Data-Mining Poultry Processing Bio-Mapping Counts of Pathogens and Indicator Organisms for Food Safety Management Decision Making
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection, Indicator Enumeration, and Pathogen Detection and Quantification
2.2. Statistical Analysis
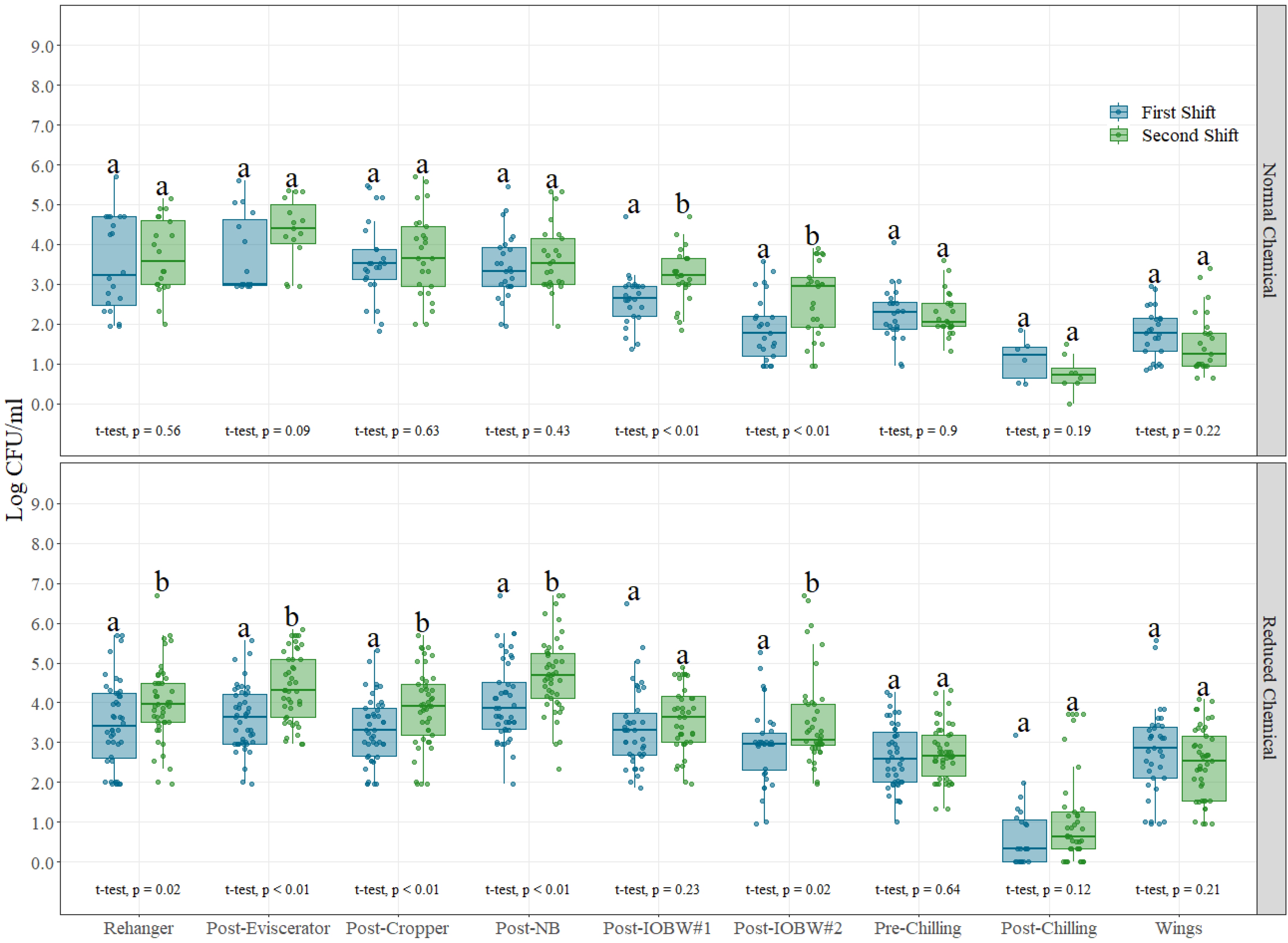
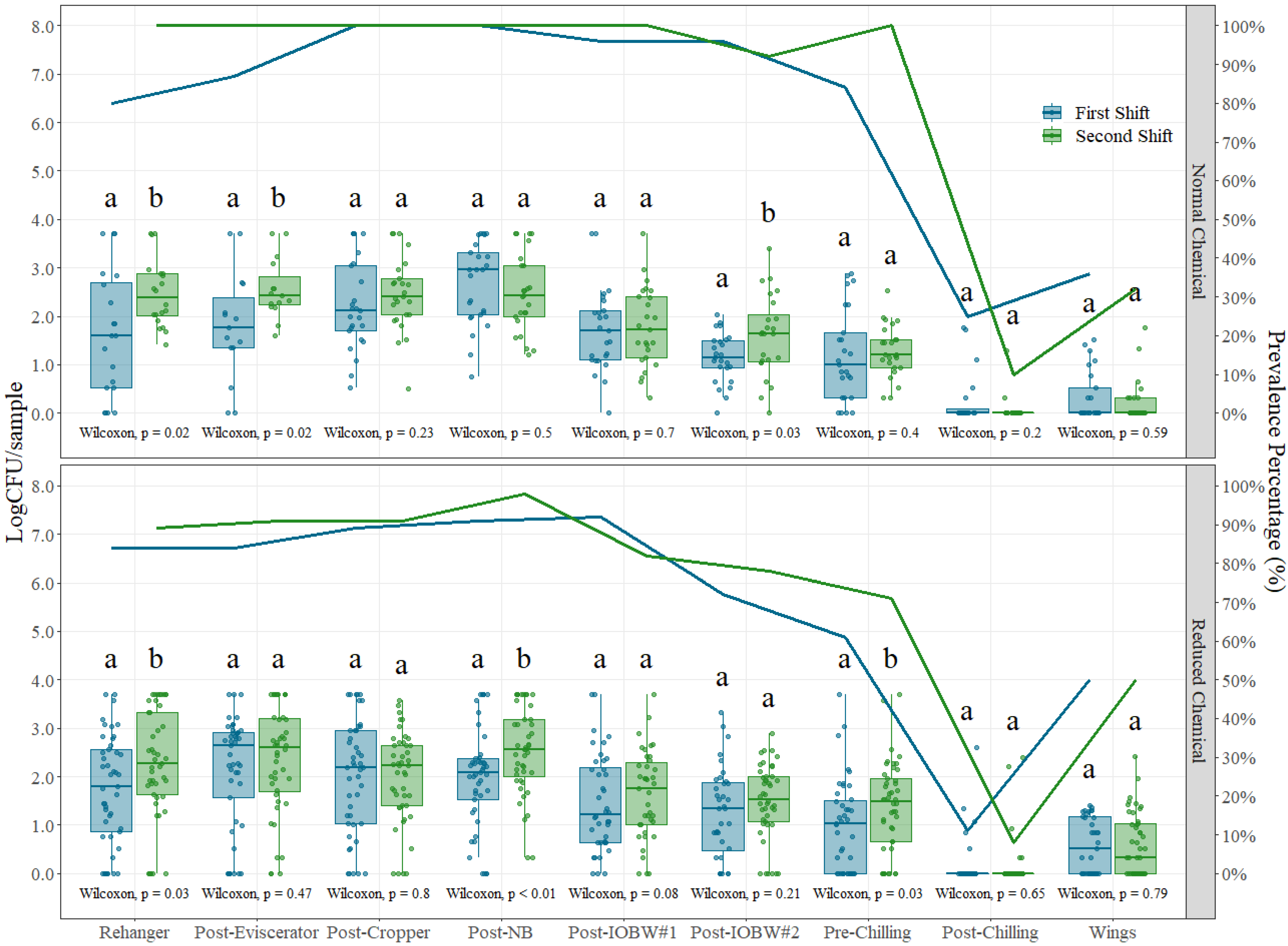
2.2.1. Shift Comparison
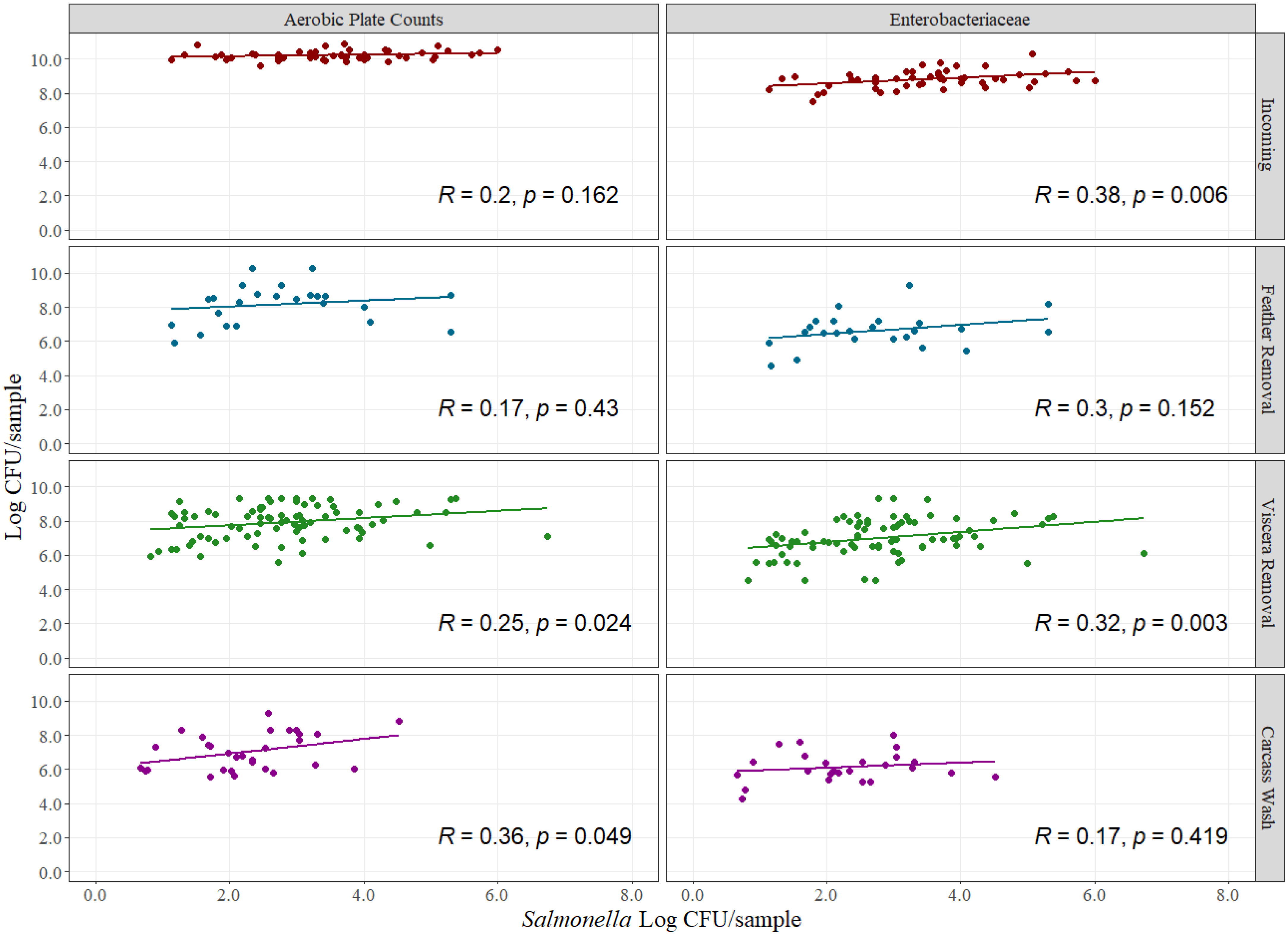
2.2.2. Indicators vs. Pathogens Correlation
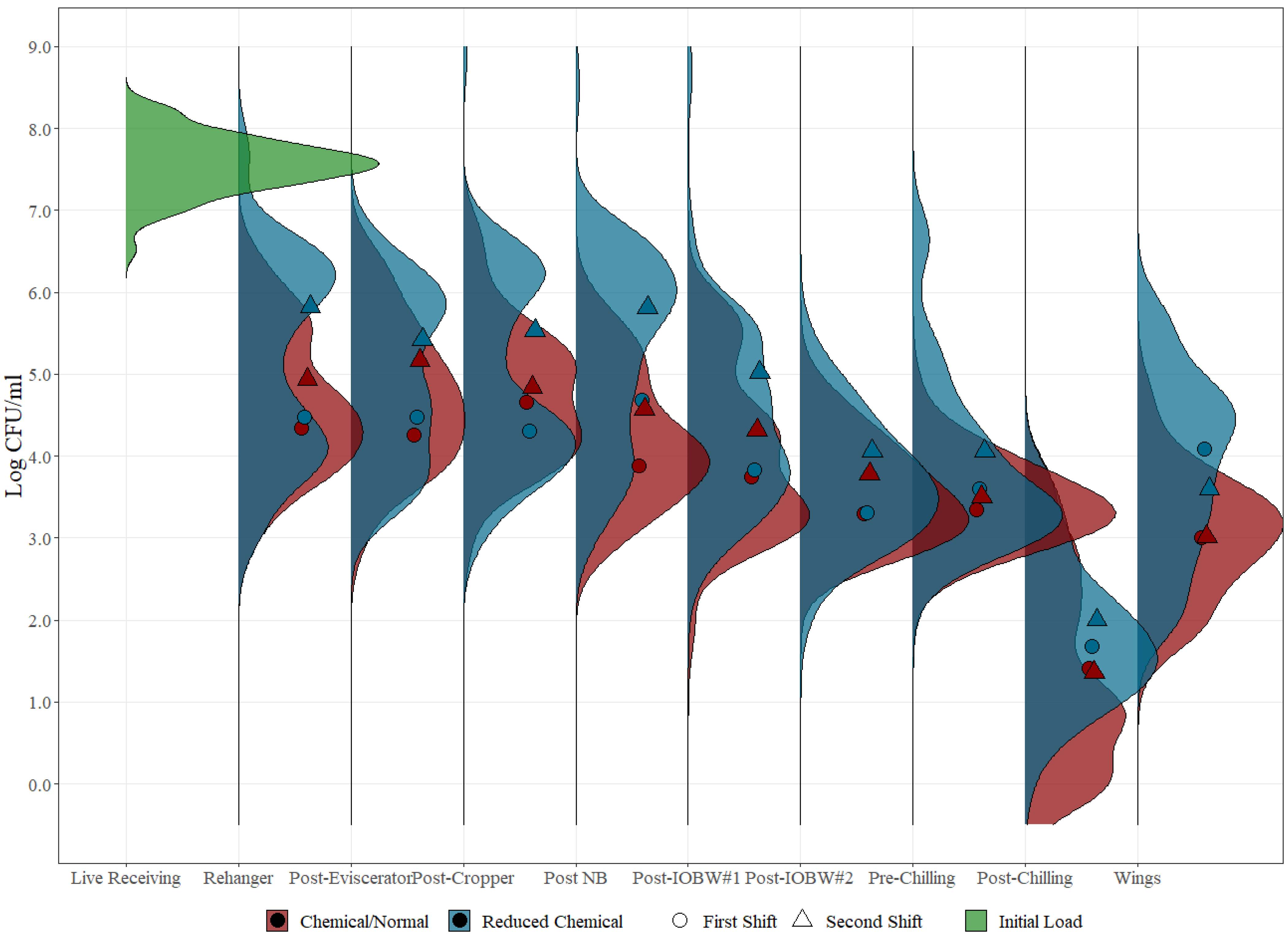
2.2.3. Indicator and Pathogen Distribution
3. Results and Discussion
- (a)
- The use of pathogen quantification can improve the use of risk assessment where interventions can target specific stages with higher loads of indicator and pathogen bacteria.
- (b)
- Non-difference between normal and reduced in chemical interventions in certain locations suggests the application of chemical interventions in strategic locations.
- (c)
- The use of prevalence as a sole measurement of food safety performance can lead to inadequate results.
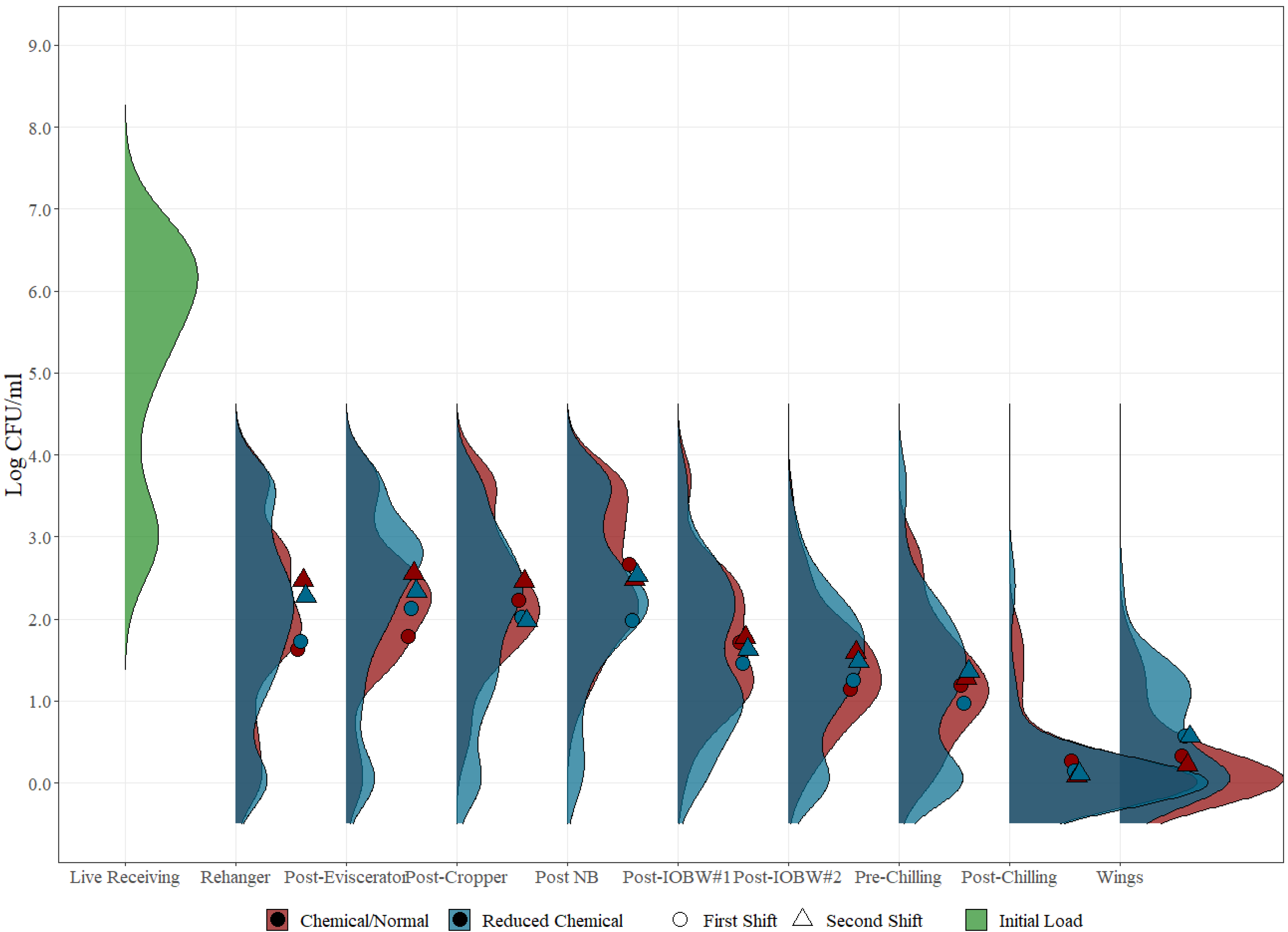
3.1. Shift Comparison
3.2. Indicator vs. Pathogen Levels Correlation
3.3. Indicator and Pathogen Distribution
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Range (Log CFU/mL) | Sampling Point | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Live Receiving | Rehanger | Post- Eviscerator | Post-Cropper | Post-NB | Post-IOBW#1 | Post-IOBW#2 | Pre-Chilling | Post-Chilling | Wings | |
| <0.0 | 0.0% (0/70) | 0.0% (0/40) | 0.0% (0/30) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/49) | 7.1% (1/14) | 0.0% (0/50) |
| 0.0–1.0 | 0.0% (0/70) | 0.0% (0/40) | 0.0% (0/30) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 14.0% (7/50) | 2.0% (1/49) | 50.0% (7/14) | 22.0% (11/50) |
| >1.0–2.0 | 0.0% (0/70) | 5.0% (2/40) | 0.0% (0/30) | 2.0% (1/50) | 4.0% (2/50) | 10.0% (5/50) | 30.0% (15/50) | 38.8% (19/49) | 42.9% (6/14) | 48.0% (24/50) |
| >2.0–3.0 | 0.0% (0/70) | 30.0% (12/40) | 20.0% (6/30) | 20.0% (10/50) | 20.0% (10/50) | 42.0% (21/50) | 24.0% (12/50) | 49.0% (24/49) | 0.0% (0/14) | 26.0% (13/50) |
| >3.0–4.0 | 0.0% (0/70) | 20.0% (8/40) | 23.3% (7/30) | 44.0% (22/50) | 50.0% (25/50) | 40.0% (20/50) | 32.0% (16/50) | 8.2% (4/49) | 0.0% (0/14) | 4.0% (2/50) |
| >4.0–5.0 | 5.7% (4/70) | 40.0% (16/40) | 33.3% (10/30) | 18.0% (9/50) | 18.0% (9/50) | 8.0% (4/50) | 0.0% (0/50) | 2.0% (1/49) | 0.0% (0/14) | 0.0% (0/50) |
| >5.0–6.0 | 34.3% (24/70) | 5.0% (2/40) | 23.3% (7/30) | 16.0% (8/50) | 8.0% (4/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/49) | 0.0% (0/14) | 0.0% (0/50) |
| >6.0–7.0 | 52.9% (37/70) | 0.0% (0/40) | 0.0% (0/30) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/49) | 0.0% (0/14) | 0.0% (0/50) |
| >7.0 | 7.1% (5/70) | 0.0% (0/40) | 0.0% (0/30) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/49) | 0.0% (0/14) | 0.0% (0/50) |
Appendix B
| Range (Log CFU/mL) | Sampling Point | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Live Receiving | Rehanger | Post- Eviscerator | Post-Cropper | Post-NB | Post-IOBW#1 | Post-IOBW#2 | Pre-Chilling | Post-Chilling | Wings | |
| <0.0 | 0.0% (0/70) | 0.0% (0/89) | 0.0% (0/90) | 0.0% (0/90) | 0.0% (0/90) | 0.0% (0/80) | 0.0% (0/80) | 0.0% (0/90) | 1.2% (1/86) | 0.0% (0/80) |
| 0.0–1.0 | 0.0% (0/70) | 0.0% (0/89) | 0.0% (0/90) | 0.0% (0/90) | 0.0% (0/90) | 0.0% (0/80) | 1.3% (1/80) | 0.0% (0/90) | 10.5% (9/86) | 6.3% (5/80) |
| >1.0–2.0 | 0.0% (0/70) | 6.7% (6/89) | 1.1% (1/90) | 7.8% (7/90) | 1.1% (1/90) | 2.5% (2/80) | 7.5% (6/80) | 0.0% (0/90) | 51.2% (44/86) | 21.3% (17/80) |
| >2.0–3.0 | 0.0% (0/70) | 14.6% (13/89) | 14.4% (13/90) | 17.8% (16/90) | 6.7% (6/90) | 27.5% (22/80) | 40.0% (32/80) | 13.3% (12/90) | 24.4% (21/86) | 35.0% (28/80) |
| >3.0–4.0 | 0.0% (0/70) | 38.2% (34/89) | 35.6% (32/90) | 42.2% (38/90) | 31.1% (28/90) | 41.3% (33/80) | 32.5% (26/80) | 51.1% (46/90) | 12.8% (11/86) | 32.5% (26/80) |
| >4.0–5.0 | 5.7% (4/70) | 30.3% (27/89) | 30.0% (27/90) | 21.1% (19/90) | 32.2% (29/90) | 25.0% (20/80) | 11.3% (9/80) | 22.2% (20/90) | 0.0% (0/86) | 2.5% (2/80) |
| >5.0–6.0 | 34.3% (24/70) | 9.0% (8/89) | 18.9% (17/90) | 11.1% (10/90) | 22.2% (20/90) | 2.5% (2/80) | 5.0% (4/80) | 7.8% (7/90) | 0.0% (0/86) | 2.5% (2/80) |
| >6.0–7.0 | 52.9% (37/70) | 1.1% (1/89) | 0.0% (0/90) | 0.0% (0/90) | 6.7% (6/90) | 1.3% (1/80) | 2.5% (2/80) | 5.6% (5/90) | 0.0% (0/86) | 0.0% (0/80) |
| >7.0 | 7.1% (5/70) | 0.0% (0/89) | 0.0% (0/90) | 0.0% (0/90) | 0.0% (0/90) | 0.0% (0/80) | 0.0% (0/80) | 0.0% (0/90) | 0.0% (0/86) | 0.0% (0/80) |
Appendix C
| Range (Log CFU/mL) | Sampling Point | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Live Receiving | Rehanger | Post- Eviscerator | Post-Cropper | Post-NB | Post-IOBW#1 | Post-IOBW#2 | Pre-Chilling | Post-Chilling | Wings | |
| <0.0 | 0.0% (0/70) | 0.0% (0/40) | 0.0% (0/30) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 23.3% (7/30) | 0.0% (0/50) |
| 0.0–1.0 | 0.0% (0/70) | 0.0% (0/40) | 0.0% (0/30) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 26.7% (8/30) | 0.0% (0/50) |
| >1.0–2.0 | 0.0% (0/70) | 0.0% (0/40) | 0.0% (0/30) | 0.0% (0/50) | 0.0% (0/50) | 2.0% (1/50) | 0.0% (0/50) | 0.0% (0/50) | 16.7% (5/30) | 6.0% (3/50) |
| >2.0–3.0 | 0.0% (0/70) | 2.5% (1/40) | 0.0% (0/30) | 2.0% (1/50) | 6.0% (3/50) | 50.0% (25/50) | 14.0% (7/50) | 16.0% (8/50) | 23.3% (7/30) | 42.0% (21/50) |
| >3.0–4.0 | 0.0% (0/70) | 22.5% (9/40) | 23.3% (7/30) | 20.0% (10/50) | 34.0% (17/50) | 26.0% (13/50) | 62.0% (31/50) | 74.0% (37/50) | 10.0% (3/30) | 44.0% (22/50) |
| >4.0–5.0 | 0.0% (0/70) | 45.0% (18/40) | 43.3% (13/30) | 38.0% (19/50) | 38.0% (19/50) | 22.0% (11/50) | 24.0% (12/50) | 10.0% (5/50) | 0.0% (0/30) | 8.0% (4/50) |
| >5.0–6.0 | 0.0% (0/70) | 22.5% (9/40) | 23.3% (7/30) | 34.0% (17/50) | 22.0% (11/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/30) | 0.0% (0/50) |
| >6.0–7.0 | 2.86% (2/70) | 7.5% (3/40) | 10.0% (3/30) | 6.0% (3/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/30) | 0.0% (0/50) |
| >7.0 | 97.14% (68/70) | 0.0% (0/40) | 0.0% (0/30) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/30) | 0.0% (0/50) |
Appendix D
| Range (Log CFU/mL) | Sampling Point | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Live Receiving | Rehanger | Post- Eviscerator | Post-Cropper | Post-NB | Post-IOBW#1 | Post-IOBW#2 | Pre-Chilling | Post-Chilling | Wings | |
| <0.0 | 0.0% (0/70) | 0.0% (0/90) | 0.0% (0/90) | 0.0% (0/90) | 0.0% (0/90) | 0.0% (0/80) | 0.0% (0/80) | 0.0% (0/90) | 1.2% (1/86) | 0.0% (0/80) |
| 0.0–1.0 | 0.0% (0/70) | 0.0% (0/90) | 0.0% (0/90) | 0.0% (0/90) | 0.0% (0/90) | 0.0% (0/80) | 0.0% (0/80) | 0.0% (0/90) | 22.1% (19/86) | 0.0% (0/80) |
| >1.0–2.0 | 0.0% (0/70) | 0.0% (0/90) | 0.0% (0/90) | 0.0% (0/90) | 0.0% (0/90) | 0.0% (0/80) | 0.0% (0/80) | 0.0% (0/90) | 51.2% (44/86) | 2.5% (2/80) |
| >2.0–3.0 | 0.0% (0/70) | 0.0% (0/90) | 0.0% (0/90) | 0.0% (0/90) | 1.1% (1/90) | 3.8% (3/80) | 18.8% (15/80) | 13.3% (12/90) | 24.4% (21/86) | 22.5% (18/80) |
| >3.0–4.0 | 0.0% (0/70) | 17.8% (16/90) | 25.6% (23/90) | 24.4% (22/90) | 17.7% (16/90) | 36.3% (29/80) | 51.3% (41/80) | 51.1% (46/90) | 12.8% (11/86) | 27.5% (22/80) |
| >4.0–5.0 | 0.0% (0/70) | 30.0% (27/90) | 26.7% (24/90) | 33.3% (30/90) | 22.2% (20/90) | 31.3% (25/80) | 26.3% (21/80) | 22.2% (20/90) | 0.0% (0/86) | 35.0% (28/80) |
| >5.0–6.0 | 0.0% (0/70) | 17.8% (16/90) | 30.0% (27/90) | 15.6% (14/90) | 28.9% (26/90) | 25.0% (20/80) | 3.8% (3/80) | 7.8% (7/90) | 0.0% (0/86) | 12.5% (10/80) |
| >6.0–7.0 | 2.86% (2/70) | 31.1% (28/90) | 17.8% (16/90) | 25.6% (23/90) | 28.9% (26/90) | 3.0% (2/80) | 0.0% (0/80) | 5.6% (5/90) | 0.0% (0/86) | 0.0% (0/80) |
| >7.0 | 97.14% (68/70) | 3.3% (3/90) | 0.0% (0/90) | 1.1% (1/90) | 1.1% (0/90) | 1.3% (1/80) | 0.0% (0/80) | 0.0% (0/90) | 0.0% (0/86) | 0.0% (0/80) |
Appendix E
| Range (Log CFU/Sample) | Sampling Point | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Live Receiving | Rehanger | Post- Eviscerator | Post-Cropper | Post-NB | Post-IOBW#1 | Post-IOBW#2 | Pre-Chilling | Post-Chilling | Wings | |
| Negative | 5.7% (4/70) | 57.5% (23/40) | 53.3% (16/30) | 72.0% (36/50) | 84.0% (42/50) | 88.0% (44/50) | 90.0% (45/50) | 96.0% (48/50) | 100.0% (0/50) | 90.0% (45/50) |
| 0.3–1.0 | 21.43% (15/70) | 30.0% (12/40) | 20.0% (6/30) | 12.0% (6/50) | 14.0% (7/50) | 12.0% (6/50) | 10.0% (5/50) | 4.0% (2/50) | 0.0% (0/50) | 8.0% (4/50) |
| >1.0–2.0 | 8.6% (6/70) | 7.5% (3/40) | 13.3% (4/30) | 2.0% (1/50) | 2.0% (1/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 2.0% (1/50) |
| >2.0–3.0 | 12.9% (9/70) | 2.5% (1/40) | 3.3% (1/30) | 2.0% (1/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) |
| >3.0–4.0 | 5.7% (4/70) | 0.0% (0/40) | 10.0% (3/30) | 2.0% (1/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) |
| >4.0–5.0 | 12.9% (9/70) | 2.5% (1/40) | 0.0% (0/30) | 2.0% (1/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) |
| >5.0–6.0 | 10.0% (7/70) | 0.0% (0/40) | 0.0% (0/30) | 2.0% (1/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) |
| >6.0 | 0.0% (0/70) | 0.0% (0/40) | 0.0% (0/30) | 2.0% (1/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) |
Appendix F
| Range (Log CFU/Sample) | Sampling Point | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Live Receiving | Rehanger | Post- Eviscerator | Post-Cropper | Post-NB | Post-IOBW#1 | Post-IOBW#2 | Pre-Chilling | Post-Chilling | Wings | |
| Negative | 5.7% (4/70) | 54.4% (49/90) | 60.0% (54/90) | 64.4% (58/90) | 66.7% (60/90) | 68.8% (55/80) | 83.8% (67/80) | 76.7% (69/90) | 98.9% (89/90) | 88.8% (71/80) |
| 0.3–1.0 | 21.43% (15/70) | 24.4% (22/90) | 14.4% (13/90) | 16.7% (15/90) | 10.0% (9/90) | 18.8% (15/80) | 11.3% (9/80) | 10.0% (9/90) | 1.1% (1/90) | 7.5% (6/80) |
| >1.0–2.0 | 8.6% (6/70) | 4.4% (4/90) | 3.3% (3/90) | 5.6% (5/90) | 4.4% (4/90) | 1.3% (1/80) | 2.5% (2/80) | 4.4% (4/90) | 0.0% (0/90) | 0.0% (0/80) |
| >2.0–3.0 | 12.9% (9/70) | 7.8% (7/90) | 10.0% (9/90) | 6.7% (6/90) | 13.3% (12/90) | 6.3% (5/80) | 2.5% (2/80) | 6.7% (6/90) | 0.0% (0/90) | 1.3% (1/80) |
| >3.0–4.0 | 28.6% (20/70) | 5.6% (5/90) | 10.0% (9/90) | 4.4% (4/90) | 4.4% (4/90) | 3.8% (3/80) | 0.0% (0/80) | 2.2% (2/90) | 0.0% (0/90) | 2.5% (2/80) |
| >4.0–5.0 | 12.9% (9/70) | 1.1% (1/90) | 2.2% (2/90) | 2.2% (2/90) | 1.1% (1/90) | 1.3% (1/80) | 0.0% (0/80) | 0.0% (0/90) | 0.0% (0/90) | 0.0% (0/80) |
| >5.0–6.0 | 10.0% (7/70) | 2.2% (2/90) | 0.0% (0/90) | 0.0% (0/90) | 0.0% (0/90) | 0.0% (0/80) | 0.0% (0/80) | 0.0% (0/90) | 0.0% (0/90) | 0.0% (0/80) |
| >6.0 | 0.0% (0/70) | 0.0% (0/90) | 0.0% (0/90) | 0.0% (0/90) | 0.0% (0/90) | 0.0% (0/80) | 0.0% (0/80) | 0.0% (0/90) | 0.0% (0/90) | 0.0% (0/80) |
Appendix G
| Range (Log CFU/mL) | Sampling Point | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Live Receiving | Rehanger | Post- Eviscerator | Post-Cropper | Post-NB | Post-IOBW#1 | Post-IOBW#2 | Pre-Chilling | Post-Chilling | Wings | |
| Negative | 0.0% (0/70) | 10.0% (4/40) | 6.7% (2/30) | 0.0% (0/50) | 0.0% (0/50) | 2.0% (1/50) | 4.1% (2/50) | 8.0% (4/50) | 82.5% (33/40) | 66.0% (33/50) |
| 0.3–1.0 | 0.0% (0/70) | 10.0% (4/40) | 3.3% (1/30) | 6.0% (3/50) | 6.0% (3/50) | 12.0% (6/50) | 22.4% (11/50) | 30.0% (15/50) | 7.5% (3/40) | 20.0% (10/50) |
| >1.0–2.0 | 0.0% (0/70) | 25.0% (10/40) | 26.7% (8/30) | 28.0% (14/50) | 28.0% (14/50) | 46.0% (23/50) | 59.2% (29/50) | 48.0% (24/50) | 10.0% (4/40) | 14.0% (7/50) |
| >2.0–3.0 | 17.1% (12/70) | 40.0% (16/40) | 43.3% (13/30) | 42.0% (21/50) | 42.0% (21/50) | 34.0% (17/50) | 12.2% (6/50) | 14.0% (7/50) | 0.0% (0/40) | 0.0% (0/50) |
| >3.0–4.0 | 5.7% (4/70) | 15.0% (6/40) | 20.0% (6/30) | 24.0% (12/50) | 24.0% (12/50) | 6.0% (3/50) | 2.0% (1/50) | 0.0% (0/50) | 0.0% (0/40) | 0.0% (0/50) |
| >4.0–5.0 | 10.0% (7/70) | 0.0% (0/40) | 0.0% (0/30) | 6.0% (3/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/40) | 0.0% (0/50) |
| >5.0–6.0 | 25.7% (18/70) | 0.0% (0/40) | 0.0% (0/30) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/40) | 0.0% (0/50) |
| >6.0 | 41.4% (29/70) | 0.0% (0/40) | 0.0% (0/30) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/50) | 0.0% (0/40) | 0.0% (0/50) |
Appendix H
| Range (Log CFU/mL) | Sampling Point | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Live Receiving | Rehanger | Post- Eviscerator | Post-Cropper | Post-NB | Post-IOBW#1 | Post-IOBW#2 | Pre-Chilling | Post-Chilling | Wings | |
| Negative | 0.0% (0/70) | 13.3% (12/90) | 11.2% (10/89) | 10.0% (0/90) | 4.5% (4/89) | 7.6% (6/79) | 16.3% (13/80) | 26.1% (23/88) | 89.6% (86/96) | 43.9% (36/82) |
| 0.0–1.0 | 0.0% (0/70) | 6.7% (6/90) | 5.6% (5/89) | 6.7% (6/90) | 6.7% (6/89) | 21.5% (17/79) | 12.5% (10/80) | 13.6% (12/88) | 5.2% (5/96) | 24.4% (20/82) |
| >1.0–2.0 | 0.0% (0/70) | 26.7% (24/90) | 15.7% (14/89) | 25.6% (23/90) | 21.3% (19/89) | 35.4% (28/79) | 48.8% (39/80) | 42.0% (37/88) | 2.1% (2/96) | 30.5% (25/82) |
| >2.0–3.0 | 17.1% (12/70) | 30.0% (27/90) | 42.7% (38/89) | 40.0% (36/90) | 41.6% (37/89) | 30.4% (24/79) | 20.0% (16/80) | 13.6% (12/88) | 3.1% (3/96) | 1.2% (1/82) |
| >3.0–4.0 | 5.7% (4/70) | 23.3% (21/90) | 24.7% (22/89) | 17.8% (16/90) | 25.8% (23/89) | 5.1% (4/79) | 2.5% (2/80) | 4.5% (4/88) | 0.0% (0/96) | 0.0% (0/82) |
| >4.0–5.0 | 10.0% (7/70) | 0.0% (0/90) | 0.0% (0/89) | 0.0% (0/90) | 0.0% (0/89) | 0.0% (0/79) | 0.0% (0/80) | 0.0% (0/88) | 0.0% (0/96) | 0.0% (0/82) |
| >5.0–6.0 | 25.7% (18/70) | 0.0% (0/90) | 0.0% (0/89) | 0.0% (0/90) | 0.0% (0/89) | 0.0% (0/79) | 0.0% (0/80) | 0.0% (0/88) | 0.0% (0/96) | 0.0% (0/82) |
| >6.0 | 41.4% (29/70) | 0.0% (0/90) | 0.0% (0/89) | 0.0% (0/90) | 0.0% (0/89) | 0.0% (0/79) | 0.0% (0/80) | 0.0% (0/88) | 0.0% (0/96) | 0.0% (0/82) |
References
- Weaver, M. Industry and Trade Summary Poultry Industry; U.S. International Trade in Goods and Services: Washington, DC, USA, 2014; pp. 1–89.
- De Villena, J.F.; Vargas, D.A.; López, R.B.; Chávez-Velado, D.R.; Casas, D.E.; Jiménez, R.L.; Sanchez-Plata, M.X. Bio-Mapping Indicators and Pathogen Loads in a Commercial Broiler Processing Facility Operating with High and Low Antimicrobial Intervention Levels. Foods 2022, 11, 775. [Google Scholar] [CrossRef] [PubMed]
- United States Department of Agriculture National Agricultural Statistics Service Poultry-Production and Value Poultry-Production and Value. Available online: https://www.nass.usda.gov/Publications/Todays_Reports/reports/plva0421.pdf (accessed on 5 January 2023).
- United States Department of Agriculture Economic Research Service Poultry & Eggs. Available online: https://www.ers.usda.gov/topics/animal-products/poultry-eggs (accessed on 9 January 2023).
- United States Departement of Agriculture Economic Research Service Per Capita Meat Consumption in the United States in 2021 and 2031, by Type. Available online: https://www.usda.gov/sites/default/files/documents/USDA-Agricultural-Projections-to-2031.pdf (accessed on 9 January 2023).
- Centers for Disease Control and Prevention. Estimates of Foodborne Illness in the United States; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2011. [Google Scholar]
- Umaraw, P.; Prajapati, A.; Verma, A.K.; Pathak, V.; Singh, V.P. Control of campylobacter in poultry industry from farm to poultry processing unit: A review. Crit. Rev. Food Sci. Nutr. 2017, 57, 659–665. [Google Scholar] [CrossRef] [PubMed]
- Chaney, W.E.; Englishbey, A.K.; Stephens, T.P.; Applegate, S.F.; Sanchez-Plata, M.X. Application of a Commercial Salmonella Real-Time PCR Assay for the Detection and Quantitation of Salmonella enterica in Poultry Ceca. J. Food Prot. 2022, 85, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention Salmonella. Available online: https://www.cdc.gov/salmonella/index.html (accessed on 9 January 2023).
- Centers for Disease Control and Prevention Campylobacter. Available online: https://www.cdc.gov/campylobacter/faq.html (accessed on 9 January 2023).
- Interagency Food Safety Analytics Collaboration. Foodborne Illness Source Attribution Estimates for 2018 for Salmonella, Escherichia coli O157, Listeria monocytogenes, and Campylobacter Using Multi-Year Outbreak Surveillance Data, United States. 2020. Available online: https://www.cdc.gov/foodsafety/ifsac/annual-reports.html. (accessed on 12 January 2023).
- Forgey, S.J. Development, Verification, and Validation of a RT-PCR Based Methodology for Salmonella Quantification as a Tool for Integrated Food Safety Management in Poultry from Live Production to Final product; Texas Tech University: Lubbock, TX, USA, 2020. [Google Scholar]
- Food Safety and Inspection Service Salmonella Verification testing Program Monthly Posting. Available online: https://www.fsis.usda.gov/science-data/data-sets-visualizations/microbiology/salmonella-verification-testing-program-monthly#:~:text=FSIS%20continuously%20samples%20(up%20to,establishment's%20process%20control%20over%20time (accessed on 9 January 2023).
- Wideman, N.; Bailey, M.; Bilgili, S.F.; Thippareddi, H.; Wang, L.; Bratcher, C.; Sanchez-Plata, M.; Singh, M. Evaluating best practices for Campylobacter and Salmonella reduction in poultry processing plants. Poult. Sci. 2016, 95, 306–315. [Google Scholar] [CrossRef] [PubMed]
- Hayama, Y.; Yamamoto, T.; Kasuga, F.; Tsutsui, T. Simulation Model for Campylobacter Cross-Contamination During Poultry Processing at Sluaghterhouses. Zoonoses Public Health 2011, 58, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; De Reu, K.; Gabriël, S.; Mattheus, W.; De Zutter, L.; Rasschaert, G. Salmonella prevalence and persistence in industrialized poultry slaughterhouses. Poult. Sci. 2021, 100, 100991. [Google Scholar] [CrossRef] [PubMed]
- Borges, K.A.; Cisco, I.C.; Furian, T.Q.; Tedesco, D.C.; Rodrigues, L.B.; do Nascimento, V.P.; dos Santos, L.R. Detection and quantification of Campylobacter spp. In Brazilian poultry processing plants. J. Infect. Dev. Ctries. 2020, 14, 109–113. [Google Scholar] [CrossRef] [PubMed]
- United States Department of Agriculture. The Nationwide Microbiological Baseline Data Collection Program: Raw Chicken Parts Survey; United States Department of Agriculture: Washington, DC, USA, 2012. [Google Scholar]
- Riyana, I.; Rahimi, B. The Importance of Data Science in the Fashion Industry. Available online: https://www.whichplm.com/the-importance-of-data-science-in-the-fashion-industry/ (accessed on 6 January 2023).
- Obe, T.; Nannapaneni, R.; Schilling, W.; Zhang, L.; McDaniel, C.; Kiess, A. Prevalence of Salmonella enterica on poultry processing equipment after completion of sanitization procedures. Poult. Sci. 2020, 99, 4539–4548. [Google Scholar] [CrossRef] [PubMed]
- Potter, B.D.; Marcy, J.A.; Owens, C.M.; Slavik, M.F.; Goodwin, H.L.; Apple, J.K. Impact of performance-based sanitation systems on microbiological characteristics of poultry processing equipment and carcasses as compared with traditional sanitation systems. J. Appl. Poult. Res. 2012, 21, 669–678. [Google Scholar] [CrossRef]
- Olsen, J.E.; Brown, D.J.; Madsen, M.; Bisgaard, M. Cross-contamination with Salmonella on a broiler slaughterhouse line demonstrated by use of epidemiological markers. J. Appl. Microbiol. 2003, 94, 826–835. [Google Scholar] [CrossRef] [PubMed]
- Botta, C.; Ferrocino, I.; Pessione, A.; Cocolin, L.; Rantsiou, K. Spatiotemporal Distribution of the Environmental Microbiota in Food Processing Plants as Impacted by Cleaning and Sanitizing Procedures: The Case of Slaughterhouses and Gaseous Ozone. Appl. Environ. Microbiol. 2020, 86, e01861-20. [Google Scholar] [CrossRef] [PubMed]
- Rasschaert, G.; Houf, K.; Godard, C.; Wildemauwe, C.; Pastuszczak-Frąk, M.; De Zutter, L. Contamination of carcasses with Salmonella during poultry slaughter. J. Food Prot. 2008, 71, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Møretrø, T.; Langsrud, S. Residential Bacteria on Surfaces in the Food Industry and Their Implications for Food Safety and Quality. Compr. Rev. Food Sci. Food Saf. 2017, 16, 1022–1041. [Google Scholar] [CrossRef] [PubMed]
- Food Safety and Inspection Service Performing the Pre-Operational Sanitation Standard Operating Procedures Verification Task. Available online: https://www.fsis.usda.gov/sites/default/files/media_file/2020-07/5000.4.pdf (accessed on 9 January 2023).
- Kotula, K.L.; Pandya, Y. Bacterial contamination of broiler chickens before scalding. J. Food Prot. 1995, 58, 1326–1329. [Google Scholar] [CrossRef] [PubMed]
- Rouger, A.; Tresse, O.; Zagorec, M. Bacterial contaminants of poultry meat: Sources, species, and dynamics. Microorganisms 2017, 5, 50. [Google Scholar] [CrossRef] [PubMed]
- Tortorello, M.L. Indicator Organisms for Safety and Quality—Uses and Methods.pdf. J. AOAC Int. 2003, 86, 1208. [Google Scholar] [CrossRef] [PubMed]
- Saini, P.K.; Marks, H.M.; Dreyfuss, M.S.; Evans, P.; Victor Cook, L.; Dessai, U. Indicator organisms in meat and poultry slaughter operations: Their potential use in process control and the role of emerging technologies. J. Food Prot. 2011, 74, 1387–1394. [Google Scholar] [CrossRef] [PubMed]
- Schaffner, D.W.; Smith-Simpson, S. Indicator Organisms in Meat; Elsevier Inc.: Amsterdam, The Netherlands, 2014. [Google Scholar]
- Mead, G.C. Fecal indicator organisms for red meat and poultry. In Microbiological Analysis of Red Meat, Poultry and Eggs; Mead, G.C., Ed.; Woodhead Publishing Series in Food Science; Technology and Nutrition: Cambridge, UK, 2007; pp. 83–100. ISBN 9781845690595. [Google Scholar]
- National Research Council. An Evaluation of the Role of Microbiological Criteria for Foods and Food Ingredients; National Academy Press: Washington, DC, USA, 1985; ISBN 0309034973. [Google Scholar]
- Vargas, D.A.; Rodríguez, K.M.; Betancourt-Barszcz, G.K.; Ajcet-Reyes, M.I.; Dogan, O.B.; Randazzo, E.; Sánchez-Plata, M.X.; Brashears, M.M.; Miller, M.F. Bio-Mapping of Microbial Indicators to Establish Statistical Process Control Parameters in a Commercial Beef Processing Facility. Foods 2022, 11, 133. [Google Scholar] [CrossRef] [PubMed]
- Roccato, A.; Mancin, M.; Barco, L.; Cibin, V.; Antonello, K.; Cocola, F.; Ricci, A. Usefulness of indicator bacteria as potential marker of Campylobacter contamination in broiler carcasses. Int. J. Food Microbiol. 2018, 276, 63–70. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vargas, D.A.; De Villena, J.F.; Larios, V.; Bueno López, R.; Chávez-Velado, D.R.; Casas, D.E.; Jiménez, R.L.; Blandon, S.E.; Sanchez-Plata, M.X. Data-Mining Poultry Processing Bio-Mapping Counts of Pathogens and Indicator Organisms for Food Safety Management Decision Making. Foods 2023, 12, 898. https://doi.org/10.3390/foods12040898
Vargas DA, De Villena JF, Larios V, Bueno López R, Chávez-Velado DR, Casas DE, Jiménez RL, Blandon SE, Sanchez-Plata MX. Data-Mining Poultry Processing Bio-Mapping Counts of Pathogens and Indicator Organisms for Food Safety Management Decision Making. Foods. 2023; 12(4):898. https://doi.org/10.3390/foods12040898
Chicago/Turabian StyleVargas, David A., Juan F. De Villena, Valeria Larios, Rossy Bueno López, Daniela R. Chávez-Velado, Diego E. Casas, Reagan L. Jiménez, Sabrina E. Blandon, and Marcos X. Sanchez-Plata. 2023. "Data-Mining Poultry Processing Bio-Mapping Counts of Pathogens and Indicator Organisms for Food Safety Management Decision Making" Foods 12, no. 4: 898. https://doi.org/10.3390/foods12040898
APA StyleVargas, D. A., De Villena, J. F., Larios, V., Bueno López, R., Chávez-Velado, D. R., Casas, D. E., Jiménez, R. L., Blandon, S. E., & Sanchez-Plata, M. X. (2023). Data-Mining Poultry Processing Bio-Mapping Counts of Pathogens and Indicator Organisms for Food Safety Management Decision Making. Foods, 12(4), 898. https://doi.org/10.3390/foods12040898






