Regulator of G-Protein Signalling 9: A New Candidate Gene for Sweet Food Liking?
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Data Collection
2.3. Genotyping and Imputation
2.4. Genome-Wide Association Study and Meta-Analysis
2.5. Replication Analysis
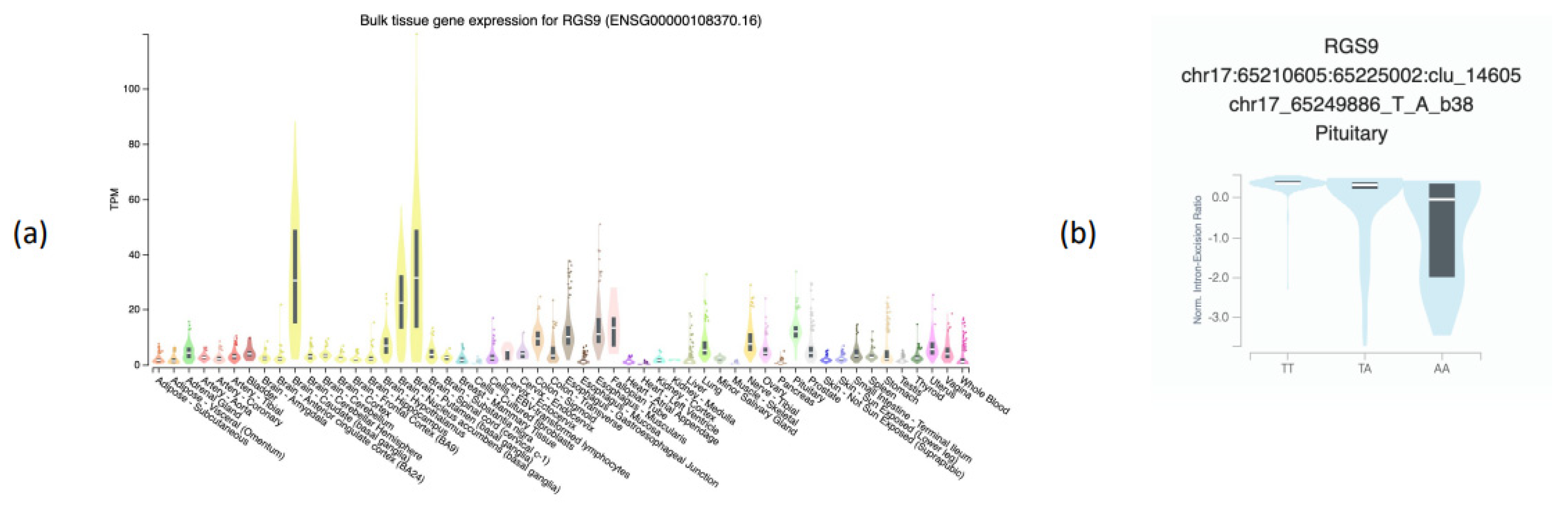
2.6. Genotype-Tissue Expression (GTEx) Analysis
2.7. Association of the Strongest Result with Other Phenotypes
2.8. Association of the Sweet Liking Group with Known Genes/SNPs
3. Results
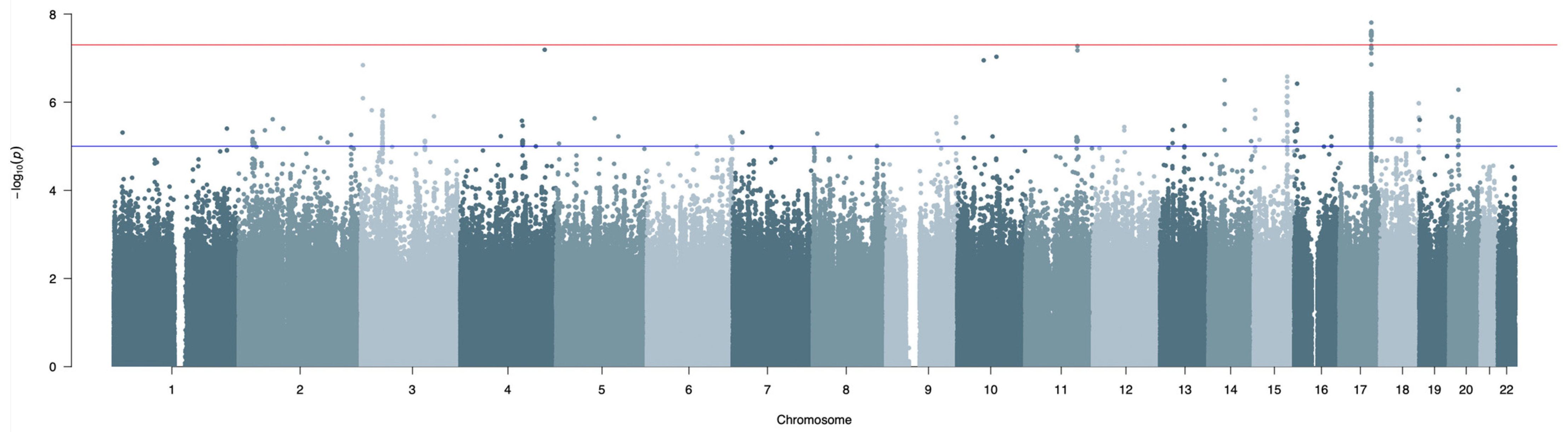
3.1. GWAS and Meta-Analysis Results
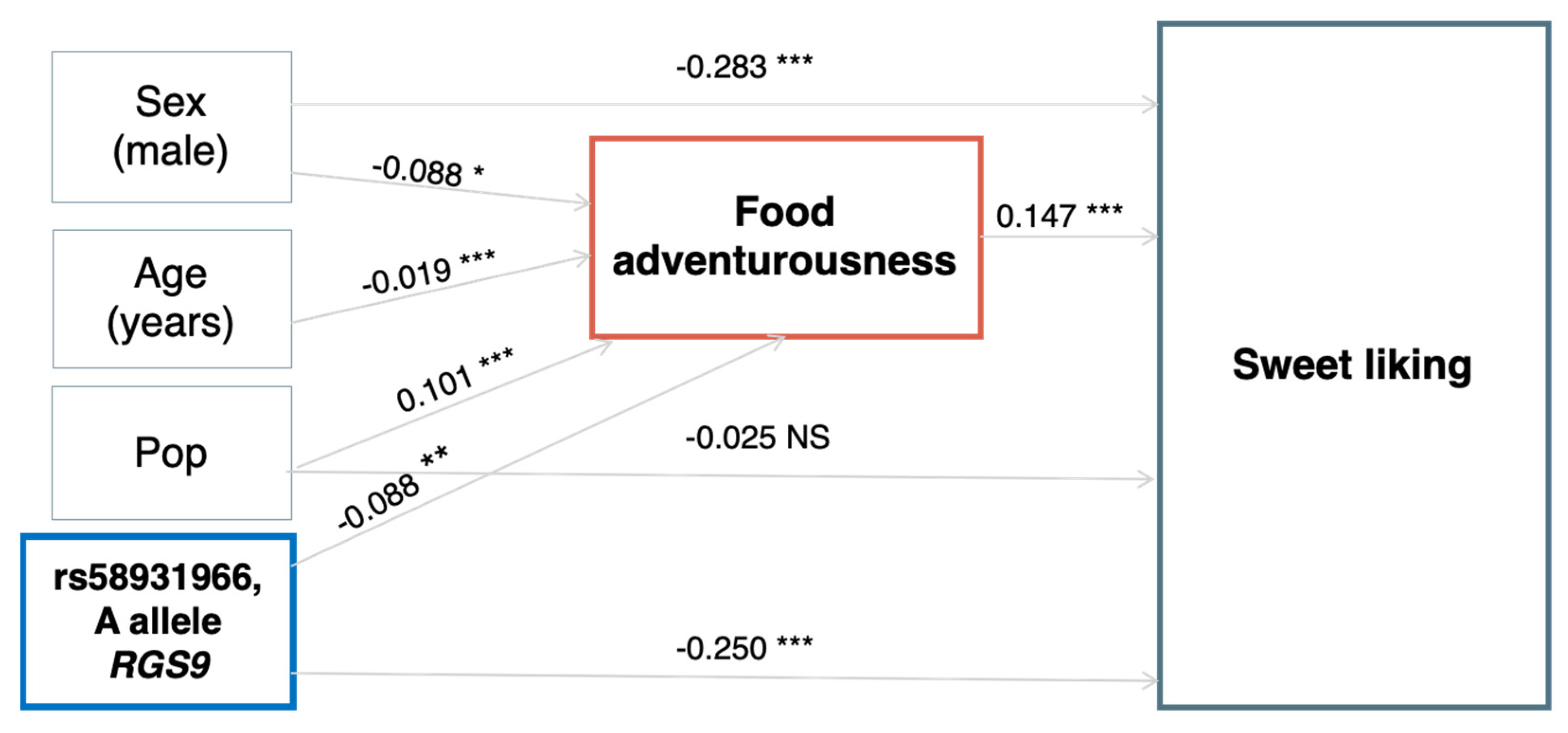
3.2. Association of RGS9 rs58931966 with Other Health and Eating-Related Psychological Traits
3.3. Association of the Sweet Liking Group with Other Known Genes/SNPs
4. Discussion
Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Viguiliouk, E.; Glenn, A.J.; Nishi, S.K.; Chiavaroli, L.; Seider, M.; Khan, T.; Bonaccio, M.; Iacoviello, L.; Mejia, S.B.; Jenkins, D.J.A.; et al. Associations between dietary pulses alone or with other legumes and cardiometabolic disease outcomes: An umbrella review and updated systematic review and meta-analysis of prospective cohort studies. Adv. Nutr. 2019, 10 (Suppl. 4), S308–S319. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; De Souza, R.J.; Choo, V.L.; Ha, V.; Cozma, A.I.; Chiavaroli, L.; Mirrahimi, A.; Mejia, S.B.; Di Buono, M.; Bernstein, A.M.; et al. Effects of dietary pulse consumption on body weight: A systematic review and meta-analysis of randomized controlled trials. Am. J. Clin. Nutr. 2016, 103, 1213–1223. [Google Scholar] [CrossRef]
- Becerra-Tomás, N.; Paz-Graniel, I.; Tresserra-Rimbau, A.; Martínez-González, M.; Barrubés, L.; Corella, D.; Muñoz-Martínez, J.; Romaguera, D.; Vioque, J.; Alonso-Gómez, Á.M.; et al. Fruit consumption and cardiometabolic risk in the PREDIMED-plus study: A cross-sectional analysis. Nutr. Metab. Cardiovasc. Dis. 2021, 31, 1702–1713. [Google Scholar] [CrossRef] [PubMed]
- Ramne, S.; Alves Dias, J.; González-Padilla, E.; Olsson, K.; Lindahl, B.; Engström, G.; Ericson, U.; Johansson, I.; Sonestedt, E. Association between added sugar intake and mortality is nonlinear and dependent on sugar source in 2 Swedish population–based PROSPECTIVE COHORTS. Am. J. Clin. Nutr. 2019, 109, 411–423. [Google Scholar] [CrossRef]
- Malik, V.S.; Hu, F.B. The role of sugar-sweetened beverages in the global epidemics of obesity and chronic diseases. Nat. Rev. Endocrinol. 2022, 18, 205–218. [Google Scholar] [CrossRef]
- Kumar, S.; Kelly, A.S. Review of childhood obesity: From epidemiology, etiology, and comorbidities to clinical assessment and treatment. Mayo. Clin. Proc. 2017, 92, 251–265. [Google Scholar] [CrossRef] [PubMed]
- Drozdz, D.; Alvarez-Pitti, J.; Wójcik, M.; Borghi, C.; Gabbianelli, R.; Mazur, A.; Herceg-čavrak, V.; Lopez-Valcarcel, B.G.; Brzeziński, M.; Lurbe, E.; et al. Obesity and cardiometabolic risk factors: From childhood to adulthood. Nutrients 2021, 13, 4176. [Google Scholar] [CrossRef]
- Ordovas, J.M.; Ferguson, L.R.; Tai, E.S.; Mathers, J.C. Personalised nutrition and health. BMJ 2018, 361, bmj.k2173. [Google Scholar] [CrossRef]
- Berridge, K.C. Food reward: Brain substrates of wanting and liking. Neurosci. Biobehav. Rev. 1996, 20, 1–25. [Google Scholar] [CrossRef]
- Feeney, E.; O’Brien, S.; Scannell, A.; Markey, A.; Gibney, E.R. Genetic variation in taste perception: Does it have a role in healthy eating? Proc. Nutr. Soc. 2011, 70, 135–143. [Google Scholar] [CrossRef]
- Jayasinghe, S.N.; Kruger, R.; Walsh, D.C.I.; Cao, G.; Rivers, S.; Richter, M.; Breier, B.H. Is sweet taste perception associated with sweet food liking and intake? Nutrients 2017, 9, 750. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Mohebbi, M.; Seo, H.S.; Hummel, T. Sensitivity to Sweetness correlates to elevated reward brain responses to sweet and high-fat food odors in young healthy volunteers. Neuroimage 2020, 208, 116413. [Google Scholar] [CrossRef]
- Shin, A.C.; Townsend, R.L.; Patterson, L.M.; Berthoud, H.R. “Liking” and “Wanting” of sweet and oily food stimuli as affected by high-fat diet-induced obesity, weight loss, leptin, and genetic predisposition. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011, 301, R1267–R1280. [Google Scholar] [CrossRef]
- Robino, A.; Concas, M.P.; Catamo, E.; Gasparini, P. A brief review of genetic approaches to the study of food preferences: Current knowledge and future directions. Nutrients 2019, 11, 1735. [Google Scholar] [CrossRef]
- Tuorila, H. Hedonic responses to falvor and their implications for food acceptance. Trends Food Sci. Technol. 1996, 7, 453–456. [Google Scholar] [CrossRef]
- Iatridi, V.; Armitage, R.M.; Yeomans, M.R.; Hayes, J.E. Effects of sweet-liking on body composition depend on age and lifestyle: A challenge to the simple sweet-liking-obesity hypothesis. Nutrients 2020, 12, 2702. [Google Scholar] [CrossRef] [PubMed]
- Moskowitz, H.R.; Jacobs, B.E.; Lazar, N. Product response segmentation and the analysis of individual differences in liking. J. Food Qual. 1985, 8, 169–181. [Google Scholar] [CrossRef]
- Armitage, R.M.; Iatridi, V.; Yeomans, M.R. Understanding sweet-liking phenotypes and their implications for obesity: Narrative review and future directions. Physiol. Behav. 2021, 235, 113398. [Google Scholar] [CrossRef] [PubMed]
- Beauchamp, G.K.; Moran, M. Acceptance of sweet and salty tastes in 2-year-old children. Appetite 1984, 5, 291–305. [Google Scholar] [CrossRef]
- Bacon, A.W.; Miles, J.S.; Schiffman, S.S. Effect of race on perception of fat alone and in combination with sugar. Physiol. Behav. 1994, 55, 603–606. [Google Scholar] [CrossRef]
- Eulalia, C.; Luciano, N.; Paolo, G.; Antonietta, R. Are taste variations associated with the liking of sweetened and unsweetened coffee? Physiol. Behav. 2022, 244, 113655. [Google Scholar] [CrossRef] [PubMed]
- Dalton, M.; Finlayson, G. Psychobiological examination of liking and wanting for fat and sweet taste in trait binge eating females. Physiol. Behav. 2014, 136, 128–134. [Google Scholar] [CrossRef]
- Weafer, J.; Lyon, N.; Hedeker, D.; de Wit, H. Sweet taste liking is associated with subjective response to amphetamine in women but not men. Psychopharmacology 2017, 234, 3185–3194. [Google Scholar] [CrossRef]
- Chamoun, E.; Mutch, D.M.; Allen-Vercoe, E.; Buchholz, A.C.; Duncan, A.M.; Spriet, L.L.; Haines, J.; Ma, D.W.L.; Guelph Family Health Study. A review of the associations between single nucleotide polymorphisms in taste receptors, eating behaviors, and health. Crit. Rev. Food Sci. Nutr. 2018, 58, 194–207. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Costanzo, A.; Evans, M.D.M.; Archer, N.S.; Nowson, C.; Duesing, K.; Keast, R. Expression of the candidate fat taste receptors in human fungiform papillae and the association with fat taste function. Br. J. Nutr. 2018, 120, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Kim, U.; Wooding, S.; Ricci, D.; Jorde, L.B.; Drayna, D. Worldwide haplotype diversity and coding sequence variation at human bitter taste receptor loci. Hum. Mutat. 2005, 26, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Habberstad, C.; Drake, I.; Sonestedt, E. Variation in the sweet taste receptor gene and dietary intake in a Swedish middle-aged population. Front. Endocrinol. 2017, 8, 348. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Keast, R.S.J.; Roura, E. Salivary leptin and TAS1R2/TAS1R3 polymorphisms are related to sweet taste sensitivity and carbohydrate intake from a buffet meal in healthy young adults. Br. J. Nutr. 2017, 118, 763–770. [Google Scholar] [CrossRef]
- Dias, A.G.; Eny, K.M.; Cockburn, M.; Chiu, W.; Nielsen, D.E.; Duizer, L.; El-Sohemy, A. Variation in the TAS1R2 gene, sweet taste perception and intake of sugars. Lifestyle Genom. 2015, 8, 81–90. [Google Scholar] [CrossRef]
- Diószegi, J.; Mohammad Kurshed, A.A.; Pikó, P.; Kósa, Z.; Sándor, J.; Ádány, R. Association of single nucleotide polymorphisms with taste and food preferences of the hungarian general and roma populations. Appetite 2021, 164, 105270. [Google Scholar] [CrossRef]
- Hwang, L.D.; Lin, C.; Gharahkhani, P.; Cuellar-Partida, G.; Ong, J.S.; An, J.; Gordon, S.D.; Zhu, G.; Macgregor, S.; Lawlor, D.A.; et al. New insight into human sweet taste: A genome-wide association study of the perception and intake of sweet substances. Am. J. Clin. Nutr. 2019, 109, 1724–1737. [Google Scholar] [CrossRef]
- Pirastu, N.; Kooyman, M.; Traglia, M.; Robino, A.; Willems, S.M.; Pistis, G.; Amin, N.; Sala, C.; Karssen, L.C.; Van Duijn, C.; et al. A genome-wide association study in isolated populations reveals new genes associated to common food likings. Rev. Endocr. Metab. Disorders 2016, 17, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Latimer, L.A.; Pope, L.; Wansink, B. Food neophiles: Profiling the adventurous eater. Obesity 2015, 23, 1577–1581. [Google Scholar] [CrossRef]
- Thondre, P.S. Food-based ingredients to modulate blood glucose. Adv. Food Nutr. Res. 2013, 70, 181–227. [Google Scholar] [CrossRef]
- Podboi, I.C.R.; Stephenson, S.; Pilic, L.; Graham, C.A.-M.; King, A.; Mavrommatis, Y. Dietary intake and TCF7L2 Rs7903146 T allele are associated with elevated blood glucose levels in healthy individuals. Lifestyle Genom. 2021, 14, 117–123. [Google Scholar] [CrossRef]
- Olsen, N.J.; Ängquist, L.; Larsen, S.C.; Linneberg, A.; Skaaby, T.; Husemoen, L.L.N.; Toft, U.; Tjønneland, A.; Halkjær, J.; Hansen, T.; et al. Interactions between genetic variants associated with adiposity traits and soft drinks in relation to longitudinal changes in body weight and waist circumference. Am. J. Clin. Nutr. 2016, 104, 816–826. [Google Scholar] [CrossRef]
- DiNicolantonio, J.J.; O’Keefe, J.H.; Wilson, W.L. Sugar addiction: Is it real? A narrative review. Br. J. Sports Med. 2018, 52, 910–913. [Google Scholar] [CrossRef]
- Concas, M.P.; Morgan, A.; Tesolin, P.; Santin, A.; Girotto, G.; Gasparini, P. Sensory capacities and eating behavior: Intriguing results from a large cohort of Italian individuals. Foods 2022, 11, 735. [Google Scholar] [CrossRef]
- Hazley, D.; McCarthy, S.N.; Stack, M.; Walton, J.; McNulty, B.A.; Flynn, A.; Kearney, J.M. Food neophobia and its relationship with dietary variety and quality in Irish adults: Findings from a national cross-sectional study. Appetite 2022, 169, 105859. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, N.V.; Touger-Decker, R.; O’sullivan-Maillet, J.; Tepper, B.J. PROP Taster status and self-perceived food adventurousness influence food preferences. J. Am. Diet. Assoc. 2004, 104, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Cocca, M.; Barbieri, C.; Concas, M.P.; Robino, A.; Brumat, M.; Gandin, I.; Trudu, M.; Sala, C.F.; Vuckovic, D.; Girotto, G.; et al. A bird’s-eye view of italian genomic variation through whole-genome sequencing. Eur. J. Hum. Genet. 2019, 28, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Tepper, B.J. Nutritional implications of genetic taste variation: The role of PROP sensitivity and other taste phenotypes. Annu. Rev. Nutr. 2008, 28, 367–388. [Google Scholar] [CrossRef]
- Concas, M.P.; Minelli, A.; Aere, S.; Morgan, A.; Tesolin, P.; Gasparini, P.; Gennarelli, M.; Girotto, G. Genetic dissection of temperament personality traits in Italian isolates. Genes 2021, 13, 4. [Google Scholar] [CrossRef]
- Howie, B.N.; Donnelly, P.; Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009, 5, 1000529. [Google Scholar] [CrossRef] [PubMed]
- The 1000 Genomes Project Consortium; Altshuler, D.M.; Durbin, R.M.; Abecasis, G.R.; Bentley, D.R.; Chakravarti, A.; Clark, A.G.; Donnelly, P.; Eichler, E.E.; Flicek, P.; et al. An integrated map of genetic variation from 1092 human genomes. Nature 2012, 491, 56–65. [Google Scholar] [CrossRef]
- Aulchenko, Y.S.; Ripke, S.; Isaacs, A.; van Duijn, C.M. GenABEL: An R library for genome-wide association analysis. Bioinformatics 2007, 23, 1294–1296. [Google Scholar] [CrossRef] [PubMed]
- Concas, M.P.; Catamo, E.; Biino, G.; Toniolo, D.; Gasparini, P.; Robino, A. Factors Associated with food liking and their relationship with metabolic traits in italian cohorts. Food Qual. Prefer. 2019, 75, 64–70. [Google Scholar] [CrossRef]
- Willer, C.J.; Li, Y.; Abecasis, G.R. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010, 26, 2190–2191. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The ensembl variant effect predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef]
- GTEx Consortium; Ardlie, K.G.; DeLuca, D.S.; Segrè, A.V.; Sullivan, T.J.; Young, T.R.; Gelfand, E.T.; Trowbridge, C.A.; Maller, J.B.; Tukiainen, T.; et al. The genotype-tissue expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015, 348, 648–660. [Google Scholar] [CrossRef]
- Rosseel, Y. Lavaan: An R package for structural equation modeling. J. Stat. Softw. 2012, 48, 1–36. [Google Scholar] [CrossRef]
- Wright, S. Correlation and causation. J. Agric. Res. 1921, 20, 557–585. [Google Scholar]
- Tabachnick, B.G. Using Multivariate Statistics; Allyn and Bacon: Boston, MA, USA, 2001. [Google Scholar]
- Sobel, M.E. Asymptotic confidence intervals for indirect effects in structural equation models. Sociol. Methodol. 1982, 13, 290. [Google Scholar] [CrossRef]
- Turner, S.D. Qqman: An R package for visualizing GWAS results using Q-Q and manhattan plots. J. Open Source Softw. 2018, 3, 731. [Google Scholar] [CrossRef]
- Pruim, R.J.; Welch, R.P.; Sanna, S.; Teslovich, T.M.; Chines, P.S.; Gliedt, T.P.; Boehnke, M.; Abecasis, G.R.; Willer, C.J.; Frishman, D. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics 2010, 26, 2336–2337. [Google Scholar] [CrossRef]
- Traynor, J.R.; Neubig, R.R. Regulators of G protein signaling & drugs of abuse. Mol. Interv. 2005, 5, 30. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Alonso, M.; Woods, S.C.; Pelchat, M.; Grigson, P.S.; Stice, E.; Farooqi, S.; Khoo, C.S.; Mattes, R.D.; Beauchamp, G.K. Food reward system: Current perspectives and future research needs. Nutr. Rev. 2015, 73, 296. [Google Scholar] [CrossRef]
- Navandar, M.; Martín-García, E.; Maldonado, R.; Lutz, B.; Gerber, S.; Ruiz de Azua, I. Transcriptional signatures in prefrontal cortex confer vulnerability versus resilience to food and cocaine addiction-like behavior. Sci. Rep. 2021, 11, 9076. [Google Scholar] [CrossRef]
- Papantoni, A.; Shearrer, G.E.; Sadler, J.R.; Stice, E.; Burger, K.S. Longitudinal associations between taste sensitivity, taste liking, dietary intake and BMI in adolescents. Front. Psychol. 2021, 12, 597704. [Google Scholar] [CrossRef]
- Fernández-Carrión, R.; Sorlí, J.V.; Coltell, O.; Pascual, E.C.; Ortega-Azorín, C.; Barragán, R.; Giménez-Alba, I.M.; Alvarez-Sala, A.; Fitó, M.; Ordovas, J.M.; et al. Sweet taste preference: Relationships with other tastes, liking for sugary foods and exploratory genome-wide association analysis in subjects with metabolic syndrome. Biomedicines 2022, 10, 79. [Google Scholar] [CrossRef]
- Gao, M.; Jebb, S.A.; Aveyard, P.; Ambrosini, G.L.; Perez-Cornago, A.; Carter, J.; Sun, X.; Piernas, C. Associations between dietary patterns and the incidence of total and fatal cardiovascular disease and all-cause mortality in 116,806 individuals from the UK biobank: A prospective cohort study. BMC Med. 2021, 19, 83. [Google Scholar] [CrossRef]
- Debras, C.; Chazelas, E.; Srour, B.; Kesse-Guyot, E.; Julia, C.; Zelek, L.; Agaesse, C.; Druesne-Pecollo, N.; Galan, P.; Hercberg, S.; et al. Total and added sugar intakes, sugar types, and cancer risk: Results from the prospective NutriNet-Santé cohort. Am. J. Clin. Nutr. 2020, 112, 1267–1279. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.K. A multiple-entry, modular memory system. Psychol. Learn. Motiv. -Adv. Res. Theory 1983, 17, 81–123. [Google Scholar] [CrossRef]
- Johnson, M.K.; Kim, J.K.; Risse, G. Do alcoholic Korsakoff’s syndrome patients acquire affective reactions? J. Exp. Psychol. Learn. Mem. Cogn. 1985, 11, 22–36. [Google Scholar] [CrossRef]
- Stice, E.; Burger, K.S.; Yokum, S. Relative ability of fat and sugar tastes to activate reward, gustatory, and somatosensory regions. Am. J. Clin. Nutr. 2013, 98, 1377. [Google Scholar] [CrossRef] [PubMed]
- Roberts, C.A.; Giesbrecht, T.; Fallon, N.; Thomas, A.; Mela, D.J.; Kirkham, T.C. A systematic review and activation likelihood estimation meta-analysis of FMRI studies on sweet taste in humans. J. Nutr. 2020, 150, 1619–1630. [Google Scholar] [CrossRef] [PubMed]
- Traynor, J. Regulator of G protein-signaling proteins and addictive drugs. In Annals of the New York Academy of Sciences; Blackwell Publishing Inc.: Hoboken, NJ, USA, 2010; pp. 341–352. [Google Scholar] [CrossRef]
- Zachariou, V.; Georgescu, D.; Sanchez, N.; Rahman, Z.; DiLeone, R.; Berton, O.; Neve, R.L.; Sim-Selley, L.J.; Selley, D.E.; Gold, S.J.; et al. Essential role for RGS9 in opiate action. Proc. Natl. Acad. Sci. USA 2003, 100, 13656–13661. [Google Scholar] [CrossRef] [PubMed]
- Wolz, I.; Granero, R.; Fernández-Aranda, F. A Comprehensive model of food addiction in patients with binge-eating symptomatology: The essential role of negative urgency. Compr. Psychiatry 2017, 74, 118–124. [Google Scholar] [CrossRef]
- Jacques, A.; Chaaya, N.; Beecher, K.; Ali, S.A.; Belmer, A.; Bartlett, S. The impact of sugar consumption on stress driven, emotional and addictive behaviors. Neurosci. Biobehav. Rev. 2019, 103, 178–199. [Google Scholar] [CrossRef] [PubMed]
- Gordon, E.L.; Ariel-Donges, A.H.; Bauman, V.; Merlo, L.J. What is the evidence for “food addiction?” A systematic review. Nutrients 2018, 10, 477. [Google Scholar] [CrossRef]
- Finlayson, G. Food addiction and obesity: Unnecessary medicalization of hedonic overeating. Nat. Rev. Endocrinol. 2017, 13, 493–498. [Google Scholar] [CrossRef]
- Waugh, J.L.; Celver, J.; Sharma, M.; Dufresne, R.L.; Terzi, D.; Risch, S.C.; Fairbrother, W.G.; Neve, R.L.; Kane, J.P.; Malloy, M.J.; et al. Association between regulator of G protein signaling 9–2 and body weight. PLoS ONE 2011, 6, 27984. [Google Scholar] [CrossRef] [PubMed]
- Walker, P.D.; Jarosz, P.A.; Bouhamdan, M.; MacKenzie, R.G. Effects of gender on locomotor sensitivity to amphetamine, body weight, and fat mass in regulator of G protein signaling 9 (RGS9) knockout mice. Physiol. Behav. 2015, 138, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Kral, T.V.E. Food neophobia and its association with diet quality and weight status in children. In Food Neophobia: Behavioral and Biological Influences; Elsevier: Amsterdam, The Netherlands, 2018; pp. 287–303. [Google Scholar] [CrossRef]
- Hayes, J.E.; Pickering, G.J. Wine expertise predicts taste phenotype. Am. J. Enol. Vitic. 2012, 63, 80. [Google Scholar] [CrossRef] [PubMed]
- Falciglia, G.A.; Couch, S.C.; Gribble, L.S.; Pabst, S.M.; Frank, R. Food neophobia in childhood affects dietary variety. J. Am. Diet. Assoc. 2000, 100, 1474–1481. [Google Scholar] [CrossRef] [PubMed]

| Demographic | Discovery Sample | Replication Sample | |
|---|---|---|---|
| VBI | CAR | FVG | |
| N (M/F) | 1109 (434/675) | 373 (164/209) | 1073 (475/598) |
| Age, years | 56.1 (16.5) | 52.4 (17.1) | 50.8 (16.2) |
| Mean liking of sweet foods | 6.4 (1.5) | 6.4 (1.8) | 6.3 (1.6) |
| FA | 1.9 (1.2) | 1.8 (1.0) | 1.8 (0.9) |
| RD (n = 528) | -- | -- | 52.3 (22.6) |
| BMI, kg/m2 | 25.6 (4.4) | 26.5 (4.8) | 25.4 (4.7) |
| Blood Glucose, mg/dL | 89.0 (14.8) | 94.9 (16.9) | 93.3 (16.2) |
| Discovery Samples (CAR + VBI) | Discovery and Replication Samples (CAR + VBI + FVG) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr | Position | Near Gene | Distance | EA/OA | Mean Freq of EA | n | Effect | StdErr | p-Value | n | Effect | StdErr | p-Value |
| rs192789286 | 11 | 103927811 | DDI1 | 17,889 | A/G | 0.022 | 1482 | −1.154 | 0.212 | 5.32 × 10−8 | NA | NA | NA | NA |
| rs9896491 | 17 | 63234031 | RGS9 | 10,210 | A/G | 0.760 | 1482 | 0.360 | 0.065 | 2.90 × 10−8 | 2555 | 0.286 | 0.051 | 1.89 × 10−8 |
| rs6504288 | 17 | 63234885 | RGS9 | 11,064 | A/G | 0.239 | 1482 | −0.363 | 0.065 | 2.85 × 10−8 | 2555 | −0.292 | 0.052 | 1.40 × 10−8 |
| rs6504289 | 17 | 63235085 | RGS9 | 11,264 | A/G | 0.760 | 1482 | 0.362 | 0.065 | 2.42 × 10−8 | 2555 | 0.285 | 0.051 | 2.24 × 10−8 |
| rs57333496 | 17 | 63236332 | RGS9 | 12,511 | T/C | 0.773 | 1482 | 0.359 | 0.066 | 5.22 × 10−8 | 2555 | 0.314 | 0.053 | 2.69 × 10−9 |
| rs9916255 | 17 | 63239497 | RGS9 | 15,676 | T/C | 0.760 | 1482 | 0.362 | 0.065 | 2.78 × 10−8 | 2555 | 0.282 | 0.051 | 3.19 × 10−8 |
| rs55820790 | 17 | 63240317 | RGS9 | 16,496 | A/G | 0.774 | 1482 | 0.367 | 0.066 | 3.13 × 10−8 | 2555 | 0.321 | 0.053 | 1.44 × 10−9 |
| rs16961703 | 17 | 63240344 | RGS9 | 16,523 | A/G | 0.774 | 1482 | 0.367 | 0.066 | 3.16 × 10−8 | 2555 | 0.321 | 0.053 | 1.43 × 10−9 |
| rs7221051 | 17 | 63241713 | RGS9 | 17,892 | A/G | 0.226 | 1482 | −0.368 | 0.066 | 2.84 × 10−8 | 2555 | −0.322 | 0.053 | 1.32 × 10−9 |
| rs7221258 | 17 | 63241956 | RGS9 | 18,135 | C/G | 0.774 | 1482 | 0.369 | 0.067 | 2.95 × 10−8 | 2555 | 0.322 | 0.053 | 1.31 × 10−9 |
| rs55864812 | 17 | 63242632 | RGS9 | 18,811 | A/G | 0.774 | 1482 | 0.367 | 0.066 | 3.06 × 10−8 | 2555 | 0.321 | 0.053 | 1.38 × 10−9 |
| rs7213152 | 17 | 63243529 | RGS9 | 19,708 | T/C | 0.774 | 1482 | 0.371 | 0.067 | 2.56 × 10−8 | 2555 | 0.323 | 0.053 | 1.30 × 10−9 |
| rs7212442 | 17 | 63243586 | RGS9 | 19,765 | A/G | 0.226 | 1482 | −0.371 | 0.067 | 2.56 × 10−8 | 2555 | −0.323 | 0.053 | 1.30 × 10−9 |
| rs16961868 | 17 | 63244061 | RGS9 | 20,240 | A/G | 0.775 | 1482 | 0.372 | 0.067 | 2.57 × 10−8 | 2555 | 0.324 | 0.053 | 1.23 × 10−9 |
| rs62063085 | 17 | 63245030 | RGS9 | 21,209 | T/C | 0.226 | 1482 | −0.369 | 0.067 | 2.75 × 10−8 | 2555 | −0.323 | 0.053 | 1.25 × 10−9 |
| rs7342966 | 17 | 63245750 | RGS9 | 21,929 | T/C | 0.774 | 1482 | 0.370 | 0.067 | 2.91 × 10−8 | 2555 | 0.323 | 0.053 | 1.49 × 10−9 |
| rs58931966 | 17 | 63246004 | RGS9 | 22,183 | A/T | 0.240 | 1482 | −0.372 | 0.066 | 1.56 × 10−8 | 2555 | −0.297 | 0.051 | 7.05 × 10−9 |
| Trait | Population | SNP | Gene | EA/OA | N | Effect | StdErr | p-Value |
|---|---|---|---|---|---|---|---|---|
| FA | CAR + VBI + FVG | rs58931966 | RGS9 | A/T | 2543 | −0.065 | 0.033 | 0.0490 |
| RD | FVG | rs58931966 | RGS9 | A/T | 587 | −3.840 | 1.527 | 0.011 |
| BMI | CAR + VBI + FVG | rs58931966 | RGS9 | A/T | 2519 | 0.391 | 0.144 | 0.007 |
| Glucose | CAR + VBI + FVG | rs58931966 | RGS9 | A/T | 2444 | 1.211 | 0.489 | 0.013 |
| Gene | SNP | Already Associated Phenotype | DOI | Effect Allele | Other Allele | Effect | StdErr | p-Value | Direction |
|---|---|---|---|---|---|---|---|---|---|
| PHACTR2 | rs1416208 | Sensory perception of sweet taste | 10.1093/AJCN/NQZ043 | A | T | 0.1392 | 0.0483 | 0.00392 | +++ |
| LINC01277 | rs57083985 | Sensory perception of sweet taste | 10.1093/AJCN/NQZ043 | T | G | −0.1317 | 0.0465 | 0.004657 | −−− |
| GRID1 | rs7897266 | Aspartame | 10.1093/AJCN/NQZ043 | T | G | 0.2092 | 0.0825 | 0.01121 | +++ |
| TAS1R3 | rs35424002 | Sweet perception | 10.1111/j.1750-3841.2012.02852.x | A | G | 0.2284 | 0.0926 | 0.01361 | −++ |
| RP11-401I19.2 | rs306356 | Intake of total sugars | 10.1093/AJCN/NQZ043 | A | C | 0.2847 | 0.121 | 0.01863 | −++ |
| TAS1R2 | rs7534618 | Sweet perception | 10.1111/j.1750-3841.2012.02852.x | T | G | 0.1066 | 0.0466 | 0.02214 | +++ |
| ANO3 | rs75941298 | Sensory perception of sweet taste | 10.1093/AJCN/NQZ043 | A | G | −0.172 | 0.0775 | 0.02643 | −−− |
| CAPN13 | rs115354913 | Intake of total sugars | 10.1093/AJCN/NQZ043 | A | G | 0.2712 | 0.1224 | 0.02677 | +++ |
| SSBP4 | rs8106096 | Intake of total sugars | 10.1093/AJCN/NQZ043 | C | G | −0.104 | 0.0498 | 0.03675 | −−− |
| AC074019.1 | rs62202380 | Intake of total sugars | 10.1093/AJCN/NQZ043 | T | C | −0.2426 | 0.122 | 0.04675 | +−− |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Graham, C.A.-M.; Spedicati, B.; Pelliccione, G.; Gasparini, P.; Concas, M.P. Regulator of G-Protein Signalling 9: A New Candidate Gene for Sweet Food Liking? Foods 2023, 12, 1739. https://doi.org/10.3390/foods12091739
Graham CA-M, Spedicati B, Pelliccione G, Gasparini P, Concas MP. Regulator of G-Protein Signalling 9: A New Candidate Gene for Sweet Food Liking? Foods. 2023; 12(9):1739. https://doi.org/10.3390/foods12091739
Chicago/Turabian StyleGraham, Catherine Anna-Marie, Beatrice Spedicati, Giulia Pelliccione, Paolo Gasparini, and Maria Pina Concas. 2023. "Regulator of G-Protein Signalling 9: A New Candidate Gene for Sweet Food Liking?" Foods 12, no. 9: 1739. https://doi.org/10.3390/foods12091739
APA StyleGraham, C. A.-M., Spedicati, B., Pelliccione, G., Gasparini, P., & Concas, M. P. (2023). Regulator of G-Protein Signalling 9: A New Candidate Gene for Sweet Food Liking? Foods, 12(9), 1739. https://doi.org/10.3390/foods12091739






