In Vitro Inhibitory Effects of Polyphenols from Flos sophorae immaturus on α-Glucosidase: Action Mechanism, Isothermal Titration Calorimetry and Molecular Docking Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Preparation of the FSI Extract
2.3. Determination of α-Glucosidase Inhibitory Activity
2.4. Omission Experiment
2.5. Interaction Assay
2.6. α-Glucosidase Inhibition Type and Kinetic Analysis
2.7. Fluorescence Measurements
2.8. Thermodynamic Parameters
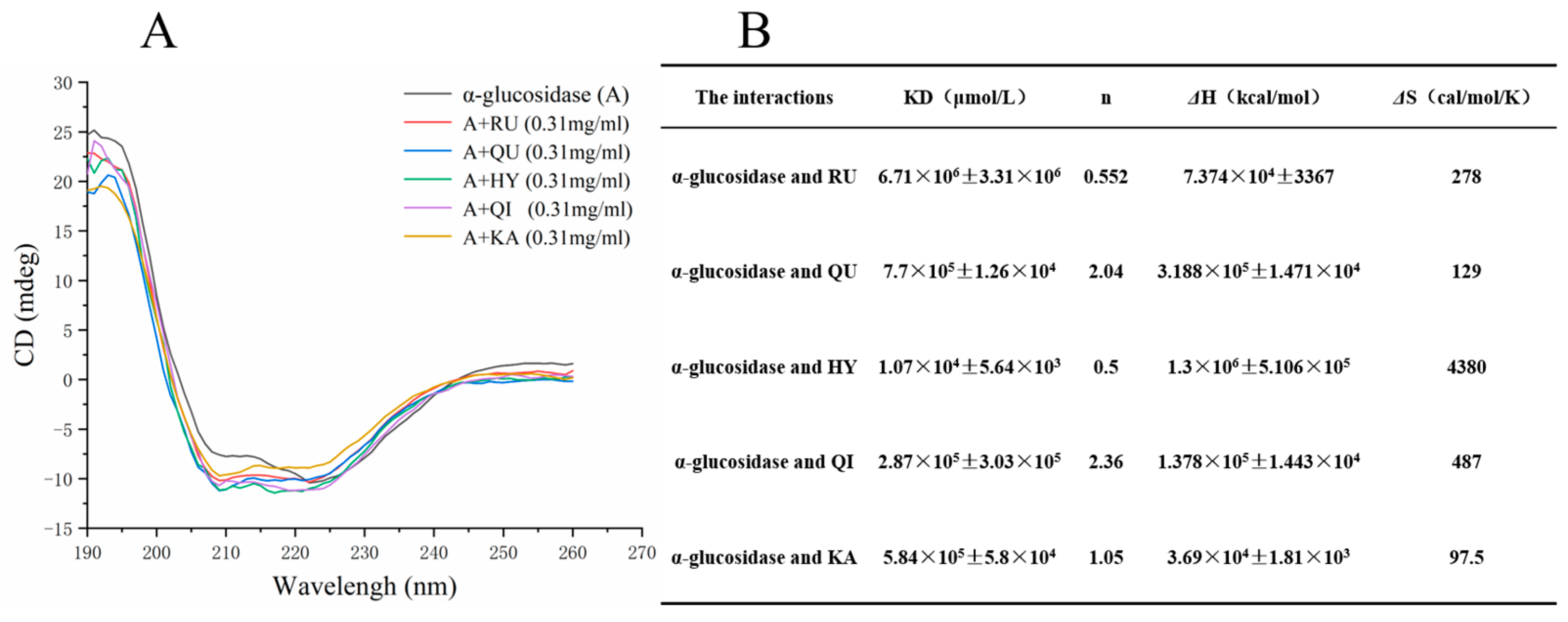
2.9. Circular Dichroism (CD) Analysis
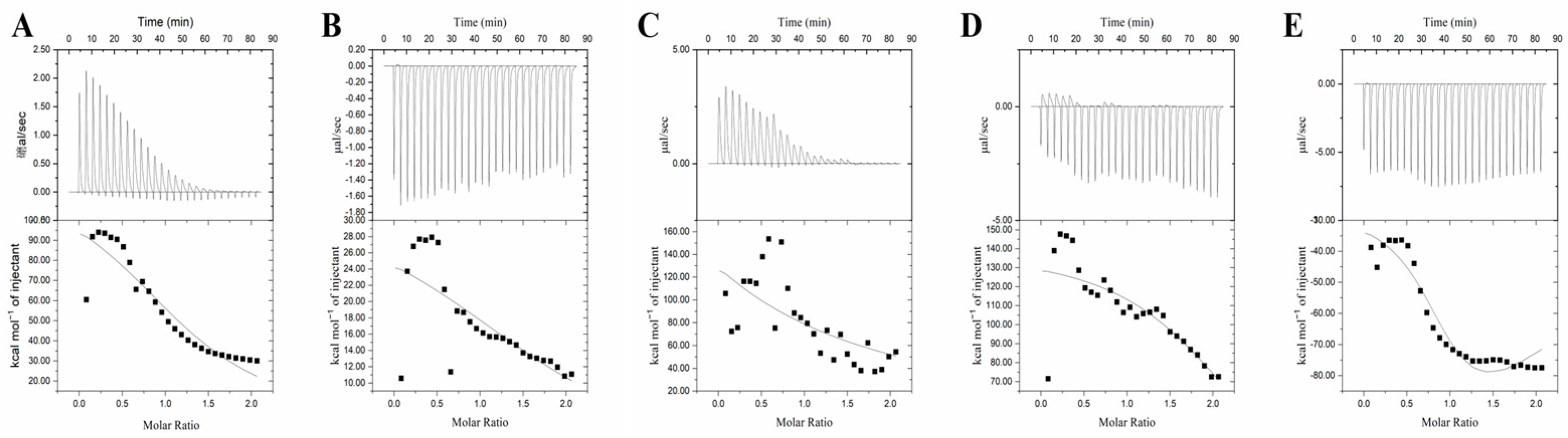
2.10. Isothermal Titration Calorimetry (ITC) Analysis
2.11. Molecular Docking
2.12. Data Analysis
3. Results and Discussion
3.1. Key α-Glucosidase Inhibitors Were Identified by IC50 Value and Omission Test of Polyphenolic Compounds in FSI
3.2. Interactions Analysis
3.3. Inhibition Types and Constants
3.4. Fluorescence Quenching
3.5. Thermodynamic Parameters
3.6. Circular Dichroism (CD) Analysis
3.7. Isothermal Titration Calorimetry (ITC) Analysis
3.8. Molecular Docking Analysis
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gong, Y.; Fan, L.; Wang, L.; Li, J. Flos sophorae immaturus: Phytochemistry, bioactivities, and its potential applications. Food Rev. Int. 2021, 1–19. [Google Scholar] [CrossRef]
- He, X.; Bai, Y.; Zhao, Z.; Wang, X.; Fang, J.; Huang, L.; Zeng, M.; Zhang, Q.; Zhang, Y.; Zheng, X. Local and traditional uses, phytochemistry, and pharmacology of Sophora japonica L.: A review. J. Ethnopharmacol. 2016, 187, 160–182. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Li, J.; Li, J.; Fan, L.; Wang, L. Effect of ultrasound-assisted freeze-dried on microstructure, bioactive substances, and antioxidant activity of Flos sophorae immaturus. Food Biosci. 2022, 49, 101913. [Google Scholar] [CrossRef]
- Miao, M.S.; Li, Y.; Wang, T.; Shao, S.; Feng, S.X.; Miao, Y.Y. Effect of Total Flavonoids of Flos Sophorae on Glucose Levels, Serum Lipid and Antioxidation Ability in Diabetic Rats Model. Adv. J. Food Sci. Technol. 2016, 12, 663–666. [Google Scholar] [CrossRef]
- Ji, Y.; Liu, D.; Jin, Y.; Zhao, J.; Zhao, J.; Li, H.; Li, L.; Zhang, H.; Wang, H. In vitro and in vivo inhibitory effect of anthocyanin-rich bilberry extract on α-glucosidase and α-amylase. LWT-Food Sci. Technol. 2021, 145, 111484. [Google Scholar] [CrossRef]
- Cao, H.; Ou, J.; Chen, L.; Zhang, Y.; Szkudelski, T.; Delmas, D.; Daglia, M.; Xiao, J. Dietary polyphenols and type 2 diabetes: Human study and clinical trial. Crit. Rev. Food Sci. Nutr. 2019, 59, 3371–3379. [Google Scholar] [CrossRef]
- Chen, G.L.; Xu, Y.B.; Wu, J.L.; Li, N.; Guo, M.Q. Hypoglycemic and hypolipidemic effects of Moringa oleifera leaves and their functional chemical constituents. Food Chem. 2020, 333, 127478. [Google Scholar] [CrossRef]
- Bao, Y.; Xiao, J.; Weng, Z.; Lu, X.; Shen, X.; Wang, F. A phenolic glycoside from Moringa oleifera Lam. improves the carbohydrate and lipid metabolisms through AMPK in db/db mice. Food Chem. 2020, 311, 125948. [Google Scholar] [CrossRef]
- Cardullo, N.; Muccilli, V.; Saletti, R.; Giovando, S.; Tringali, C. A mass spectrometry and (1)H NMR study of hypoglycemic and antioxidant principles from a Castanea sativa tannin employed in oenology. Food Chem. 2018, 268, 585–593. [Google Scholar] [CrossRef]
- Fang, H.L.; Liu, M.L.; Li, S.Y.; Song, W.Q.; Ouyang, H.; Xiao, Z.P.; Zhu, H.L. Identification, potency evaluation, and mechanism clarification of alpha-glucosidase inhibitors from tender leaves of Lithocarpus polystachyus Rehd. Food Chem. 2022, 371, 131128. [Google Scholar] [CrossRef]
- Wang, L.; Liu, Y.; Luo, Y.; Huang, K.; Wu, Z. Quickly screening for potential alpha-glucosidase inhibitors from Guava Leaves Tea by bioaffinity ultrafiltration coupled with HPLC-ESI-TOF/MS method. J. Agric. Food Chem. 2018, 66, 1576–1582. [Google Scholar] [CrossRef]
- Li, J.; Gong, Y.; Li, J.; Fan, L. In vitro xanthine oxidase inhibitory properties of Flos sophorae immaturus and potential mechanisms. Food Biosci. 2022, 47, 101711. [Google Scholar] [CrossRef]
- Xiao, Y.; Yang, C.; Xu, H.; Zhang, J.; Zhang, L. Study on the change of flavonoid glycosides to aglycones during the process of steamed bread containing tartary buckwheat flour and antioxidant, α-glucosidase inhibitory activities evaluation in vitro. LWT-Food Sci. Technol. 2021, 145, 111527. [Google Scholar] [CrossRef]
- Li, J.; Gong, Y.; Li, J.; Fan, L. In vitro inhibitory effects of polyphenols from Tartary buckwheat on xanthine oxidase: Identification, inhibitory activity, and action mechanism. Food Chem. 2022, 379, 132100. [Google Scholar] [CrossRef]
- Yu, Q.; Fan, L.; Duan, Z. Five individual polyphenols as tyrosinase inhibitors: Inhibitory activity, synergistic effect, action mechanism, and molecular docking. Food Chem. 2019, 297, 124910. [Google Scholar] [CrossRef]
- Cao, J.; Yan, S.; Xiao, Y.; Han, L.; Sun, L.; Wang, M. Number of galloyl moiety and intramolecular bonds in galloyl-based polyphenols affect their interaction with alpha-glucosidase. Food Chem. 2022, 367, 129846. [Google Scholar] [CrossRef]
- Li, X.; Bai, Y.; Ji, H.; Jin, Z. The binding mechanism between cyclodextrins and pullulanase: A molecular docking, isothermal titration calorimetry, circular dichroism and fluorescence study. Food Chem. 2020, 321, 126750. [Google Scholar] [CrossRef]
- Wang, Q.; Li, R.; Li, N.; Jia, Y.; Wang, Y.; Chen, Y.; Panichayupakaranant, P.; Chen, H. The antioxidant activities, inhibitory effects, kinetics, and mechanisms of artocarpin and alpha-mangostin on alpha-glucosidase and alpha-amylase. Int. J. Biol. Macromol. 2022, 213, 880–891. [Google Scholar] [CrossRef]
- Sun, L.; Warren, F.J.; Gidley, M.J.; Guo, Y.; Miao, M. Mechanism of binding interactions between young apple polyphenols and porcine pancreatic alpha-amylase. Food Chem. 2019, 283, 468–474. [Google Scholar] [CrossRef]
- Oboh, G.; Ademosun, A.O.; Ayeni, P.O.; Omojokun, O.S.; Bello, F. Comparative effect of quercetin and rutin on α-amylase, α-glucosidase, and some pro-oxidant-induced lipid peroxidation in rat pancreas. Comp. Clin. Pathol. 2014, 24, 1103–1110. [Google Scholar] [CrossRef]
- Ulusoy, H.G.; Sanlier, N. A minireview of quercetin: From its metabolism to possible mechanisms of its biological activities. Crit. Rev. Food Sci. Nutr. 2019, 60, 3290–3303. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Ferruzzi, M.G.; Hamaker, B.R. Structural requirements of flavonoids for the selective inhibition of alpha-amylase versus alpha-glucosidase. Food Chem. 2022, 370, 130981. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Liu, S.; Yao, L.; Wang, L.; Li, C. Free and bound phenolics of buckwheat varieties: HPLC characterization, antioxidant activity, and inhibitory potency towards alpha-glucosidase with molecular docking analysis. Antioxidants 2019, 8, 606. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Huang, L.; Sun, C.; Zhao, D.; Tang, H. Studies on the structure-activity relationship and interaction mechanism of flavonoids and xanthine oxidase through enzyme kinetics, spectroscopy methods and molecular simulations. Food Chem. 2020, 323, 126807. [Google Scholar] [CrossRef]
- Huang, Y.; Wu, P.; Ying, J.; Dong, Z.; Chen, X.D. Mechanistic study on inhibition of porcine pancreatic alpha-amylase using the flavonoids from dandelion. Food Chem. 2021, 344, 128610. [Google Scholar] [CrossRef]
- Avwioroko, O.J.; Anigboro, A.A.; Otuechere, C.A.; Atanu, F.O.; Dairo, O.F.; Oyetunde, T.T.; Ilesanmi, O.B.; Apiamu, A.; Ejoh, A.S.; Olorunnisola, D.; et al. α-Amylase inhibition, anti-glycation property and characterization of the binding interaction of citric acid with α-amylase using multiple spectroscopic, kinetics and molecular docking approaches. J. Mol. Liq. 2022, 360, 119454. [Google Scholar] [CrossRef]
- Skrt, M.; Benedik, E.; Podlipnik, C.; Ulrih, N.P. Interactions of different polyphenols with bovine serum albumin using fluorescence quenching and molecular docking. Food Chem. 2012, 135, 2418–2424. [Google Scholar] [CrossRef]
- Brandao, E.; Santos Silva, M.; Garcia-Estevez, I.; Mateus, N.; de Freitas, V.; Soares, S. Molecular study of mucin-procyanidin interaction by fluorescence quenching and Saturation Transfer Difference (STD)-NMR. Food Chem. 2017, 228, 427–434. [Google Scholar] [CrossRef]
- Joye, I.J.; Davidov-Pardo, G.; Ludescher, R.D.; McClements, D.J. Fluorescence quenching study of resveratrol binding to zein and gliadin: Towards a more rational approach to resveratrol encapsulation using water-insoluble proteins. Food Chem. 2015, 185, 261–267. [Google Scholar] [CrossRef]
- Zheng, Y.; Tian, J.; Yang, W.; Chen, S.; Liu, D.; Fang, H.; Zhang, H.; Ye, X. Inhibition mechanism of ferulic acid against alpha-amylase and alpha-glucosidase. Food Chem. 2020, 317, 126346. [Google Scholar] [CrossRef]
- Kayukawa, C.T.M.; de Oliveira, M.A.S.; Kaspchak, E.; Sanchuki, H.B.S.; Igarashi-Mafra, L.; Mafra, M.R. Effect of tannic acid on the structure and activity of Kluyveromyces lactis beta-galactosidase. Food Chem. 2019, 275, 346–353. [Google Scholar] [CrossRef]
- Yuksel, Z.; Avci, E.; Erdem, Y.K. Characterization of binding interactions between green tea flavanoids and milk proteins. Food Chem. 2010, 121, 450–456. [Google Scholar] [CrossRef]
- Zhang, J.; Ding, W.; Tang, Z.; Kong, Y.; Liu, J.; Cao, X. Identification of the effective alpha-amylase inhibitors from Dalbergia odorifera: Virtual screening, spectroscopy, molecular docking, and molecular dynamic simulation. Spectrochim Acta A Mol Biomol Spectrosc. 2022, 280, 121448. [Google Scholar] [CrossRef]
- Kayukawa, C.T.M.; Oliveira, M.A.S.; Kaspchak, E.; Sanchuki, H.B.S.; Igarashi-Mafra, L.; Mafra, M.R. Quillaja bark saponin effects on Kluyveromyces lactis beta-galactosidase activity and structure. Food Chem. 2020, 303, 125388. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, J.L.; Shen, L.H.; Feng, L.J.; Zhou, Q. Inhibition mechanism of diacylated anthocyanins from purple sweet potato (Ipomoea batatas L.) against alpha-amylase and alpha-glucosidase. Food Chem. 2021, 359, 129934. [Google Scholar] [CrossRef]
- Cardullo, N.; Muccilli, V.; Pulvirenti, L.; Cornu, A.; Pouysegu, L.; Deffieux, D.; Quideau, S.; Tringali, C. C-glucosidic ellagitannins and galloylated glucoses as potential functional food ingredients with anti-diabetic properties: A study of alpha-glucosidase and alpha-amylase inhibition. Food Chem. 2020, 313, 126099. [Google Scholar] [CrossRef]
- Arroyo-Maya, I.J.; McClements, D.J. Application of ITC in foods: A powerful tool for understanding the gastrointestinal fate of lipophilic compounds. Biochim Biophys Acta 2016, 1860, 1026–1035. [Google Scholar] [CrossRef]
- He, J.; Li, S.; Xu, K.; Tang, B.; Yang, H.; Wang, Q.; Li, H. Binding properties of the natural red dye carthamin with human serum albumin: Surface plasmon resonance, isothermal titration microcalorimetry, and molecular docking analysis. Food Chem. 2017, 221, 650–656. [Google Scholar] [CrossRef]
- Fan, Y.; He, Q.; Gan, C.; Wen, Z.; Yi, J. Investigation of binding interaction between bovine alpha-lactalbumin and procyanidin B2 by spectroscopic methods and molecular docking. Food Chem. 2022, 384, 132509. [Google Scholar] [CrossRef]
- Li, M.; Bao, X.; Zhang, X.; Ren, H.; Cai, S.; Hu, X.; Yi, J. Exploring the phytochemicals and inhibitory effects against α-glucosidase and dipeptidyl peptidase-IV in Chinese pickled chili pepper: Insights into mechanisms by molecular docking analysis. LWT-Food Sci. Technol. 2022, 162, 113467. [Google Scholar] [CrossRef]
- Tian, J.L.; Si, X.; Wang, Y.H.; Gong, E.S.; Xie, X.; Zhang, Y.; Li, B.; Shu, C. Bioactive flavonoids from Rubus Corchorifolius inhibit alpha-glucosidase and alpha-amylase to improve postprandial hyperglycemia. Food Chem. 2021, 341, 128149. [Google Scholar] [CrossRef] [PubMed]
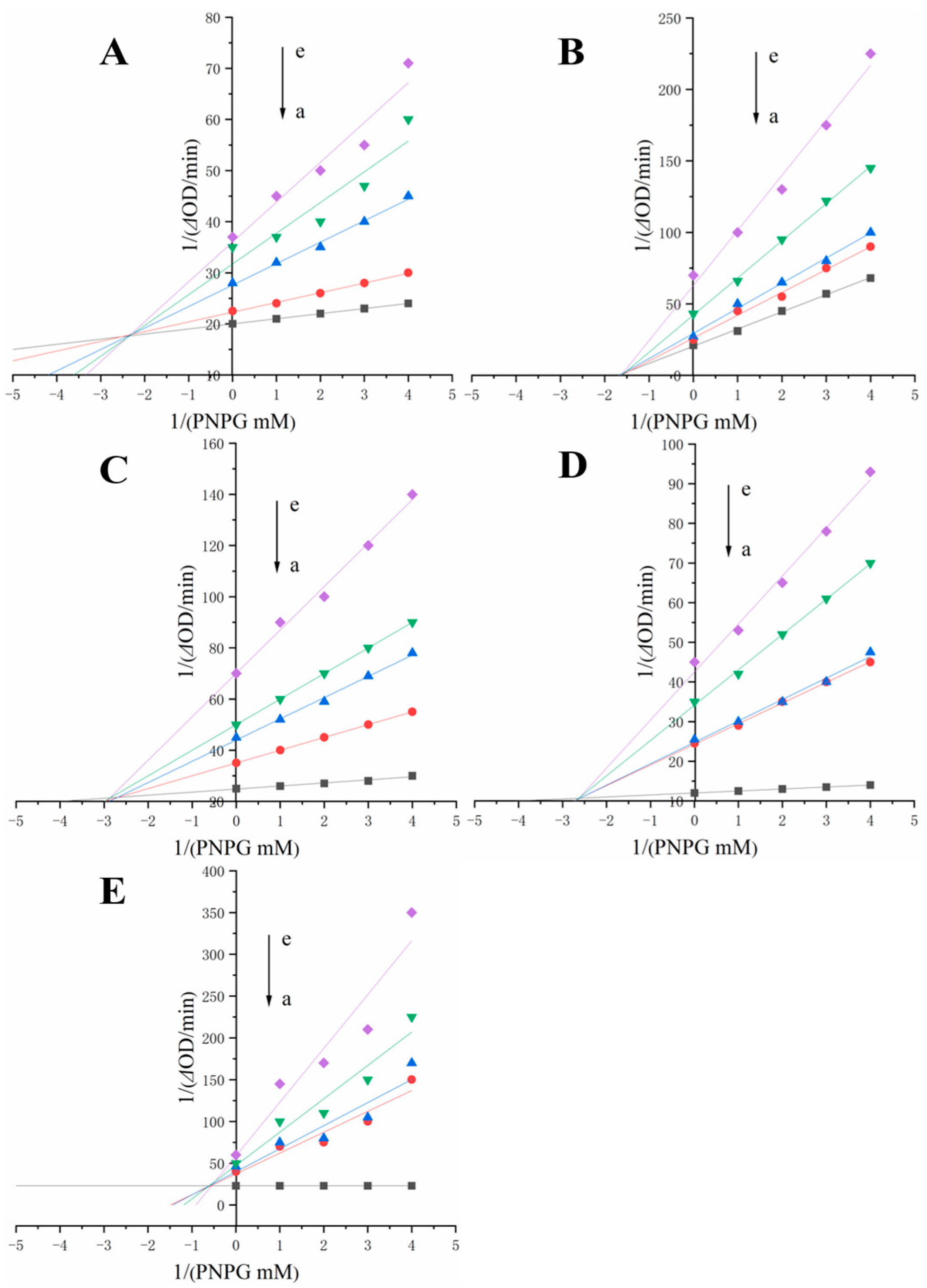


| Compounds | IC50 (mg/mL) | Ki (mM) | Kis (mM) |
|---|---|---|---|
| Protocatechuic acid | 369.00 ± 0.11 h | 17.71 | - |
| Chlorogenic acid | - | - | - |
| Rutin | 57.00 ± 0.32 f | 8.91 | - |
| Hyperoside | 12.77 ± 0.07 d | 3.79 | - |
| Kaempferol-3-O-rutinoside | - | - | - |
| Narcissoside | - | - | - |
| Quercitrin | 25.37 ± 0.13 e | 5.31 | 6.72 |
| Quercetin | 0.21 ± 0.01 a | 0.22 | 2.51 |
| Kaempferol | 0.55 ± 0.01 c | 0.67 | 5.56 |
| Isorhamnetin | 255.00 ± 0.11 g | 15.12 | - |
| Acarbose (positive control) | 0.31 ± 0.01 b | 0.57 | - |
| Omitted Compounds | α-Glucosidase Inhibition Rate of Omitted Compounds (%) | α-Glucosidase Inhibition Rate of Recombinant Solutions Omitting Different Compounds (%) | Contribution Rates of Omitted Compounds to the α-Glucosidase Inhibition Rate of Model Solution (%) |
|---|---|---|---|
| RU | 69.00 ± 0.11 | 85.81 ± 0.15 | 4.70 ± 0.13 |
| QU | 89.00 ± 0.25 | 69.92 ± 2.90 | 20.59 ± 0.26 |
| HY | 75.00 ± 0.32 | 81.90 ± 0.59 | 8.61 ± 0.16 |
| QI | 77.00 ± 0.20 | 83.79 ± 0.25 | 6.72 ± 0.08 |
| KA | 81.00 ± 0.12 | 75.89 ± 0.18 | 14.62 ± 0.17 |
| None (Model solution) | 90.55 ± 0.11 | ||
| None (Sample solution) | 90.31 ± 0.21 |
| Group | Va + Vb | Value | Interaction | |
|---|---|---|---|---|
| V*2 | Vab | V*2-Vab | ||
| 1 | RU + QU | 0.6556 | 0.3567 | 0.2989 IN |
| 2 | RU + HY | 0.6321 | 0.3356 | 0.2965 IN |
| 3 | RU + QI | 0.5527 | 0.2986 | 0.2541 IN |
| 4 | RU + KA | 0.5789 | 0.3025 | 0.2764 IN |
| 5 | QU + HY | 0.6851 | 0.3799 | 0.3052 IN |
| 6 | QU + QI | 0.6677 | 0.3657 | 0.3020 IN |
| 7 | QU + KA | 0.6956 | 0.3966 | 0.2990 SU |
| 8 | HY + QI | 0.6425 | 0.3485 | 0.2940 IN |
| 9 | HY + KA | 0.6521 | 0.3599 | 0.2922 IN |
| 10 | QI + KA | 0.6399 | 0.3567 | 0.2832 IN |
| Group | Va + Vb + Vc | V*3 | Vabc | V*3-Vabc |
| 11 | RU + QU + HY | 0.7225 | 0.4263 | 0.2962 AN |
| 12 | RU + QU + QI | 0.7100 | 0.4130 | 0.2970 AN |
| 13 | RU + QU + KA | 0.7525 | 0.4536 | 0.2989 AN |
| 14 | RU + HY + QI | 0.6977 | 0.3935 | 0.3042 IN |
| 15 | RU + HY + KA | 0.7122 | 0.4001 | 0.3121 IN |
| 16 | RU + QI + KA | 0.7055 | 0.3966 | 0.3089 IN |
| 17 | QU + HY + QI | 0.7369 | 0.4358 | 0.3011 IN |
| 18 | QU + HY + KA | 0.7768 | 0.4963 | 0.2805 IN |
| 19 | QU + QI + KA | 0.7588 | 0.4832 | 0.2756 IN |
| 20 | HY + QI + KA | 0.7322 | 0.4255 | 0.3067 IN |
| Group | Va + Vb + Vc + Vd | V*4 | Vabcd | V*4-Vabcd |
| 21 | RU + QU + HY + QI | 0.8898 | 0.5362 | 0.3536 AN |
| 22 | RU + QU + HY + KA | 0.9012 | 0.5203 | 0.3809 AN |
| 23 | RU + QU + QI + KA | 0.8922 | 0.5136 | 0.3786 IN |
| 24 | RU + HY + QI + KA | 0.8725 | 0.5001 | 0.3724 IN |
| 25 | QU + HY + QI + KA | 0.9556 | 0.5656 | 0.3900 IN |
| Compound | T (K) | KSV (×104 L mol−1) | Kq (×1012 L mol−1 s−1) | Kb (×105 L mol−1) | n | R2 | ΔH (KJ mol−1) | ΔG (KJ mol−1) | ΔS (J mol−1 K−1) |
|---|---|---|---|---|---|---|---|---|---|
| RU | 298 | 4.50 | 4.50 | 0.52 | 1.72 | 0.99 | 28.48 | −2.39 | 31.72 |
| 303 | 9.37 | 9.37 | 0.26 | 1.71 | 0.99 | −1.32 | 27.70 | ||
| 310 | 9.34 | 9.34 | 0.25 | 1.71 | 0.99 | −1.33 | 27.08 | ||
| QU | 298 | 10.71 | 10.71 | 1.46 | 2.30 | 0.99 | −5.34 | −4.46 | 8.75 |
| 303 | 7.52 | 7.52 | 1.44 | 2.35 | 0.99 | −4.64 | 8.01 | ||
| 310 | 5.55 | 5.55 | 1.42 | 2.34 | 0.99 | −4.94 | 6.87 | ||
| HY | 298 | 12.43 | 12.43 | 0.77 | 1.91 | 0.99 | 8.06 | −7.49 | 1.41 |
| 303 | 8.96 | 8.96 | 0.79 | 1.57 | 0.99 | −5.41 | 5.48 | ||
| 310 | 11.17 | 11.17 | 0.71 | 1.80 | 0.99 | −7.04 | 0.09 | ||
| QI | 298 | 7.45 | 7.45 | 0.62 | 1.72 | 0.99 | −40.30 | −61.22 | 181.73 |
| 303 | 7.87 | 7.87 | 0.68 | 1.71 | 0.99 | −59.72 | 176.69 | ||
| 310 | 5.45 | 5.45 | 0.69 | 1.71 | 0.99 | −59.69 | 176.58 | ||
| KA | 298 | 22.23 | 22.23 | 1.22 | 1.67 | 0.99 | −18.49 | −69.32 | 208.92 |
| 303 | 49.33 | 49.33 | 1.26 | 1.83 | 0.99 | −68.62 | 203.15 | ||
| 310 | 42.46 | 42.46 | 1.25 | 1.86 | 0.99 | −69.23 | 200.53 |
| The Interactions | KD (μmol/L) | n | ΔH (kcal/mol) | ΔS (cal/mol/K) |
|---|---|---|---|---|
| α-glucosidase and RU | 1.24 × 105 ± 6.5 × 104 | 1.34 | 1.212 × 105 ± 2.145 × 104 | 430 |
| α-glucosidase and QU | 7.7 × 105 ± 1.26 × 104 | 2.04 | −3.188 × 105 ± 1.471 × 104 | 129 |
| α-glucosidase and HY | 1.07 × 104 ± 5.64 × 103 | 0.5 | 1.3 × 106 ± 5.106 × 105 | 4380 |
| α-glucosidase and QI | 2.87 × 105 ± 3.03 × 104 | 2.36 | −1.378 × 105 ± 1.443 × 104 | 487 |
| α-glucosidase and KA | 5.84 × 105 ± 5.8 × 104 | 1.05 | −3.69 × 104 ± 1.81 × 103 | 97.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gong, Y.; Li, J.; Li, J.; Wang, L.; Fan, L. In Vitro Inhibitory Effects of Polyphenols from Flos sophorae immaturus on α-Glucosidase: Action Mechanism, Isothermal Titration Calorimetry and Molecular Docking Analysis. Foods 2023, 12, 715. https://doi.org/10.3390/foods12040715
Gong Y, Li J, Li J, Wang L, Fan L. In Vitro Inhibitory Effects of Polyphenols from Flos sophorae immaturus on α-Glucosidase: Action Mechanism, Isothermal Titration Calorimetry and Molecular Docking Analysis. Foods. 2023; 12(4):715. https://doi.org/10.3390/foods12040715
Chicago/Turabian StyleGong, Yuhong, Jun Li, Jinwei Li, Li Wang, and Liuping Fan. 2023. "In Vitro Inhibitory Effects of Polyphenols from Flos sophorae immaturus on α-Glucosidase: Action Mechanism, Isothermal Titration Calorimetry and Molecular Docking Analysis" Foods 12, no. 4: 715. https://doi.org/10.3390/foods12040715
APA StyleGong, Y., Li, J., Li, J., Wang, L., & Fan, L. (2023). In Vitro Inhibitory Effects of Polyphenols from Flos sophorae immaturus on α-Glucosidase: Action Mechanism, Isothermal Titration Calorimetry and Molecular Docking Analysis. Foods, 12(4), 715. https://doi.org/10.3390/foods12040715






