7.2. Docking Experiment
2.7.3. Post-Docking Analysis, Modeling, and Statistical Analysis
3. Results
3.1. Hemp Seed Extraction
3.2. GC–MS Analysis
3.3. Preliminary Antibacterial Activity
3.4. Determination of Minimal Inhibitory Concentration (MIC) and Minimal Bactericidal Concentration (MBC)
3.5. Screening and IC50 on α-Glucosidase Inhibition of Hemp Seed Oils and Extracts
3.6. Combination Study on α-Glucosidase Inhibitory of Hemp Seed Extract and Acarbose
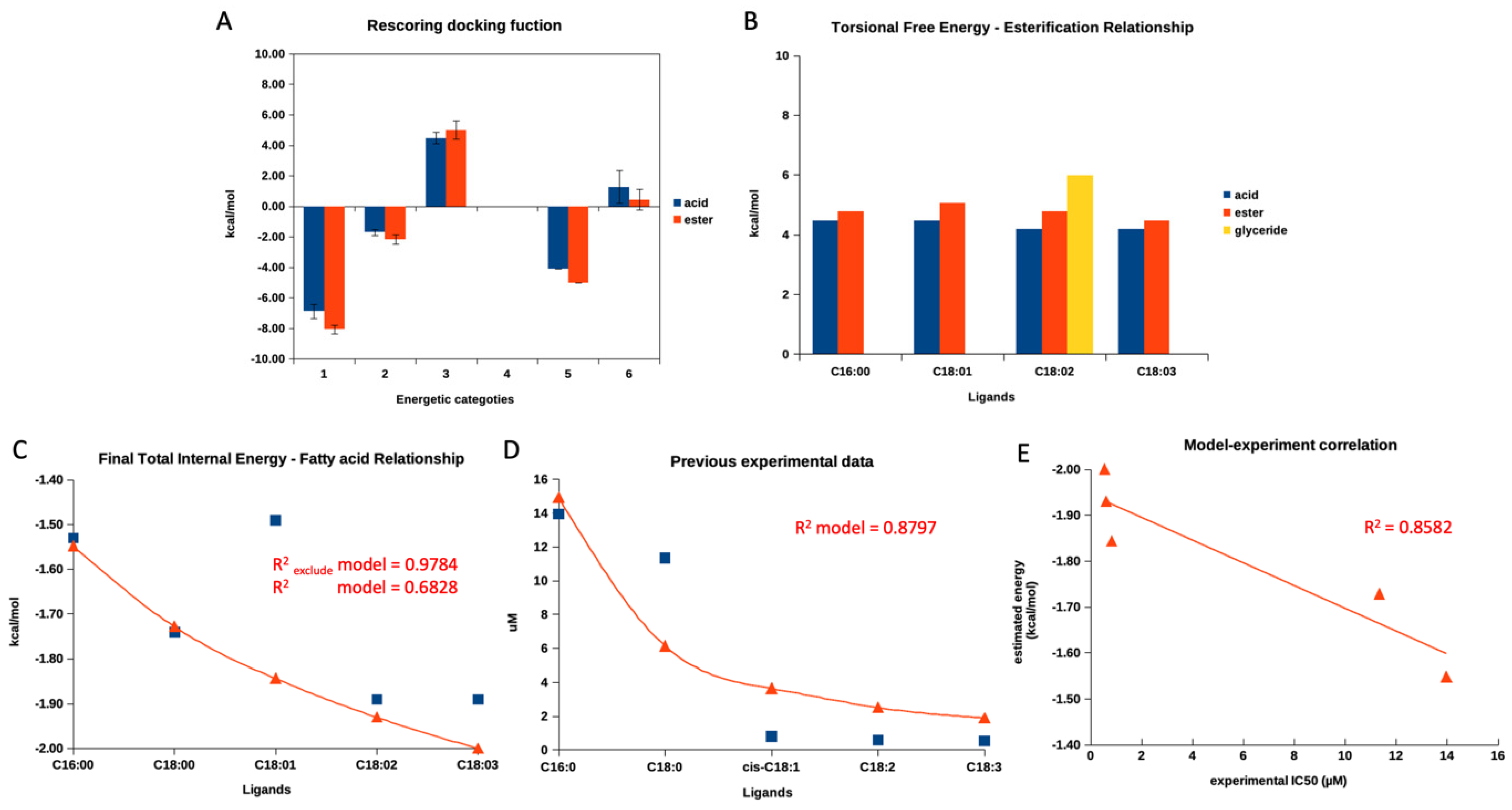
3.7. Molecular Docking and a Mathematical Model
3.7.1. Molecular Docking and Mathematical Model of Free Fatty Acids and Esters
3.7.2. Molecular Docking of THC and THCV
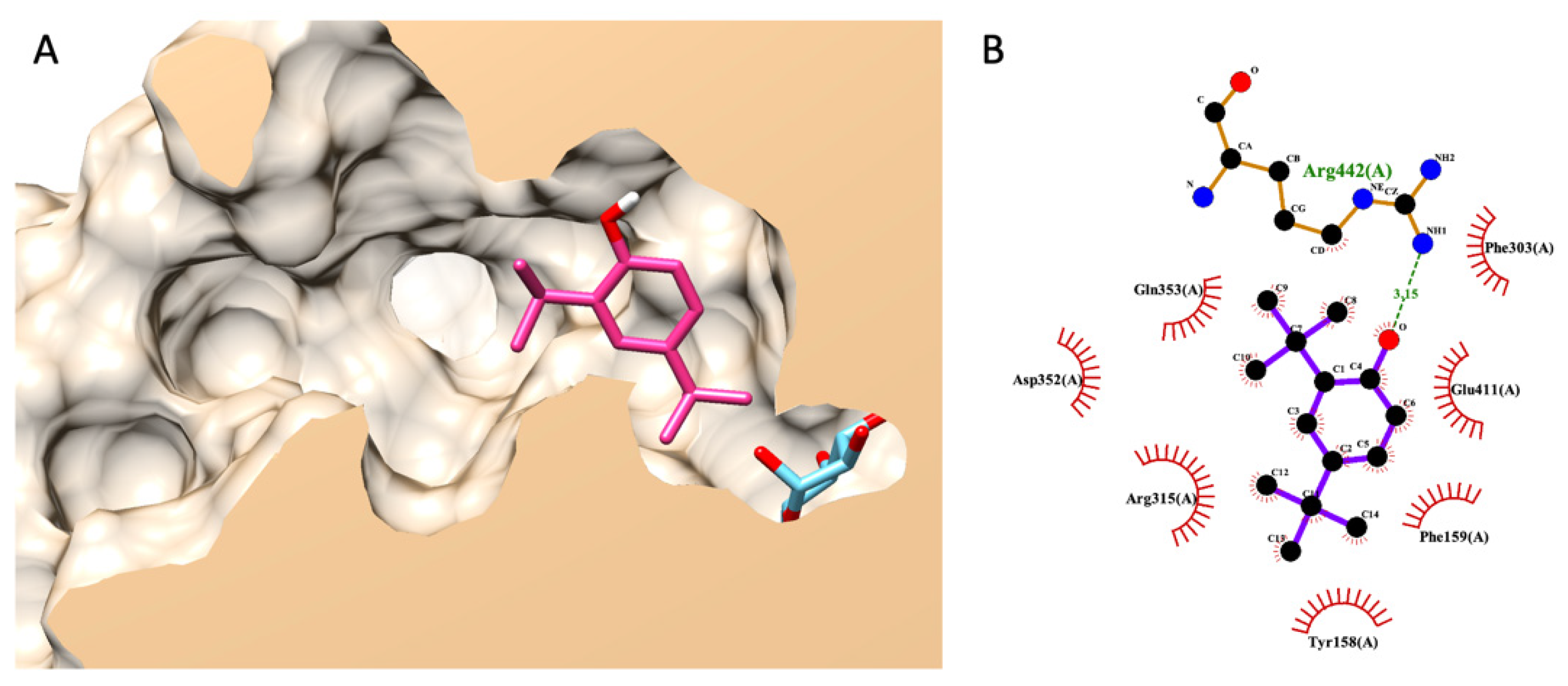
3.7.3. Molecular Docking of DTBP
3.7.4. Molecular Docking of Guanosine: Proposing a Possible Allosteric Binding Site
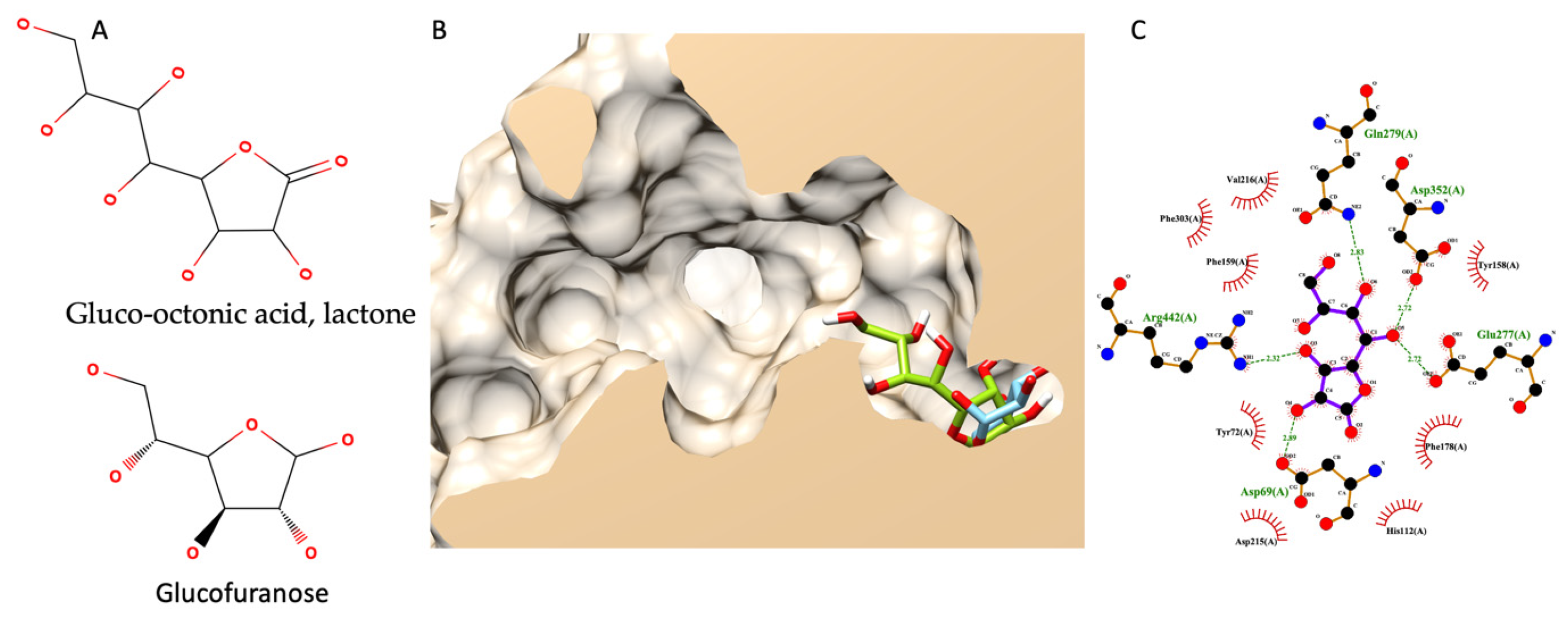
3.7.5. Molecular Docking of Gluco-Octonic Acid Lactone, Proposing a Competitive Behavior
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anwar, F.; Latif, S.; Ashraf, M. Analytical Characterization of Hemp (Cannabis sativa) Seed Oil from Different Agro-ecological Zones of Pakistan. J. Am. Oil Chem. 2006, 83, 323–329. [Google Scholar] [CrossRef]
- Saadati, M.; Bagheri, A. Respondent Driven Sampling Method Compared with Other Sampling Methods of Hidden Populations. Iran. J. Epidemiol. 2016, 12, 63–74. [Google Scholar]
- European Commission. Council Directive 2002/57/EC of 13 June 2002 on the Marketing of Seed of Oil and Fibre Plants. Off. J. Eur. Communities 2002, 74–97. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32002L0057 (accessed on 30 October 2023).
- European Parliament and the Council of the European Union. Regulation (EU) 2021/2115 of the European Parliament and of the Council. Off. J. Eur. Union 2021, 435/1–435/186. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32021R2115 (accessed on 30 October 2023).
- Wimalasiri, E.M.; Wijesekara Mudiyanselage, A.U.K.M.W.; Madhuwanthi, P.I.; Ranasinghe, P.; Jahanshiri, E. Uncovering the Potential and Handicaps of Non-drug Hemp Cultivation in South and Southeast Asia. Rev. Agric. Sci. 2023, 11, 121–136. [Google Scholar] [CrossRef] [PubMed]
- Deferne, J.L.; Pate, D.W. Hemp Seed Oil: A Source of Valuable Essential Fatty Acids. J. Int. Hemp. Assoc. 1996, 3, 4–7. [Google Scholar]
- Rao, V.S.N.; Menezes, A.M.S.; Viana, G.S.B. Effect of Myrcene on Nociception in Mice. J. Pharm. Pharmacol. 1990, 42, 877–878. [Google Scholar] [CrossRef] [PubMed]
- Gurr, M.I.; Harwood, J.L.; Frayn, K.N. Lipid Biochemistry; Springer: Berlin/Heidelberg, Germany, 2002. [Google Scholar] [CrossRef]
- Martinenghi, L.D.; Jønsson, R.; Lund, T.; Jenssen, H. Isolation, Purification, and Antimicrobial Characterization of Cannabidiolic Acid and Cannabidiol from Cannabis sativa L. Biomolecules 2020, 10, 900. [Google Scholar] [CrossRef]
- Ostapczuk, K.; Apori, S.O.; Estrada, G.; Tian, F. Hemp Growth Factors and Extraction Methods Effect on Antimicrobial Activity of Hemp Seed Oil: A Systematic Review. Separations 2021, 8, 183. [Google Scholar] [CrossRef]
- Wang, C.-Y.; Chen, Y.-W.; Hou, C.-Y. Antioxidant and Antibacterial Activity of Seven Predominant Terpenoids. Int. J. Food Prop. 2019, 22, 230–238. [Google Scholar] [CrossRef]
- Mikulcová, V.; Kašpárková, V.; Humpolíček, P.; Buňková, L. Formulation, Characterization and Properties of Hemp Seed Oil and Its Emulsions. Molecules 2017, 22, 700. [Google Scholar] [CrossRef]
- Brazilian Diabetes Society (SBD). There Are 13.4 Million People with Diabetes in Brazil. 2013. Available online: http://www.diabetes.org.br/sala-denoticias/2364-sao-134-milhoes-de-pessoas-portadoras-de-diabetes-no-brasilbetes-no-Brasil (accessed on 30 October 2023).
- Li, D.Q.; Qian, Z.M.; Li, S.P. Inhibition of Three Selected Beverage Extracts on Alpha-glucosidase and Rapid Identification of Their Active Compounds using HPLC-DAD-MS/MS and Biochemical Detection. J. Agric. Food Chem. 2010, 58, 6608–6613. [Google Scholar] [CrossRef] [PubMed]
- Farinon, B.; Molinari, R.; Costantini, L.; Merendino, N. The Seed of Industrial Hemp (Cannabis sativa L.): Nutritional Quality and Potential Functionality for Human Health and Nutrition. Nutrients 2020, 12, 1935. [Google Scholar] [CrossRef] [PubMed]
- Zengin, G.; Menghini, L.; Di Sotto, A.; Mancinelli, R.; Sisto, F.; Carradori, S.; Cesa, S.; Fraschetti, C.; Filippi, A.; Angiolella, L. Chromatographic Analyses, in vitro Biological Activities, and Cytotoxicity of Cannabis sativa L. Essential Oil: A Multidisciplinary Study. Molecules 2018, 23, 3266. [Google Scholar] [CrossRef] [PubMed]
- Hourfane, S.; Mechqoq, H.; Bekkali, A.Y.; Rocha, J.M.; El Aouad, N. A Comprehensive Review on Cannabis sativa Ethnobotany, Phytochemistry, Molecular Docking and Biological Activities. Plants 2023, 12, 1245. [Google Scholar] [CrossRef] [PubMed]
- M07–a9; Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. Clinical and Laboratory Standards Institute: Wayne, IL, USA, 2012.
- Sarker, S.D.; Nahar, L.; Kumarasamy, Y. Microtitre Plate-Based Antibacterial Assay Incorporation Resazurin as An Indicator of Cell Growth, and Its Application in the In Vitro Antibacterial Screening of Phytochemicals. Methods 2007, 42, 321–324. [Google Scholar] [CrossRef]
- Dej-Adisai, S.; Pitakbut, T. Determination of α-Glucosidase Inhibitory Activity from Selected Fabaceae Plants. Pak. J. Pharm. Sci. 2015, 28, 1679–1683. [Google Scholar] [PubMed]
- Chou, T.C. Drug Combination Studies and Their Synergy Quantification using The Chou-Talalay Method. Cancer Res. 2010, 70, 440–446. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open Chemical Toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Yamamoto, K.; Miyake, H.; Kusunoki, M.; Osaki, S. Crystal Structures of Isomaltase from Saccharomyces cerevisiae and in Complex with Its Competitive Inhibitor Maltose. FEBS J. 2010, 277, 4205–4214. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Sakulkeo, O.; Wattanapiromsakul, C.; Pitakbut, T.; Dej-adisai, S. Alpha-Glucosidase Inhibition and Molecular Docking of Isolated Compounds from Traditional Thai Medicinal Plant, Neuropeltis racemosa Wall. Molecules 2022, 27, 639. [Google Scholar] [CrossRef] [PubMed]
- Phoopha, S.; Wattanapiromsakul, C.; Pitakbut, T.; Dej-adisai, S. Chemical Constituents of Litsea elliptica and Their Alpha-Glucosidase Inhibition with Molecular Docking. Pharmacogn. Mag. 2020, 16, 327. [Google Scholar]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple Ligand–Protein Interaction Diagrams for Drug Discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, R.; Nie, K.; Liu, C.; Deng, L.; Wang, F. Construction of Fatty Acid Derivatives from Rubber Seed Oil as α-Glucosidase Inhibitors Based on Rubber Seed Oil. Bioresour. Bioprocess. 2022, 9, 23. [Google Scholar] [CrossRef]
- Ogasawara, M.; Yoshii, K.; Wada, J.; Yamamoto, Y.; Inouye, K. Identification of Guanine, Guanosine, and Inosine for α-Amylase Inhibitors in the Extracts of the Earthworm Eisenia fetida and Characterization of Their Inhibitory Activities against Porcine Pancreatic α-Amylase. Enzym. Microb. Technol. 2020, 142, 109693. [Google Scholar] [CrossRef]
- Kalita, D.; Holm, D.G.; LaBarbera, D.V.; Petrash, J.M.; Jayanty, S.S. Inhibition of α-Glucosidase, α-Amylase, and Aldose Reductase by Potato Polyphenolic Compounds. PLoS ONE 2018, 13, e0191025. [Google Scholar] [CrossRef]
- Callaway, J.C. Hempseed as A nutritional Resource: An overview. Euphytica 2004, 140, 65–72. [Google Scholar] [CrossRef]
- Sokoła-Wysoczanska, E.; Wysoczański, T.; Wagner, J.; Czyz, K.; Bodkowski, R.; Lochynski, S.; Patkowska-Sokoła, B. Polyunsaturated Fatty Acids and Their Potential Therapeutic Role in Cardiovascular System Disorders—A review. Nutrients 2018, 10, 1561. [Google Scholar] [CrossRef]
- Blasi, F.; Tringaniello, C.; Verducci, G.; Cossignani, L. Bioactive Minor Components of Italian and Extra-European Hemp Seed Oils. LWT 2022, 158, 113167. [Google Scholar] [CrossRef]
- Irakli, M.; Tsaliki, E.; Kalivas, A.; Kleisiaris, F.; Sarrou, E.; Cook, C.M. Effect of Genotype and Growing Year on The nutritional, Phytochemical, and Antioxidant Properties of Industrial Hemp (Cannabis sativa L) Seeds. Antioxidants 2019, 8, 491. [Google Scholar] [CrossRef] [PubMed]
- Galasso, I.; Russo, R.; Mapelli, S.; Ponzoni, E.; Brambilla, I.M.; Battelli, G.; Reggiani, R. Variability in Seed Traits in a Collection of Cannabis sativa L. Genotypes. Front. Plant Sci. 2016, 7, 688. [Google Scholar] [CrossRef] [PubMed]
- Taaifi, Y.; Benmoumen, A.; Belhaj, K.; Aazza, S.; Abid, M.; Azeroual, E.; Elamrani, A.; Mansouri, F.; Caid, H.S. Seed Composition of Non-industrial Hemp (Cannabis sativa L.) Varieties from Four Regions in Northern Morocco. Int. J. Food Sci. Technol. 2021, 56, 5931–5947. [Google Scholar] [CrossRef]
- Gutiérrez-Luna, K.; Ansorena, D.; Astiasarán, I. Fatty Acid Profile, Sterols, and Squalene Content Comparison between Two Conventional (olive oil and linseed oil) and Three non-conventional Vegetable Oils (echium oil, hempseed oil, and moringa oil). J. Food Sci. 2022, 87, 1489–1499. [Google Scholar] [CrossRef]
- Liang, J.; Appukuttan Aachary, A.; Hollader, U.T. Hemp Seed Oil: Minor Components and Oil Quality. Lipid Technol. 2015, 27, 231–233. [Google Scholar] [CrossRef]
- Montserrat-DeLaPaz, S.; Marín-Aguilar, F.; García-Giménez, M.D.; Fernández-Arche, M.A. Hemp (Cannabis sativa L.) Seed Oil: Analytical and Phytochemical Characterization of the Unsaponifiable Fraction. J. Agric. Food Chem. 2014, 62, 1105–1110. [Google Scholar] [CrossRef]
- Fiume, M.M.; Bergfeld, W.F.; Belsito, D.V.; Hill, R.A.; Klaassen, C.D.; Liebler, D.C.; Marks, J.G.; Jr Shank, R.C.; Slaga, T.J.; Snyder, P.W.; et al. Safety Assessment of Monoglyceryl Monoesters as Used in Cosmetics. Int. J. Toxicol. 2020, 39, 93S–126S. [Google Scholar] [CrossRef]
- Jin, S.; Lee, M.Y. The Ameliorative Effect of Hemp Seed Hexane Extracts on the Propionibacterium acnes-Induced Inflammation and Lipogenesis in Sebocytes. PLoS ONE 2018, 13, e0202933. [Google Scholar] [CrossRef]
- Hayes, M.L.; Berkovitz, B.K. The Reduction of Fissure Caries in Wistar Rats by a Soluble Salt of Nonanoinic Acid. Arch. Oral Biol. 1979, 24, 663–666. [Google Scholar] [CrossRef]
- Sangkanu, S.; Rukachaisirikul, V.; Suriyachadkun, C.; Phongpaichit, S. Evaluation of Antibacterial Potential of Mangrove Sediment-Derived Actinomycetes. Microb. Pathog. 2017, 112, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Adisakwattana, S.; Jiphimai, P.; Prutanopajai, P.; Chanathong, B.; Sapwarobol, S.; Ariyapitipan, T. Evaluation of Alphaglucosidase, Alpha-amylase and Protein Glycation Inhibitory Activities of eEdible Plants. Int. J. Food Sci. Nutr. 2010, 61, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Mulimani, V.H.; Supriya, D. Effect of Heat Treatments on Alpha-amylase Inhibitor Activity in Sorghum (Sorghum bicolour L.). Plant Foods Hum. Nutr. 1993, 44, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Amalraj, S.; Gurav, S.S.; Kalaskar, M.G.; Maroyi, A.; Ayyanar, M. Hypoglycemic, Anti-inflammatory, and Anti-aging Potential of Canthium coromandelicum (Burm.f.) Alston Leaf Extracts: In vitro and in silico ADMET studies. S. Afr. J. Bot. 2023, 161, 377–387. [Google Scholar] [CrossRef]
- Sansenya, S.; Payaka, A.; Mansalai, P. Biological Activity and Inhibition Potential against α-Glucosidase and α-Amylase of 2,4-Di-Tert-Butylphenol from Bamboo Shoot Extract by In Vitro and In Silico Studies. Process Biochem. 2023, 126, 15–22. [Google Scholar] [CrossRef]
- Artanti, N.; Tachibana, S.; Kardono, L.B.S.; Sukiman, H. Isolation of Alpha-Glucosidase Inhibitors Produced by an Endophytic Fungus, Colletotrichum Sp. TSC13 from Taxus Sumatrana. Pak. J. Biol. Sci. 2012, 15, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Van Chen, T.; Thong, C.L.T.; Tran-Trung, H.; Nhung, N.T.A.; Triet, N.T. Phytochemical Constituents Isolated From Distichochlamys citrea M.F. Newman Rhizomes and Their Antioxidant and α-Glucosidase Inhibitory Activities. Nat. Prod. Commun. 2023, 18, 1934578X231177878. [Google Scholar] [CrossRef]
- Knockenhauer, K.E.; Copeland, R.A. The Importance of Binding Kinetics and Drug–Target Residence Time in Pharmacology. Br. J. Pharmacol. 2023. [Google Scholar] [CrossRef]
- Suttithumsatid, W.; Shah, M.A.; Bibi, S.; Panichayupakaranant, P. α-Glucosidase Inhibitory Activity of Cannabidiol, Tetrahydrocannabinol and Standardized Cannabinoid Extracts from Cannabis Sativa. Curr. Res. Food Sci. 2022, 5, 1091–1097. [Google Scholar] [CrossRef]
- Reddy, V.A.; Leong, S.H.; Jang, I.-C.; Rajani, S. Metabolic Engineering of Nicotiana benthamiana to Produce Cannabinoid Precursors and Their Analogues. Metabolites 2022, 12, 1181. [Google Scholar] [CrossRef]



| Chemicals | % of the Total of Each Chemical Constituent in Each Sample | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HS-TH-1-O | HS-TH-1-M-H | HS-TH-1-M-E | HS-TH-2-O | HS-TH-2-M-H | HS-TH-2-M-E | HS-FS-1-O | HS-FS-1-M-H | HS-FS-1-M-E | HS-FS-2-O | HS-FS-2-M-H | HS-FS-2-M-E | |
| Palmitic acid | 2.05 | 0.28 | 9.94 | 2.74 | 7.56 | 4.43 | - | 10.21 | 5.58 | - | - | 1.60 |
| Ethyl palmitoleate | - | - | - | - | - | 1.56 | - | - | - | - | - | - |
| Palmitic acid, ethyl ester | - | 2.25 | 4.51 | - | 1.52 | 7.14 | - | - | 8.57 | - | 1.18 | - |
| α-Linolenic acid (omega-3) | - | - | - | - | - | - | - | - | - | - | - | 1.52 |
| Oleic Acid | - | - | - | - | - | - | 3.51 | - | - | - | - | - |
| Linoleic acid (omega-6) | 20.09 * | 22.93 * | 35.13 * | 86.53 * | 66.24 * | 34.08 * | - | 17.63 * | 15.04 | - | - | 1.43 |
| trans-Oleic acid | 16.42 | - | - | - | - | - | - | - | 5.36 | - | - | - |
| Linoleic acid ethyl ester | - | 7.89 | 10.36 | - | - | 20.61 | - | 1.58 | 11.49 | - | 1.08 | - |
| Linolenic acid, ethyl ester | - | - | 12.27 | - | - | - | - | - | - | - | - | - |
| Ethyl Oleate | - | - | - | - | - | 13.02 | - | - | 10.72 | - | - | - |
| Stearic acid | - | - | - | - | - | - | - | 1.73 | - | - | - | - |
| Stearic acid ethyl ester | - | - | 3.92 | - | - | 2.37 | - | - | 2.65 | - | - | - |
| 2-Pentylfuran | - | - | - | - | - | - | - | 1.58 | - | - | - | - |
| Glycerin | - | - | - | - | - | 1.21 | - | - | - | - | - | - |
| (±)-Glycidol | - | - | - | - | - | - | - | - | - | - | - | 1.58 |
| (2R,4R)-2,4 imethyl-1-heptanol | - | - | - | - | - | - | - | 1.23 | - | - | - | - |
| 4H-Pyran-4-one, 2,3-dihydro-3,5-dihydroxy-6- methyl- | - | - | - | - | - | - | - | - | - | - | - | 3.19 |
| 2-Isopropyl-5-methyl-6-oxabicyclo[3.1.0]hexane-1-carbaldehyde | - | - | - | - | - | - | - | 1.83 | - | - | - | - |
| (2E,4E)-2,4-Decadienal | - | 6.94 | - | - | - | - | - | 8.07 | - | - | - | - |
| 5-Pentyl-2(5H)-furanone | - | - | - | - | - | - | - | 1.27 | - | - | - | - |
| 7-Ethyl-4-nonanone | - | 1.95 | - | - | - | - | - | 3.73 | - | - | - | - |
| Guanosine | - | - | - | - | - | - | - | - | - | - | - | 18.58 |
| 2,4-Di-tert-butylphenol | - | - | - | - | - | 3.19 | - | - | - | - | - | - |
| Benzoic acid, 4-ethoxy-, ethyl ester | - | 1.30 | - | - | - | - | - | 1.21 | - | - | 1.41 | - |
| Tyramine | - | - | 1.45 | - | - | - | - | - | - | - | - | - |
| Crocetane | - | - | - | - | - | - | - | - | - | - | 1.03 | - |
| Myo inositol | - | - | - | - | - | - | - | - | - | - | - | 5.84 |
| α,β-Gluco-octonic acid lactone | - | - | - | - | - | - | - | - | - | - | - | 30.39 * |
| N-(2-Furylmethyl)-2-methylanilin | - | - | - | - | - | - | - | - | - | - | - | 1.31 |
| 1-Octadecanol | - | 1.39 | - | - | - | - | - | 1.17 | - | - | 1.67 | - |
| 1-Docosanol | - | - | - | - | - | - | - | 1.06 | - | - | 1.95 | - |
| 2-Palmitoylglycerol | - | - | - | - | - | - | - | - | 1.08 | - | - | - |
| 3-Amino-2-methyl-3-(4-methylphenyl)-1-phenyl-1-propanol | - | - | - | - | - | 1.97 | - | - | - | - | - | - |
| Linoleic acid, TMS | - | - | 2.24 | 1.15 | - | - | - | - | 2.86 | - | - | - |
| 2-Monoolein | - | - | - | - | - | - | - | 2.53 | - | - | - | - |
| glyceryl-linoleate | - | - | 8.03 | - | - | - | - | - | 15.12 * | - | - | - |
| β-Monolinolein | - | 3.78 | - | - | - | 2.09 | - | - | - | - | 2.83 | - |
| Nonanoic acid, 9-(3-hexenylidenecyclopropylidene)-, 2-hydroxy-1-(hydroxymethyl)ethyl ester, (Z,Z,Z)- | - | - | 3.84 | - | - | - | - | - | 6.04 | - | - | - |
| Triterpenoid | 2.40 | - | - | - | - | - | 1.66 | - | - | 2.18 | - | - |
| Nonacosane | - | - | - | - | - | - | - | - | - | 1.77 | 1.14 | - |
| Vitamin E succinate (calcium) | 12.63 | - | - | - | - | - | 14.16 | - | - | 15.10 | 1.09 | - |
| Vitamin E | - | - | - | - | - | - | 1.25 | - | - | - | - | - |
| Campesterol | - | 1.68 | - | - | - | - | - | 1.61 | 1.04 | - | 4.81 | - |
| (3methyl,24R)-ergost-5-en-3-ol | 3.78 | - | - | - | - | - | 7.61 | - | - | 5.28 | - | - |
| Stigmasterol | - | - | - | - | - | - | 1.41 | - | - | 1.45 | - | - |
| Clionasterol | 15.35 | 6.66 | 2.75 | 1.53 | 1.44 | 1.55 | 29.07 * | 5.67 | 3.80 | 22.32 * | 13.42 * | - |
| (E)-24-Propylidenecholesterol | - | - | - | - | - | - | - | 1.73 | - | - | - | - |
| 23(Z)-ethylcholestanol | - | - | - | - | - | - | - | - | - | - | 1.05 | - |
| (3b,24Z)-Stigmasta-5,24(28)-dien-3-ol | - | - | - | - | - | - | 5.95 | - | - | 4.30 | 2.08 | - |
| Lanosterol | 1.91 | - | - | - | - | - | 5.22 | - | - | 4.29 | 2.10 | - |
| Cycloartenol | 1.85 | - | - | - | - | - | 3.48 | - | - | 1.74 | 2.03 | - |
| γ- Sitostenone | 1.69 | 2.13 | - | - | 1.35 | - | - | 1.13 | - | - | 2.81 | - |
| 9-Octadecenoic acid, (2-phenyl-1,3-dioxolan-4- yl) methyl ester, cis- | - | - | - | - | - | - | - | 1.50 | - | - | - | - |
| Phytyl linoleate | - | - | - | - | - | - | - | - | - | 2.86 | - | - |
| Palmitoleyl oleate | 2.61 | - | - | - | - | - | - | - | - | - | - | - |
| Trilinolein | - | - | - | - | - | - | - | - | - | - | 9.29 | - |
| ∆9-Tetrahydrocannabivarin (THCV) | 1.20 | - | - | - | - | - | - | - | - | 5.62 | 1.05 | 2.73 |
| 2-(6-Isopropenyl-3-methyl-2-cyclohexen-1-yl)-5-pentyl-1,3-benzenediol | - | - | - | - | - | - | - | - | 1.04 | - | - | - |
| Cannabidiol (CBD) | 0.35 | - | - | - | - | - | 13.39 | - | - | 0.53 | - | - |
| ∆9THC (Dronabino)l | 3.36 | - | - | - | - | - | - | - | - | 18.20 | 2.70 | 3.81 |
| Cannabinol (CBN) | 1.02 | 0.51 | - | - | - | - | - | - | - | - | - | - |
| Bacteria | MIC/MBC Values (µg/mL) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HS-TH-1-O | HS-TH-1-M-E | HS-TH-1-M-H | HS-TH-2-O | HS-TH-2-M-E | HS-TH-2-M-H | HS-FS-1-M-E | HS-FS-2- M-E | Oxacillin | Vancomycin | Norfloxacin | |
| SA | 2048/>2048 | 512/>2048 | 2048/>2048 | 128/>2048 | 512/>2048 | 256/>2048 | - | - | 0.125/0.50 | ND | ND |
| SE | - | 128/>2048 | - | 256/>2048 | 256/>2048 | - | 256/>2048 | 2048/>2048 | 0.125/0.50 | ND | ND |
| MRSA | - | 512/>2048 | - | 256/>2048 | 1024/>2048 | 512/>2048 | - | - | ND | 0.5/1 | ND |
| CA | - | 512/512 | - | 256/512 | 512/512 | 256/256 | 2048/2048 | 1024/1024 | 0.25/0.25 | ND | ND |
| EC | - | - | - | - | - | - | - | - | ND | ND | 2/4 |
| PA | - | - | - | - | - | - | - | - | ND | ND | 4/8 |
| No. | Sample | Extract Part | Code | % Inhibition ± SD | IC50 (µg/mL) | |
|---|---|---|---|---|---|---|
| 2 mg/mL | 0.25 mg/mL | |||||
| 1. | RPF-1 | Oil | HS-TH-1-O | 15.13 ± 2.85 | - | |
| Marc | HS-TH-1-M-H | 18.46 ± 2.75 | - | |||
| HS-TH-1-M-E | 98.51 ± 0.14 | 87.34 ± 1.23 * | 42.72 | |||
| 2. | RPF-3 | Oil | HS-TH-2-O | 16.94 ± 3.02 | - | |
| Marc | HS-TH-2-M-H | 15.97 ± 3.45 | - | |||
| HS-TH-2-M-E | 97.99 ± 0.31 | 85.58 ± 0.07 * | 33.27 | |||
| 3. | FO#2 | Oil | HS-FS-1-O | 5.66 ± 1.28 | - | |
| Marc | HS-FS-1-M-H | 20.68 ± 4.70 | - | |||
| HS-FS-1-M-E | 100.36 ± 2.4 | 75.93 ± 2.40 * | 94.09 | |||
| 4. | FO#3 | Oil | HS-FS-2-O | 14.78 ± 0.46 | - | |
| Marc | HS-FS-2-M-H | 10.95 ± 2.63 | - | |||
| HS-FS-2-M-E | 98.99 ± 0.08 | 83.83 ± 1.10 * | 67.44 | |||
| Control | Acarbose | 93.95 ± 1.07 | 47.40 ± 0.66 | 285.92 | ||
| Acarbose IC50 Value Ratio | HS-TH-2-M-E IC50 Value Ratio | ||||
|---|---|---|---|---|---|
| 0.25 | 0.50 | 1 | 2 | 4 | |
| 4 | 0.90 | 0.70 | 0.71 | 0.80 | 0.96 |
| 2 | 0.96 | 0.95 | 0.99 | 0.96 | 0.99 |
| 1 | 2.77 | 1.30 | 1.06 | 0.92 | 0.85 |
| 0.50 | 1.39 | 1.36 | 1.21 | 0.94 | 0.93 |
| 0.25 | 1.83 | 1.01 | 0.97 | 0.81 | 0.83 |
| No | Energy | THC | THCV |
|---|---|---|---|
| 1 | Final intermolecular Energy (kcal/mol) | −5.42 | −5.48 |
| vdW + Hbond + desvol Energy | −5.33 | −5.29 | |
| Electrostatic Energy | −0.09 | −0.2 | |
| 2 | Final Total Internal Energy (kcal/mol) | −0.41 | −0.20 |
| 3 | Torsional Free Energy (kcal/mol) | +1.49 | +0.89 |
| 4 | Unbound System’s Energy (kcal/mol) | 0.00 | 0.00 |
| 5 | Estimate the Free Energy of Binding (kcal/mol) | −4.34 | −4.72 |
| 6 | Estimate Ki (mM) | 0.66 | 0.34 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sangkanu, S.; Pitakbut, T.; Phoopha, S.; Khanansuk, J.; Chandarajoti, K.; Dej-adisai, S. A Comparative Study of Chemical Profiling and Bioactivities between Thai and Foreign Hemp Seed Species (Cannabis sativa L.) Plus an In-Silico Investigation. Foods 2024, 13, 55. https://doi.org/10.3390/foods13010055
Sangkanu S, Pitakbut T, Phoopha S, Khanansuk J, Chandarajoti K, Dej-adisai S. A Comparative Study of Chemical Profiling and Bioactivities between Thai and Foreign Hemp Seed Species (Cannabis sativa L.) Plus an In-Silico Investigation. Foods. 2024; 13(1):55. https://doi.org/10.3390/foods13010055
Chicago/Turabian StyleSangkanu, Suthinee, Thanet Pitakbut, Sathianpong Phoopha, Jiraporn Khanansuk, Kasemsiri Chandarajoti, and Sukanya Dej-adisai. 2024. "A Comparative Study of Chemical Profiling and Bioactivities between Thai and Foreign Hemp Seed Species (Cannabis sativa L.) Plus an In-Silico Investigation" Foods 13, no. 1: 55. https://doi.org/10.3390/foods13010055
APA StyleSangkanu, S., Pitakbut, T., Phoopha, S., Khanansuk, J., Chandarajoti, K., & Dej-adisai, S. (2024). A Comparative Study of Chemical Profiling and Bioactivities between Thai and Foreign Hemp Seed Species (Cannabis sativa L.) Plus an In-Silico Investigation. Foods, 13(1), 55. https://doi.org/10.3390/foods13010055