Abstract
Shiga-toxin-producing Escherichia coli (STEC) is typically detected on food products mainly due to cross-contamination with faecal matter. The serotype O157:H7 has been of major public health concern due to the severity of illness caused, prevalence, and management. In the food chain, the main methods of controlling contamination by foodborne pathogens often involve the application of antimicrobial agents, which are now becoming less efficient. There is a growing need for the development of new approaches to combat these pathogens, especially those that harbour antimicrobial resistant and virulent determinants. Strategies to also limit their presence on food contact surfaces and food matrices are needed to prevent their transmission. Recent studies have revealed that bacteriophages are useful non-antibiotic options for biocontrol of E. coli O157:H7 in both animals and humans. Phage biocontrol can significantly reduce E. coli O157:H7, thereby improving food safety. However, before being certified as potential biocontrol agents, the safety of the phage candidates must be resolved to satisfy regulatory standards, particularly regarding phage resistance, antigenic properties, and toxigenic properties. In this review, we provide a general description of the main virulence elements of E. coli O157:H7 and present detailed reports that support the proposals that phages infecting E. coli O157:H7 are potential biocontrol agents. This paper also outlines the mechanism of E. coli O157:H7 resistance to phages and the safety concerns associated with the use of phages as a biocontrol.
1. Introduction
Escherichia coli (E. coli) is one of the most thoroughly investigated and characterized living organisms and is widely employed as a research model [1]. Despite the fact that these organisms are known to exist as normal flora in the gastrointestinal tract of humans and other warm-blooded animals, some strains are considered to be pathogenic [2]. In general, E. coli strains linked to human illnesses can be divided into two categories, namely intestinal E. coli, also known as diarrheagenic E. coli, and extra-intestinal E. coli [3]. Based on their distinctive virulence traits, pathogenic mechanisms, and clinical symptoms, intestinal E. coli strains are further separated into six pathotypes [4] comprising enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC), enteroaggregative E. coli (EAEC), enteroinvasive E. coli (EIEC), diffusely adherence E. coli (DAEC), and enterohaemorrhagic E. coli (EHEC) [3,4]. According to Kaper et al. [4], uropathogenic E. coli, meningitis-associated E. coli, and necrotoxigenic E. coli are the three types of strains associated with extra-intestinal infections.
There are other significant EHEC serotypes, such as O104:H21 and O111:H8, that cause diseases in humans; however, strains belonging to the serotype O157:H7 are major foodborne pathogens that pose serious threats to public health globally [5,6]. These pathogens cause a number of complications ranging from mild diarrhoea to more potentially fatal haemolytic uraemic syndrome (HUS), haemorrhagic colitis (HC), and thrombotic thrombocytopenic purpura (TTP), accounting for a large proportion of kidney failure [7]. To date, South Africa has not experienced any documented food-borne outbreaks linked with E. coli O157. However, previous surveillance studies have established a significant genetic correlation between E. coli O157 strains obtained from various sources such as water, cattle, their associated meat products, and individuals suffering from diarrheal illnesses [8,9].
Furthermore, recurring and long-lasting outbreaks of foodborne illnesses which are caused by alterations in the pattern of pathogen population have a significant impact on human health and safety [10,11]. Reports from countries, such as the USA, that have more advanced public health policies reveal that Listeria monocytogenes (28%) and E. coli O157:H7 (3%) are responsible for a significant proportion (30%) of food-related deaths [12]. The transmission of E. coli O157:H7 is associated with its potential to survive in a wide range of environmental conditions and contaminate the food chain along the preharvest and postharvest production pipelines through multiple routes [13,14]. Also, it is not surprising that, on the WHO global priority pathogens (GPP) list, antimicrobial resistant E. coli O157:H7 is categorized as a critical pathogen requiring constant monitoring and surveillance [15]. Although other non-O157 serogroups, such as O26, O111, O103, O121 O45, and O145, have also been increasingly associated with foodborne illness in humans, serotype O157:H7 is predominantly known to be the causative agent of STEC infections worldwide [16]. In addition, foodborne outbreaks are increasingly associated with fresh produce, like organic fruits and vegetables [17], which are partially cooked or mainly consumed raw with no inactivation step like heating. The implementation of comprehensive strategies to prevent the transmission of E. coli O157:H7 throughout food production systems, without altering the colour, taste, texture, or nutritional characteristics of food products, at both pre and post-harvest facilities, is imperative.
Treatment of E. coli O157:H7 infections in humans and animals with antibiotics is a popular and major option [18]. However, the use of antimicrobial agents in the treatment of E. coli O157:H7 infections is controversial [19]. Certain antibiotics can stimulate E. coli O157:H7 to release toxins that can potentially worsen the clinical outcome of infected patients, e.g., through an elevated risk of HUS, and also the use of antimicrobials may contribute to the development of antibiotic-resistant strains, thereby broadening public health implications. The growing prevalence of antimicrobial resistance (AMR) contributes to the severity of E. coli O157:H7 infections.
Furthermore, E. coli O157:H7 populations with multiple drug resistance (MDR), extensive drug resistance (XDR), and pan drug resistance (PDR) are rapidly emerging [20,21]. The primary cause of this resistance is the improper and extensive use of antimicrobial drugs in both humans and animals [22]. The rise in antibiotic resistance in E. coli O157:H7 is a global health concern, resulting in high mortality and morbidity, prolonged infection durations in vulnerable persons and animals, and significant economic losses [23].
In addition, live animals, particularly cattle, harbour foodborne pathogens, such as E. coli strains that can be transmitted to other animals, that can enter the food chain and thus be transmitted to humans. There is a need to develop alternative but effective methods for reducing the quantity of pathogens in live animals [24]. Moreover, strategies to reduce faecal shedding of these organisms by animals is highly recommended [25,26]. To date, various decontamination strategies, such as the use of chemical sanitizers (sodium dichloroisocyanurate, quaternary ammonium compounds, chlorine, peracetic acid, and lactic acid), heat treatment (pasteurization), washing (water), and chilling, are commonly used during food processing to reduce the risk of pathogens entering the food chain [27,28]. These strategies have limited, but sometimes direct, microbial benefits on the food product [29].
The rise in antimicrobial resistance, as well as the difficulties associated with antibiotic use for E. coli O157:H7 infections, highlights the critical need for alternative novel approaches. Such approach should not only target E. coli O157:H7 but also the control the development and spread of antibiotic resistance. Bacteriophage-based interventions are being regarded as promising biocontrol agents in the food production systems despite the perceived shortcomings associated with phage-based control measures.
Bacteriophages (phages) are bacteria viruses that infect and kill their host (bacteria). They are the most abundant entities in the biosphere, with approximately 1031 phage particles, which is ten times more than the number of bacteria species on earth [30]. Based on their life cycle, phages are classified into two major groups, namely, lysogenic and lytic phages. The current review is focused on lytic phages due to their antibacterial properties. The process of phage infection involves phage attachment to the bacteria cell. Upon entry into the host cell (bacteria), lytic phages hijack the genetic material of the host bacteria in order to generate new virions by taking over the bacterial cell replication mechanism [31]. Subsequently, phages employ late proteins, such as lysozyme (lysin and/or endolysin), to lyse (kill) the host (bacteria) cell and thus release the mature virions. Owing to their ability to lyse the bacteria, lytic phages has attracted interest in the pharmaceutical industry. The use of lytic bacteriophages is one innovative strategy that is gradually being embraced as a green technology to combat antimicrobial resistance and the risk of illness in humans [32,33]. More importantly, the proliferation of bacteriophage-insensitive mutants (BIMs) and save our soul (S.O.S) in response to Shiga toxin production are some of the safety concerns associated with using phages as bio-control agents [34,35]. Hence, it is crucial to establish the efficacy and biosafety of these promising biocontrol and therapeutic agents. In this review, we describe the advantages of phages as an alternative antibacterial agent and how genomics has enhanced investigations aimed at assessing the safety of phage therapy, but with a direct focus on E. coli O157:H7-specific bacteriophages.
2. Evolution, Virulence and Pathogenicity of E. coli O157:H7
Non-toxigenic and less virulent E. coli O55:H7 strains are known to be ancestral cells of E. coli O157:H7 [36,37] that originated through a series of transitional phases [38]. According to the concept model, the locus of enterocyte effacement (LEE) was present in both the O55:H7 and O157:H7 strains, and it is capable of inducing diarrhoea through an attachment–effacement process [39,40,41]. Four sequential events that led to the emergence of E. coli O157:H7 include (i) acquisition of an stx operon, which is encoded in the genome of lambdoid prophages, (ii) acquisition of the rfb region encoding the O157 antigen, and (iii) loss of β-glucuronidase (GUD) activity [42,43]. Notably, the three main virulence factors of E. coli O157:H7 include (i) Shiga toxin operons, (ii) locus of enterocyte effacement, and (iii) byproducts of the F-like plasmid pO157 [44,45].
Based on the degree of its virulence, the Shiga toxin operon is divided into two major groups, namely, stx1 and stx2 genes [46]. Interesting, the Stx1 has three subtypes (Stx1a, Stx1c, and Stx1d), while Stx2 has seven subtypes (Stx2a, Stx2b, Stx2c, Stx2d, Stx2e, Stx2f, and Stx2g). It is widely acknowledged that diverse stx subtypes possess the ability to bind to multiple receptors, exhibiting varying affinities for each receptor. The primary target for Stx2a is the Gb3 receptor, whereas Stx1a can bind to both Gb3 and globotetraosylceramide (Gb4). The Stx2e subtype exhibits a broader spectrum of receptor binding capabilities, including interactions with globopentaosylceramide, pentahexosylceramides featuring Gb4-elongated core structures, and Gb4 with a preference for Gb5 [47]. Several other genes, apart from stx, have been associated with the pathogenicity and virulence of O157:H7 [48]. These genes differentiate E. coli O157:H7 from non-pathogenic strains of E. coli and are presented in Table 1.
2.1. Transmission of E. coli O157:H7 to Humans and Animals
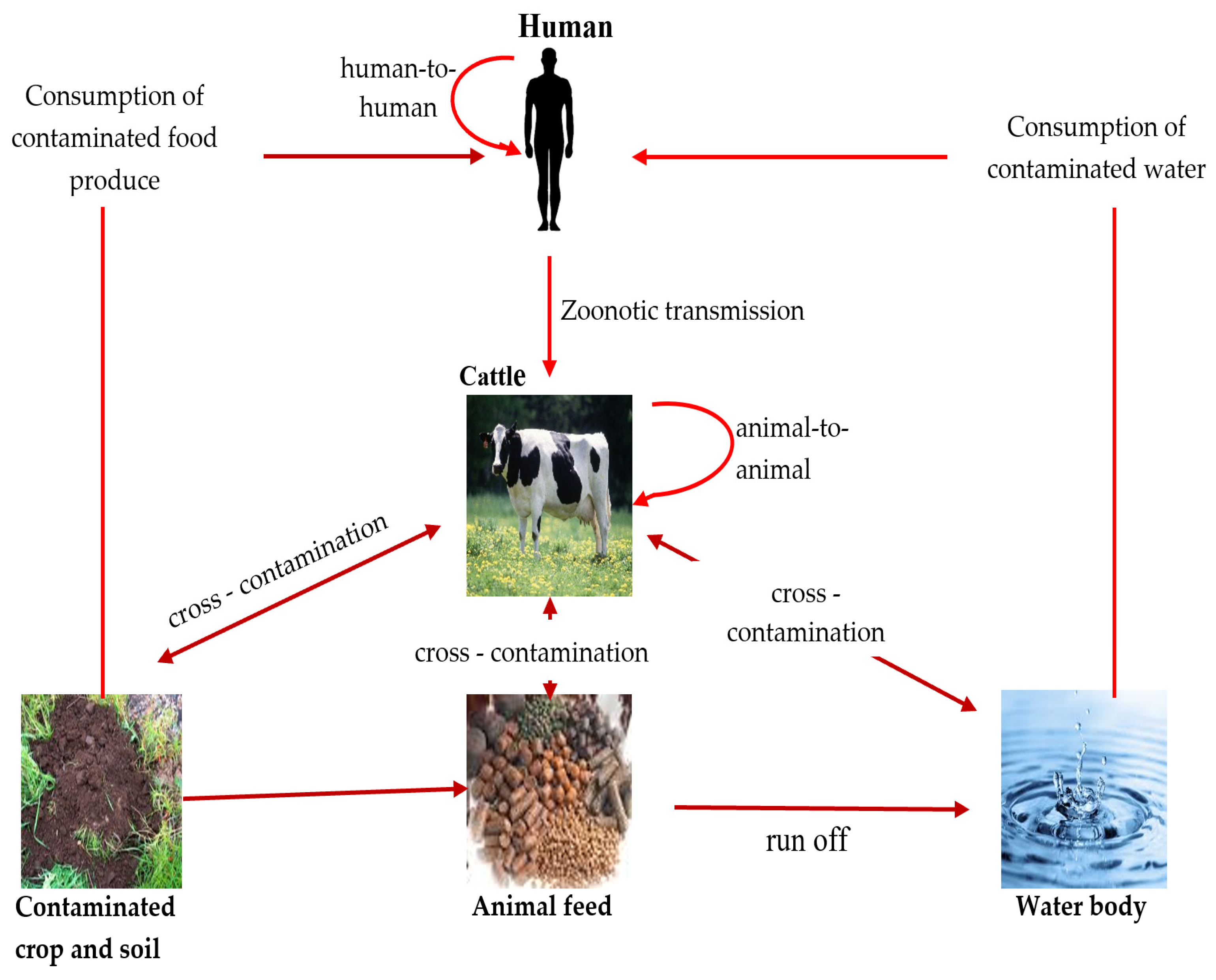
E. coli serotype O157 can be transmitted through different routes, including direct contact with animal droppings, ingestion of contaminated food and water, and transmission from one person/animal to another [5,49] (Figure 1).
Figure 1.
Transmission route of pathogenic E. coli among animal, human, and environment (source: this study—diagram prepared by authors).
2.1.1. Contaminated Food as a Transmission Vector
Contaminated food plays a crucial role in the transmission of E. coli O157:H7 to humans [50]. Food products derived from animals (beef, lamb, chicken, pork, and ground beef) are considered to be the most common source of the pathogen and the transmission of the diseases amongst humans worldwide [51]. Studies have reported that E. coli O157:H7 illnesses have also been linked to the ingestion of other food products originating from cattle, such as unpasteurized milk and other dairy products [9,52]. Furthermore, the transmission of this pathogen from the skin, intestines, and excrement of diseased animals frequently leads to the contamination of the environment, including water sources [5]. As a result, consumption of water contaminated by E. coli may lead to illness in humans.
Fresh vegetables such as alfalfa, radish sprouts, lettuce, and spinach, in addition to unpasteurized fruit juices and apple cider [53,54,55], have been connected to outbreaks of E. coli O157-related disease in humans. It is thought that these fresh vegetables become contaminated when grown in soil that was exposed to infected animal dung or polluted water [9], while fruit juices and apple ciders become contaminated due to improper manufacturing processes [55]. In general, animal reservoirs are responsible for the greatest amount of E. coli O157:H7 transmission to humans, and food-related transmission is common, making it necessary to manage the pathogens in animal-derived foods, especially beef. As a result, E. coli O157:H7 poses a major risk to public health by endangering both human health and food safety.
2.1.2. Contaminated Water as a Transmission Vector
The transmission of E. coli O157:H7 through water has been documented in various settings, including both drinking water that has been contaminated [56] and recreational water bodies [57,58]. Furthermore, studies conducted in Nigeria and South Africa demonstrated that water used for irrigation had notable influence on the dissemination of E. coli O157:H7 through the contamination of food products [59,60].
Untreated sewage released from hospitals, farms, and residential areas containing E. coli O157:H7 into nearby water bodies also increases the risk of human infections significantly [61]. Based on the evidence, E. coli O157:H7 easily survives in water and may persist for several weeks and even longer [58]. Strong rainstorms may cause sediments to be re-suspended, which could lead to an abrupt rise in E. coli O157:H7 concentrations in the water [62]. Xie et al. [58] reported the detection of E. coli O157 in urban recreational water and provided evidence of the correlation between contact with these water bodies through activities such as swimming, boating, bathing, and sailing and the development of infections. In addition, an outbreak of E. coli O157 infections occurred among a group of seven children in the United Kingdom who came in contact with recreational water at a coastal beach [63]. The pathogens originated from water in a creek that was contaminated with faeces from cows grazing upstream, and the transmission was facilitated by rainfall runoff [63]. This finding highlights the potential vulnerability of humans when in contact with recreational beach environments.
2.1.3. Person-To-Person Transmission
The transmission of E. coli O157:H7 infection most often occurs through the faecal–oral route, primarily in settings where infected individuals come in close contact with others, such as in households, daycare centres, and healthcare facilities [64,65]. Children are more vulnerable than adults due to their immunological immaturity and limited understanding of appropriate hygiene practices [66]. In numerous instances, there has been a correlation between the consumption of shared contaminated food or water and the occurrence of an outbreak, thereby emphasizing the importance of implementing strict control measures in healthcare facilities and hygiene practice amongst people living in shared apartments.
2.2. Epidemiology of E. coli O157:H7
According to Mesele and Abunna [67], E. coli O157:H7 is responsible for approximately 73,000 illnesses, 2000 hospitalizations, and 50–60 deaths annually in the United States. Reports that evaluated E. coli O157 outbreaks that occurred in the USA between 1982 and 2002 and were reported to the Centers for Disease Control and Prevention (CDC) showed 350 outbreaks, totalling 8598 cases and 1493 (17%) hospitalizations. A total of 354 (4%) presented with HUS and 40 (0.5%) died [68]. This highlights the significant public health potential of E. coli and also provides an overview of its epidemiology in the region.
The varying aetiology of E. coli O157:H7 infections explains why the pathogen has also been isolated in humans, animals, food products, and the environment, especially in the African region [69], where public health policies are not adhered to strictly. Examples of documented cases in Africa are as follows: human infection that was reported in Johannesburg, South Africa in 1990 [70]; the detection of the pathogen in individuals with haemorrhagic colitis that resulted to fatalities in Bangui, Central African Republic [71]; and the isolation of the pathogen in East African countries (Tanzania, Kenya, and Ethiopia) [72]. The prevalence rates of STEC O157:H7 reported in Africa were 7% in Morogoro, Tanzania, amongst patients suffering from diarrhoea [72], 2.3% isolated from raw cattle milk in Kwara, Nigeria [73], 1.9% amongst children with diarrhoea in Mozambique [74], and 8% in HIV infected individuals suffering from dysentery in Zimbabwe [75].

Table 1.
Key virulence factors associated with E. coli O157:H7 and their function in pathogenesis.
Table 1.
Key virulence factors associated with E. coli O157:H7 and their function in pathogenesis.
| Virulence Factor | Gene | Role in Pathogenesis | References |
|---|---|---|---|
| Shiga toxin | stx1 (a, c, and d) and stx2 (a to g) | Stx toxin is composed of 2 subunits, A and B. The A subunit is an RNA-glycosidase that interacts with the 60S rRNA, inhibiting protein synthesis, causing cell death. | [76] |
| Locus of Enterocyte Effacement (LEE) pathogenicity island (PAI) | lee | LEE-PAI encodes several genes like ehaA, ehaB, and ehaJ, which are involved in the adhesion mechanism called the attaching-and-effacing (A/E) lesion, such as type III secretion system (TTSS), an outer membrane adhesion (intimin), its translocated intimin receptor (TIR), and secreted proteins with signal transduction. | [77] |
| Intimin | eae | The eae gene is carried by the LEE-PAI and encodes an outer membrane adhesion protein called Intimin. This protein interaction with the bacterial translocated intimin receptor (Tir) leads to the attachment of the STEC cell to the host intestinal mucosa, resulting in the A/E lesions | [76,78] |
| EHEC hemolysin (EHEC-hly) | ehxA or EHEC-hlyA | EHEC-hlyA is part of an operon composed of four genes (EHEC-hlyC, EHEC-hlyA, EHEC-hlyB, and EHEC-hlyD) encoded on the pO157 plasmid. EHEC-hlyA codes the structural protein of EHEC-Hly. The proteins coded by EHEC-hlyB and EHEC-hlyD are responsible for the transport of EHEC-Hly out of the bacterial cell, and, in its turn, EHEC-hlyC product is responsible for EHEC-Hly post-translational activation. EHEC-Hly is responsible for forming pores into the cell membrane and is found as free or in association with outer membrane vesicles (OMV). Both forms target the human intestinal epithelial and microvascular endothelial cells, but in different manners. When in the free form, EHEC-Hly is responsible for lysing the cells, but when in association with OMV, induces cell apoptosis. | [79] |
| Adhesins | eibG, efa-1/lifA/toxB | Immunoglobulin-binding (Eib) G is encoded by the gene eibG, and it can bind to human IgD and IgA. It can also participate in bacterial adhesion to host epithelial cells. Toxin Efa-1 is like LifA and ToxB, and they are thought to be associated with cell adherence, lymphostatin activity, or induction of secretion of type III effectors in the STEC strain. | [78] |
| Serine protease autotransporters | espP | Encoded on the pO157 plasmid, it is important for biofilm formation and for adherence to T84 intestinal epithelial cells. | [78] |
| Long Polar Fimbriae and E. coli YcbQ laminin-binding fimbriae (ELF) | lfp and elf | Able to attach the extracellular matrix protein laminin, which contributes to colonization of the GI tract | [76] |
Adapted with permission from: Pinto et al. [80], 2023, Taylor & Francis Ltd., http://www.tandfonline.com, 1 September 2023.
2.3. Treatment for E. coli O157:H7 Infections and Antimicrobial Resistance
Antibiotics are frequently used in animal production to prevent infection and as a growth promoter. This practice has made it difficult to treat infections caused by E. coli O157:H7, as the pathogens have become resistant to antibiotics. Given the virulence potential and pathogenicity of E. coli O157:H7, it is unfortunate that there is no specific treatment for E. coli O157:H7 infection [67]. In humans, antibiotics are typically not recommended because they can increase the risk of complications, such as HUS, a serious kidney disease. Although the spread of antimicrobial resistance in E. coli O157:H7 and other pathogens is being monitored and mitigated by surveillance [81], an alternative treatment method is necessary as a strategy to combat E. coli O157:H7 and infections caused by it, and also to serve as an additive to feed stock for the prevention of sicknesses. Bacteriophage therapy may be an effective alternative to antibiotic therapy for antibiotic-resistant E. coli O157:H7, both in humans and in animal production.
3. Bacteriophages
Bacteriophages are viruses that exclusively infect bacteria. Although there were arguments regarding the discovery of phages, it has been acknowledged that the first scientists to discover the phages were William Twort and Felix d’Herelle [82,83]. Twort, an English medical bacteriologist, postulated that viruses were the causative agents of this phenomenon after observing a similar process in Micrococcus [84,85,86]. According to d’Herelle’s findings, the presence of an imperceptible microorganism was detected in stool samples obtained from dysentery patients, which were devoid of bacteria due to filtration [87]. The occurrence of clear zones observed in bacterial lawns can be attributed to the presence of “invisible germs”. D’Herelle provided the initial elucidation regarding the mechanism by which these viruses proliferate by exploiting bacteria, resulting in the death and subsequent disintegration of the bacteria. After their discovery, d’Herelle used phage preparations to treat bacterial dysentery [88]. Owing to its success in treating bacterial infection, the use phage therapy escalated in countries such as Tbilisi, Georgia, and Poland to treat conditions such as typhoid, fever, dysentery, surgical wound infections, peritonitis urinary tract infection, and septicaemia [89]. However, the use of phage therapy was halted by the discovery of the first antibiotic, penicillin, in 1940. In addition, lack of knowledge of phage therapy decreased the use of phage therapy. Despite this, Soviet countries continued to use phages to treat bacterial infections in humans.
Bacteriophages are ubiquitously found in the vicinity of their host bacteria. The phage replication cycle encompasses the processes of adsorption, penetration, nucleic acid replication, and virion assembly. Consequently, the cellular membrane undergoes lysis, leading to the release of mature virions. While it is true that certain phages can infect multiple bacterial species or strains, most phages exhibit specificity towards a particular bacterial species or strain. The determination of specificity is regulated by the existence of receptors located on the outer surface of bacterial cells, such as lipopolysaccharide (LPS), flagella, and/or other surface proteins [90]. Bacteriophages can exhibit either a lytic or lysogenic life cycle, depending on the specific life cycle they adopt after infecting a host cell. Lytic phages undergo a lytic cycle, which starts when the bacteriophage binds to specific receptors on the surface of the bacterial cell. The phage then injects its DNA into the host cell and assumes control of the cellular machinery. While using the host cell’s transcription and translation mechanisms, phage-specific proteins are synthesized, and the phage’s DNA is replicated. The components then assemble into new phage particles, and they accumulate and exert pressure on the cell’s internal environment, causing the bacterial cell to lyse or rupture. In the lytic lifecycle of a bacteriophage, the lysis process is a crucial phase. It enables the newly produced phage particles to disseminate and infect other susceptible bacterial cells, ultimately resulting in the phage population increasing. Temperate phages, on the other hand, undergo the lysogenic cycle, wherein their genetic material integrates and coexists with the host genome in a stable manner, forming a prophage. This prophage replicates along with the host cell during replication. According to Pinto et al. [80], temperate phages have the ability to transition into the lytic cycle when they encounter cellular stress.
3.1. General Properties of Bacteriophages
The attributes of interest possessed by phages as an antibacterial agent have generated considerable interest in using them as a biocontrol agent [91]. The aforementioned attributes encompass the following: firstly, phages demonstrate selective toxicity by exclusively targeting their host, such as the food-borne pathogen E. coli O157:H7, while leaving the indigenous microflora of the food substance unharmed; secondly, their low toxicity to humans stems from their predominant composition of nucleic acids and proteins; and thirdly, phages naturally exist in the environment, rendering them relatively straightforward and cost-effective to manufacture. Phages have been identified in chicken meat [92], seafood [93], fermented products [94], and different human body parts, which provides further evidence supporting the safety of phages in human applications [95]. Despite the advantageous characteristics exhibited by phages, they also possess unfavourable attributes that present obstacles in their application for biocontrol purposes [96]. Brovko et al. [96] and Bandara et al. [97] have identified several key traits associated with these phages. These include a restricted host range, the ability for virulence components to be transferred among bacterial strains, and the emergence of phage-resistant bacterial species.
Furthermore, bacteriophages have been used as a means to eliminate foodborne illnesses for several years [98]. According to O’Sullivan et al. [99], lytic phages have practical applications in ensuring food safety. Phages are commonly regarded as natural entities [100,101], in contrast to alternative food preservatives that have been associated with significant health issues such as cancer, asthma, and other ailments [102]. Jones et al. [103] posed that phages administered to food products for the purpose of eradicating a particular infection does not exhibit significant impact on non-target bacteria, like the commensal microorganisms present in the human body. There is currently no evidence to suggest that they pose any harm to mammalian cells, as they exhibit a high degree of specificity towards a particular species and so they are extensively utilized, even in food products. According to Ramos-Vivas et al. [104], they have been recognized for their efficacy in combating biofilm structures while also maintaining the sensory properties of food.
3.2. Characteristics of E. coli O157:H7 Phages
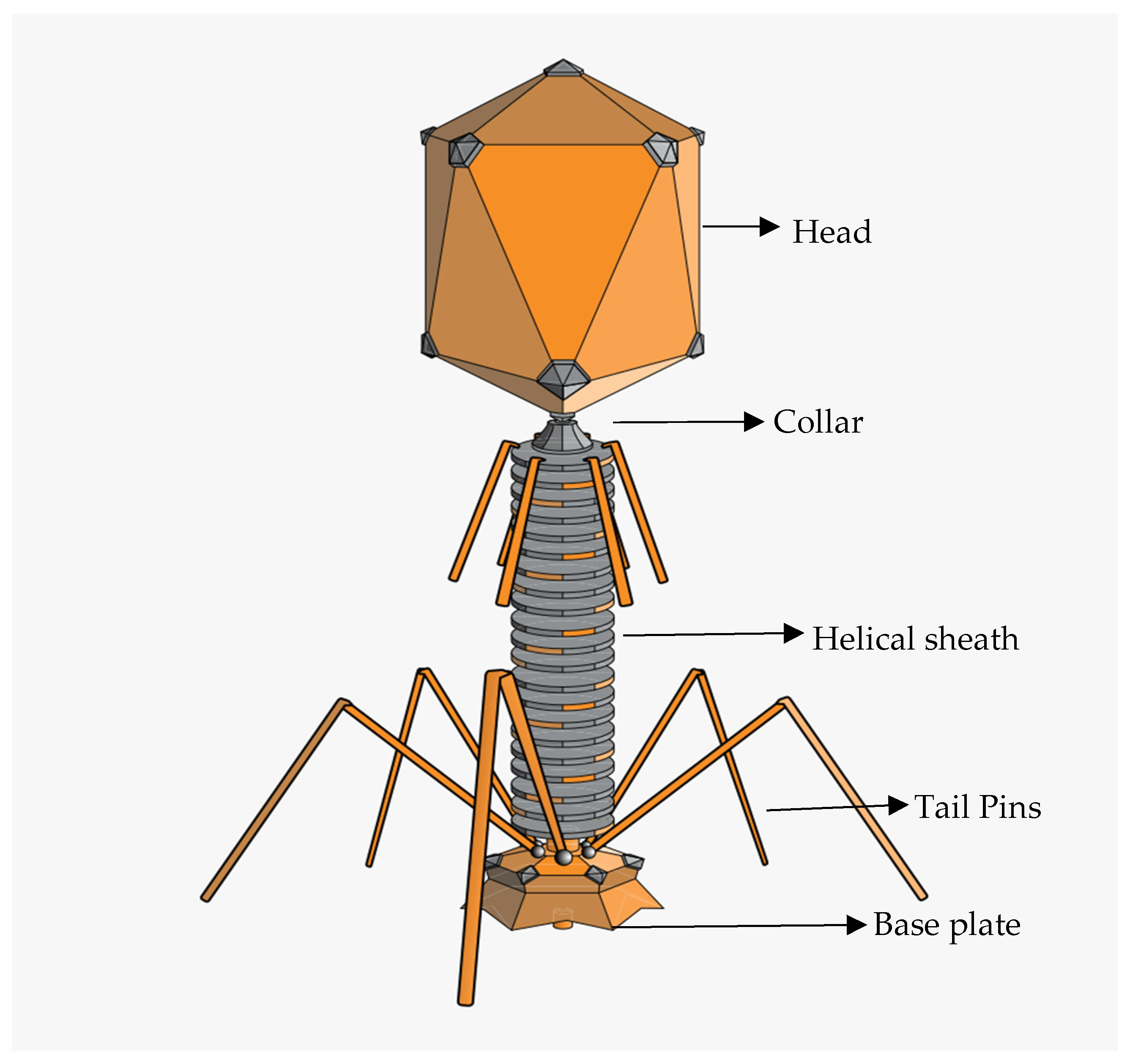
Being a pathogenic strain, E. coli phages can be targeted by various bacteriophages (phages) with different characteristics in terms of their biological, structural, and physicochemical characteristics. A typical phage particle structure consists of a three-dimensional shape made up of an icosahedral protein capsid with or without a tail and a filament [105]. Phage particles can range in size from 24 to 200 nm [106,107], with the largest being the T4 phage, which infects E. coli and is 200 nm long and 80–100 nm wide [108]. The physical properties of phages specific to E. coli O157 can vary depending on their family. They do, however, often exhibit the conventional phage structure (Figure 2), with an icosahedral head (capsid) carrying the phage’s genetic material, either DNA or RNA. The tail, which aids phage adherence to the bacterial surface and the transfer of genetic material into the host cell, varies throughout phage families and can be lengthy, flexible, or short. Certain E. coli O157 bacteriophages have unique tail fibres or tailspikes that aid in targeted binding to receptors on the surface of E. coli O157 cells.
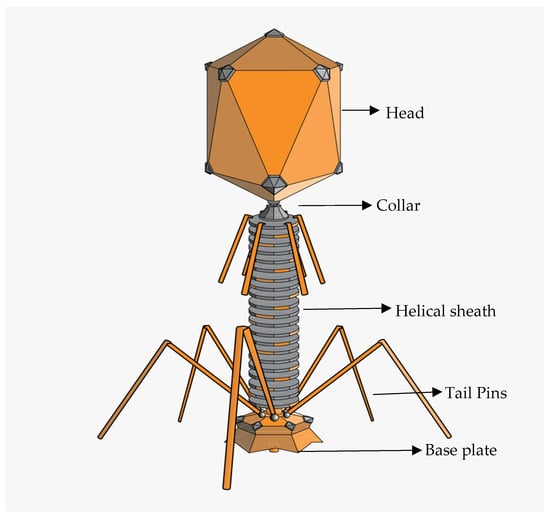
Figure 2.
Diagrammatic representation of tailed phage (https://en.m.wikipedia.org/wiki/Bacteriophage, accessed on 1 September 2023).
The International Committee on Taxonomy of Viruses (ICTV) and the Bacterial and Archaeal Subcommittee (BAVS), primarily dedicated to the study of phages, play pivotal roles in classifying viruses and assigning taxonomic names to virus groups [109]. This classification process relies on evaluating several key characteristics of viruses, including the nature of the viral genome (single-stranded or double-stranded DNA or RNA), the composition of the capsid (the protein shell) and the presence or absence of an envelope, the range of hosts a virus can infect, its pathogenicity, and its genetic sequence similarities. However, in 2022, changes were made to the taxonomy of bacterial viruses, which involved the abolishment of the morphology-based families Podoviridae, Siphoviridae, and Myoviridae, as well as the order Caudovirales, therefore establishing a binomial system of nomenclature for species [110]. All tailed bacterial and archaeal viruses with icosahedral capsids and a double-stranded DNA genome are now grouped under the class “Caudoviricetes”. With 14 new families assigned to 4 orders, the process of grouping is still ongoing; hence, most taxa have remained “unclassified” at this level.
The most common lytic phages, which includes the E. coli O157 phage, associated with human pathogens and the gut microbiota belongs to the class Caudoviricetes [111], commonly known as “tailed phages”, which contain double-stranded DNA genomes. They can belong to several families and orders, which can be confirmed via genetic similarities using online tools for phylogenetic similarities [112].
3.3. The Application of Bacteriophages in Food Production as Biocontrol Agents
In recent times, bacteriophages have demonstrated a diverse array of applications in enhancing the safety of various food products (Table 2). In the early stages of phage discovery, a pair of researchers affiliated with Michigan Agricultural College identified a substance with inhibitory properties in the liquid derived from decomposing cabbage. This substance effectively impeded the spoilage of vegetables caused by the organism Xanthomonas campestris pv. Campestris [113]. From then on, bacteriophages have been systematically evaluated for their efficacy in combating various bacterial species responsible for plant spoilage, with experiments conducted in controlled laboratory conditions as well as real-world environments [114]. In a study conducted by Das et al. [115], it was demonstrated that grapevines artificially infected with Xylella fastidiosa, the causal agent of Pierce’s disease, were treated with high-concentration combination of four lytic phages. This treatment was administered three weeks after the initial inoculation of the pathogen, it was observed that the phage cocktail was able to significantly lower the level of Xylella fastidiosa. Bacterial pathogens that pose a potential threat to agricultural animals can lead to adverse alterations in the quality of food products like meat and unpasteurized milk, thereby resulting in production losses. Therefore, the investigation into the efficacy of phages in the treatment of animal infections and in the enhancement of animal health and growth has become popular.
In addition to the analysis of food samples, various bacteriophages have also been evaluated in environmental samples associated with food processing facilities. Biofilms play a crucial role in the context of the food industry. In their study, Dalmasso et al. [116] conducted an assessment of the potential anti-biofilm properties exhibited by three recently discovered bacteriophages. The efficacy of the three bacteriophages in combating biofilms were assessed individually as well as in combination. The use of a three-phage cocktail has been identified as a highly effective strategy for the management of biofilms.

Table 2.
An overview of E. coli O157:H7-specific phages, their application on food products and food contact surfaces, their mode of administration, and the obtained results.
Table 2.
An overview of E. coli O157:H7-specific phages, their application on food products and food contact surfaces, their mode of administration, and the obtained results.
| Phages | Product Tested | Methodology | Results | References |
|---|---|---|---|---|
| BEC8 cocktail (38, 39, 41, CEV2, AR1, 42, ECA1, and ECB7) | Spinach leaves and romaine lettuce. | Bacteria were spot inoculated onto the leaves and allowed to dry for 1 h in a biosafety cabinet. BEC8, Transcinnamaldehyde, or TSB were applied on the top of the leaf inoculated previously. Positive controls were prepared by mixing the bacterial inoculum with BEC8 or TC without drying. | Both BEC8 and Transcinnamaldehyde were able to reduce the cell counts at different MOIs and temperatures. The loss of viability of high inoculum levels of E. coli O157:H7 was less than 1 log CFU at all conditions with the exception of 3 log CFU after 24 h at room temperature and 37 °C for both leafy greens. The combination of both agents resulted in an increase in the antimicrobial effect. | [117] |
| Phage OSY- SP | Green bell pepper and spinach | Both matrices were spot inoculated with E. coli. The phage was applied by rinsing the food products with phage lysate (for optimization) and phages suspended in PBS before storing. A rinse of 2 min was used for spinach leaves; however, due to the migration of cells in the pepper, a 5 min rinse was then selected for this kind of matrix. | Cell reduction was observed during refrigerated storage conditions. E. coli O157:H7 was reduced by 2.4–3.0 log CFU/g on cut green pepper (5 min rinse) and 3.4–3.5 log CFU/g on spinach leaves (2 min rinse) during 72 h storage. The rinse treatment with phages was successful in both fresh produces tested. | [118] |
| A cocktail composed by the phages e11/2 and e4/1c | Cattle hide | The phage cocktail was administered using a hand-held spray bottle. As negative control, no wash treatment was performed. | The phage cocktail was more efficient when applied to the cattle hide and left for 1 h. There was a 1.5 log10 CFU/cm2 reduction in E. coli O157:H7 colonies compared to the colonies recovered on samples treated with water only. | [119] |
| BEC8 cocktail | Sterilized hard surfaces (stainless steel chips, ceramic tile chips, high density polyethylene chips—HDPEC). | Bacteria were spotted on the chip and dried in a biosafety cabinet. BEC8 or TSB were applied on the chip surface previously inoculated. MOIs used were 1, 10, and 100. Positive controls were prepared by mixing the bacterial inoculum with BEC8 or TC without drying. | Phage cocktail could inactivate the bacterial mixture with higher performance rating from low to high MOIs, low to high temperatures, and shorter to longer periods of exposure. With a reduction of at least one log CFU in the number of the E. coli O157:H7 cells, and at approximately 104 CFU, no survival was detected. | [120] |
| Wild-type T4 phage | Raw beef | A total of 25 g of meat sample was inoculated with diluted culture bacteria overnight and allowed to attach for 10 min at room temperature. Phages immobilized on cellulose membranes were used to cover the contaminated surface of the meat. | The use of immobilized phages resulted in the reduction of 1 log unit after 6 and 9 days, and below detection limit on days 12 and 15 at 4 °C. | [121] |
| Cocktail composed of phages DT1 to DT6 | Milk and meat | Sterile, commercial, reconstituted milk supplemented with CaCl2 was inoculated with each bacterial strain (one per batch). Each batch was divided into 2: one treated with phage cocktail and other to be the control. Meat pieces (1 cm, 0.4 cm thick) were spotted with bacterial strains and allowed to attach for 10 min at room temperature. Then, phage cocktail was added to each meat piece. Controls were prepared by adding TMG buffer. | Phage cocktail could reduce the amount of E. coli strains tested in milk with reduction values reaching up to 3.4 log10 CFU/mL at 24 °C and 3.6 log10 CFU/mL at 37 °C after 24 h. In meat, cell inactivation was achieved at 24 °C (2.6–4.0 log10 CFU/mL) and 37 °C (3.0–3.8 log10 CFU/mL) as well. A higher inactivation value for O157:H7 STEC (1.55 ± 0.35 log10 CFU/mL) was observed at 4 °C after 6 days. | [122] |
| Ecoshield | Lettuce | Overnight cultures were applied to fresh cut lettuce. Phages were applied by immersion or spraying in combination with sodium hypochlorite solution. | Spraying of phages on lettuce surface reduced the E. coli O157:H7 populations (2.22 log CFU/cm2) compared with control treatments (4.10 log CFU/cm2), while immersion of lettuce in suspensions containing high concentrations of EcoShield™ (9.8 log PFU/mL) resulted in the deposition of high concentrations (7.8 log PFU/cm2) of bacteriophages on the surface of fresh cut lettuce, thereby contributing to the efficacy of the lytic phages on lettuce. | [123] |
| FAHEc1 | UHT milk; ready-to-eat meat; raw beef | Phage FAHEc1 was treated with UV light before being used on food products (even losing viability, phages are capable of lysing bacterial cells). UHT milk was inoculated with E. coli O157:H7 and phages. The raw beef inoculation was at 37 °C and was used to simulate the phage application right before slathering in carcasses. | In comparison to UHT milk and raw beef, there was a greater reduction produced by viable phages with a reduction value of 4.5 log10 CFU piece−1 after 24 h incubation. In milk, UVP produced a 2–2.5 log10 CFU reduction, while in raw beef, UVP produced a reduction of 1.75–2.5 log10 CFU. | [124] |
| Phages T5, T1, T4, and O1 | Beef | Beef portions were placed in a sterile petri plate and E. coli O157 was added. After 10 min (for samples to dry) phages (individually or in cocktail) were added and compared at different time and temperature. PBS was used as control. | Effect of temperature and time showed a reduction in E. coli O157 numbers by 3.2 log10 CFU/cm2 compared to phage-free controls at 4 °C after 144 h, the same reduction was observed at 22 °C and 37 °C but after 6 and 3 h, respectively. | [125] |
| phiEco1, phiEco2, phiEco3, phiEco5, phiEco6 and phiS1 | Oyster | Bacteria grown overnight were added to the oysters and allowed to attach for 1 h at 37 °C. Phage suspension was added, and the oyster meat was then incubated at 3 °C for 2 days followed by an incubation at 37 °C for 2 h. | High concentration of phages could reduce all bacteria strains. The reduction was observed when bacteria were present in single or combined manners. Phages were able to reduce E. coli (ATCC BAA-196) concentration from 8.5 ± 3.5 × 107 CFU/g oyster meat to 2.0 ± 1.5 × 106 CFU/g oyster meat even after 50 h of incubation. | [126] |
| E. coli O157:H7 phage isolated by Cui et al. (2018) | Lettuce Cucumber and carrot | The different vegetables were immersed in E. coli suspensions for 30 min and then placed at 37 °C for 24 h. The vegetables were then sequentially treated with Cold Nitrogen Plasma (CNP) and phages. | The sequential treatment led to a reduction in cell viability to 1.21 log10 CFU/g on the third day and no viable bacteria was detected on the ninth day. The results were independent of temperature (4, 12, or 25 °C). | [127] |
| E coli O157:H7 phage | Spinach; spinach harvester blade | Spinach extract was prepared using 25 g fresh spinach leaves. The Blades were inoculated with the extract or 10% TSB (control). Two Different conditions were used at a static temperature (22 °C) or dynamic temperature (30 °C 12 h, 20 °C 12 h), to stimulate day and night temperatures in California. Blades inoculated with E. coli were treated with phages. | phages could successfully eliminate E. coli O157:H7 population on the blades after 2 h exposure with a 4.5 log CFU reduction. After 24 and 48 h incubation at 30 °C, growth was significantly higher (6.09 and 6.37 log CFU/mL) than when incubated at 22 °C (4.84 and 5.68 log10 CFU/mL) respectively. | [128] |
| Phages phiJLA23, phiKP26, phiC119, and phiE142 | Tomatoes | A phage cocktail containing 109 PFU/mL of each phage. Microencapsulated phages were also prepared containing a polymer mixture consisting of 10% solids (modified starch and maltodextrin), 60% of SM buffer and 30% of phage cocktail. Tomatoes were divided into 3 groups: 1st group was inoculated with E. coli O157:H7; 2nd group was inoculated with microencapsulated cocktail phage and sprayed with the bacteria host, and the 3rd was not inoculated (control). | After 120 h at 4 °C, microencapsulated phages could significantly reduce E. coli O157:H7 concentrations from 5.2 Log10 CFU/tomatoes at 0 h to 2.3 Log10 CFU/tomatoes. Results obtained showed that microencapsulated phages are more stable under stress factors than the free phages. | [129] |
3.4. Safety Attributes of Bacteriophages for Biocontrol of E. coli O157:H7
Recent studies have revealed a global resurgence of interest in phages as potential therapeutic agents for clinically relevant bacterial infections, especially those caused by multi-drug-resistant strains [130,131]. This renewed interest in phages is attributed to some advantages they have over conventional antibiotics.
Phages have the ability to selectively target and kill specific bacterial groups while preserving commensal microbiomes, which results in a reduced number of bacteria cells being subjected to selection pressure [132]. This specificity reduces the risk of phage-based biocontrol causing damage to beneficial bacteria or non-target organisms in the environment, thereby enhancing its safety profile. Aside from the fact that phages can replicate at infection sites and penetrate biofilms [133], extensive research has demonstrated that phages are generally non-toxic and harmless for human consumption [134] unlike antibiotics, which can lead to the development of antibiotic-resistant bacteria if mis-used.
Furthermore, phages are self-limiting; their population tends to decrease as the amount of their host bacteria decreases [135]. They ultimately degrade in the environment, thereby reducing concerns regarding persistence and accumulation over time. This property helps prevent the overgrowth of phages when they are used as a biocontrol, thereby minimizing their effect on the bacterial community. Finally, bacteriophages can be found in a variety of environments, including water sources, sediments, and the digestive tracts of animals [136], hence isolating them is not a difficult task in terms of their availability.
3.5. Bacteriophage Therapy and Phage-Based Products for the Control of E. coli O157:H7
Phage therapy is a method that uses phages to effectively treat bacterial infections in medicine; this is made possible by the abundance of phages in nature, their simplicity in isolation, and the efficiency with which they kill bacteria, especially when used in controlled laboratory research. The emergence of AMR has made it possible to investigate phages as an antibiotic alternative. E. coli phages have been created for oral administration and utilised in randomised trials [137,138]. Currently, researchers are attempting to employ phages in a dried powder form that is considerably more stable and easier to store, transport, and administer. Despite the research on phage therapy, there are currently no phage therapy products licenced for human use in the EU or the US. To manage E. coli O157:H7 in the food sector, phage-based products have been developed to be used as biocontrol of bacterial diseases and have been authorised by the FDA as “generally considered safe.” Examples of phage-based anti-E. coli O157:H7 products include the following; EcoShieldTM, originally designed for application in “red meat parts” [139], and Secure Shield E1, a product manufactured by FINK TEC GmbH, a German company, that is specifically designed for application on the external areas of beef carcasses [140]. Every product has obtained approval from the Food and Drug Administration (FDA). Several food products have recently received approval for the utilization of phage mixes from OmniLytics (Sandy, UT), Micreos’ PhageGuard E., and Intralytix’s EcoShield PXTM to mitigate the risk of infection caused by E. coli O157:H7 and other pathogenic E. coli strains [141] (Table 3).

Table 3.
Approved phage-based products for E. coli O157:H7.
The FDA has granted approval for the phage products to be classified as generally recognized as safe (GRAS) food additives. The GRAS categorization mentioned above is derived from the regulatory framework established by the Federal Food, Drug, and Cosmetic Act (FD&C Act) and the regulations outlined in Title 21 of the Code of Federal Regulations (21 CFR). In order to introduce novel chemicals into the human food supply, particularly those intended for use as food additives, it is necessary to adhere to the regulatory guidelines outlined in 21 CFR part 170, subpart E [142]. This procedure has been used for the purpose of certifying bacteriophages, predominantly in the United States.
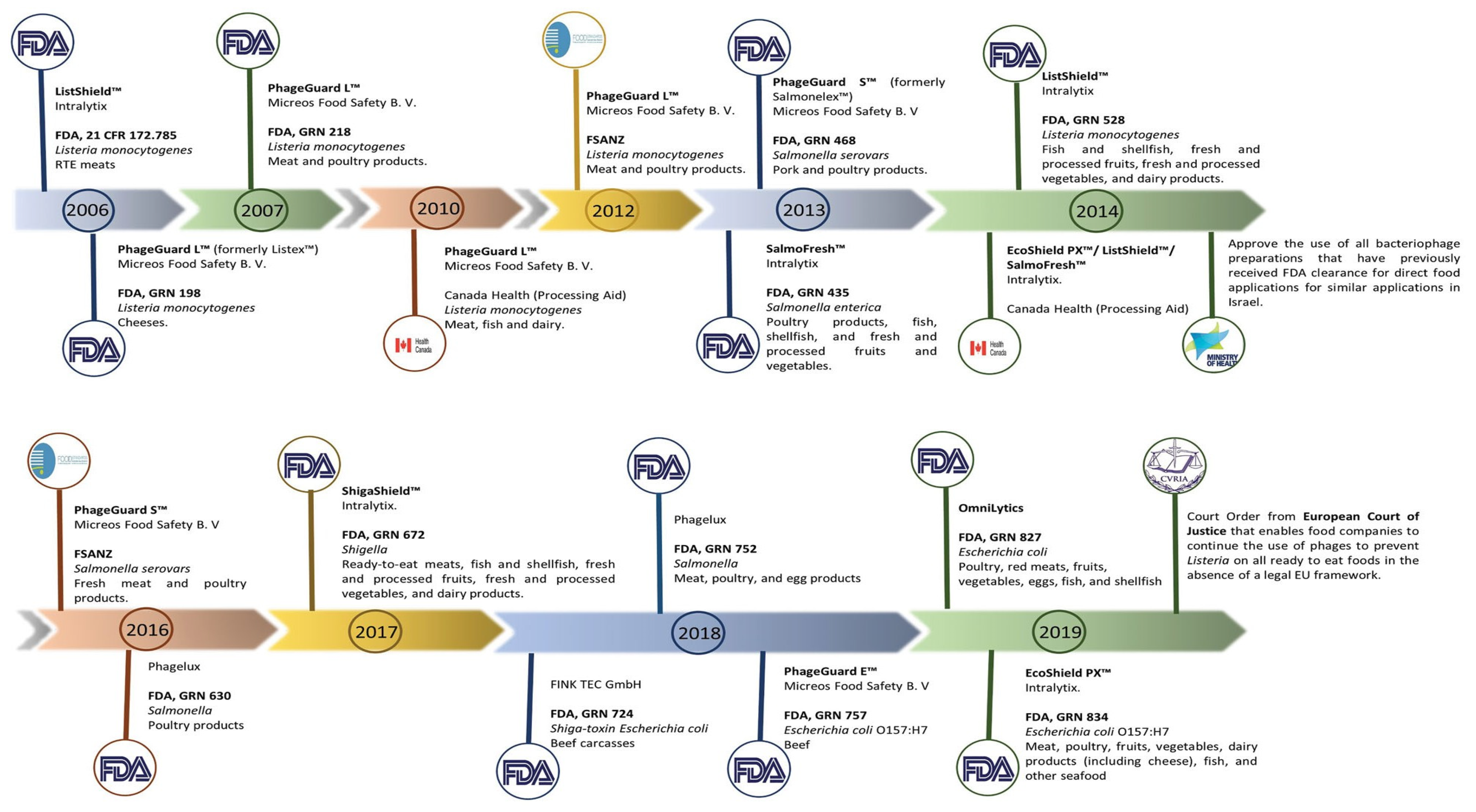
Other nations have also granted approval for the use of bacteriophages to ensure the safety of food products. In 2014, the Israeli Ministry of Health’s National Food Service authorized the utilization of bacteriophages, which had previously received approval from the FDA, for similar applications. This decision was made in accordance with the regulations outlined in the document titled “Guidelines: use of bacteriophages (bacteria-killing viruses) in food” [143]. In Canada, the authorization for the use of phage-based products as food processing aids has been granted. These products include PhageGuard LTM (previously known as ListexTM), ListShieldTM, SalmoFreshTM, and EcoShieldTM, among others [143] (Figure 3).
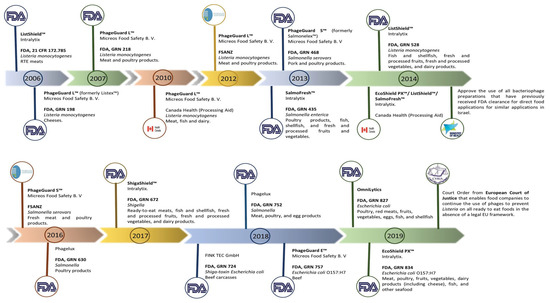
Figure 3.
Phage-based products that have already received approval around the world. The manufacturer, the target microorganisms, and the food matrix where the product is intended to be applied are all listed in the product description. The FDA, FSANZ, and the companies’ news websites were used to create the timeline. Adapted with permission from: Pinto et al. [80], 2023, Taylor & Francis Ltd., http://www.tandfonline.com, 1 September 2023.
4. Challenges Faced in the Application of E. coli O157:H7 Phages
There are a number of difficulties and factors to take into account when using phages as a biocontrol for E. coli O157:H7 either on food products or as a medication in humans and animals. Phages are very specific to individual bacterial strains [144]. Since E. coli O157:H7 strains can vary genetically and develop resistance to phages over time, it can be difficult to find phages that effectively target these strains. As a result, a combination of different phages in the form of a cocktail may be necessary for effective biocontrol of this pathogen. E. coli O157:H7 has a low infectious dosage (10 to 100 CFU/mL) and can survive in low-pH conditions, such as the acidic environment of the stomach and acidic food, which is one of its stress resistance mechanisms [145]. It can be therefore difficult to deliver phages to the site of infection because they may need to endure stomach acid and other digestive processes in order to reach the intestinal E. coli O157:H7 in the gut. For phages to be considered effective at inhibiting E. coli O157:H7, they must possess acid resistance in order to survive in the acidic environment of the stomach. Furthermore, phage products can be expensive to create, produce, and distribute due to their sensitivity to environmental factors like pH and temperature [146]; their quality and effectiveness can be compromised if they are transported and stored in unsuitable environmental conditions. The issue of storage and transportation has made it difficult for patients situated in low-resource environments to have access to them. Additionally, research has shown that phage therapy may be more effective when combined with other treatments, such as antibiotics [147,148]. Since its usage in humans could raise ethical concerns, it is crucial to carefully consider the best sequence and combination of therapies that will not cause further complications in infected patients. Inadequate clinical data also pose a challenge in the application of E. coli O157:H7 phages; while there have been some case reports and small clinical studies on phage therapy [149], more large-scale, randomised controlled trials are required to establish the safety and efficacy of phages as a biocontrol agent in treating E. coli O157:H7 infections.
Phage Resistance by E. coli O157:H7
The evolution of phage resistance in E. coli O157:H7 is a continuous and natural process. As a result of repeated use of phages against E. coli O157:H7, in vitro experiments have shown that E. coli O157:H7 can develop phage resistance [150]. To avoid phage infection, they employ a variety of processes that involve the following techniques.
Phage inability to attach: E. coli O157:H7 may alter the architecture of its cell surfaces to restrict phages from attaching to its surface [151]. To attach to particular receptors on the bacterial cell wall, phages often use receptor-binding proteins [152]. Phage attachment may be prevented by changes to these receptors. E. coli O157:H7 can change its surface characteristics, such as the lipopolysaccharide (LPS) layer, thereby making it challenging for phages to adhere to the bacterial cell [153].
CRISPR-Cas Systems: Some E. coli O157:H7 strains have Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated (CRISPR-Cas) systems, which are a form of adaptive immunity against phages. CRISPR-Cas systems play a significant role in limiting phage infection and proliferation as an important bacteriophage resistance mechanism [154]. By using these systems, bacteria can collect and preserve genetic material from earlier phage contacts and use this information to locate and eliminate the phage DNA when there is a contact with the same phage.
Restriction–Modification (R-M) Systems: E. coli O157:H7 and other organisms may have R-M systems. These systems are made up of enzymes that protect bacterial cells by breaking down foreign DNA (including phage DNA) that enters the cell. They recognize and cleave phage DNA sequences on the recognition site [155], thereby inhibiting phage genetic material from multiplying.
Abortive Infection: The infected E. coli strain employs abortive infection mechanisms, resulting in self-destruction prior to the completion of the phage’s replication cycle [156]. This behaviour effectively inhibits the phage epidemic from spreading to neighbouring cells, thus ensuring the safety of the bacterial colony.
Understanding the mechanisms of phage resistance is critical in phage therapy. The mechanisms described above are strategies adopted by numerous bacterial populations to protect themselves from phage predation, and they can differ amongst strains of E. coli O157:H7. Due to the pressure exerted by phages, the pathogen can develop resistance to phages over time. As a result, phage cocktails can be used as a method to target multiple mechanisms or in combination with other treatment options to overcome bacterial resistance.
5. Determining the Safety of E. coli O157:H7 Specific Bacteriophages as a Biocontrol Agent
To maximise the potential of phage treatment of E. coli O157:H7 infections, phage candidates must be safe and capable of serving as biocontrol agents without having negative effects on people or the environment.
Firstly, phage’s specificity for E. coli O157:H7 must be ensured by isolating and properly characterising candidate phage; afterwards, phages should be screened for the existence of genes linked to virulence and antibiotic resistance within the phage’s genome simply because genes that encode toxins and allergens must not be present in phages intended for biocontrol as they can be detrimental to humans and animals [108]. New generation sequencing techniques like the Whole Genome Sequencing (WGS) can be used to analyse the phage genome and determine the presence of these genes.
WGS is a method of typing that relies on sequencing of the entire genome of an isolate, allowing for the identification of variations at the level of individual nucleotides [157]. Based on the findings of the study conducted by Lee et al. [158], WGS was used to determine the absence of virulence factors, toxins, antibiotic resistance, and potential allergen-coding genes in the genome of the isolated phage KFS-EC. The phage which was obtained from wastewater samples from a slaughterhouse in Korea exhibits specificity exclusively towards E. coli O157:H7 and was considered to be suitable as a biocontrol agent. In addition, as described by Liao et al. [159], WGS can be used to determine the presence of lysogenic elements that may potentially enable the dissemination of undesirable genes among bacterial communities. Having confirmed the lack of such elements, the phages can then be considered to be safe for use as a biocontrol. It is vital to highlight that WGS supports the “One Health” concept, which considers the interrelated nature of human, animal, and environmental health. This is demonstrated by the impact WGS has on public health responses, as its usage can quickly pinpoint the safety of phage candidates and support the implementation of efficient control measures to stop the spread of antimicrobial resistant microbes [160].
Having considered the genomic screening of phages, conducting animal studies to assess the safety of the phages in the target host organisms (e.g., livestock) is essential before a large scale production. Studies on the effectiveness of phages in treating experimentally infected animals [161,162] have demonstrated that using mice, chickens, or sheep in biocontrol trials will allow for the monitoring of any negative effects on animal health [163]. Clinical trial studies can evaluate the safety of phage therapy in people, including possible allergic reactions or other adverse effects. They can also determine the right concentration and dosage of phages required to control E. coli O157:H7 in people.
Due to the sensitive nature and ethical issues associated with phage application, stakeholders like farmers, food processors, and healthcare professionals should obtain training on how to handle phages safely and responsibly in the real-world settings. Furthermore, while ensuring phage purity and quality during preparation, there is a possibility of generated residues and contaminants causing environmental hazards, which could pose a health risk. It is crucial to monitor the potential environmental impact of using phages, such as phage persistence in the environment and potential ecological disruptions.
In the event that genetic modifications in the host bacterium impart phage resistance, such phages are not suitable for use as a biocontrol. Therefore, phage safety concerns require constant monitoring and surveillance. For example, it is important to use appropriate testing methods (such as microbiological assays) to quantify pathogen levels before and after phage application, monitor the development of phage resistance, regularly evaluate the effectiveness of phage treatments in controlling target pathogens like E. coli O157:H7, and monitor the safety and efficacy of phage products once they are on the market.
6. Conclusions and Future Perspectives
In the current era of antimicrobial resistance and the pursuit of alternative methods to combat E. coli O157:H7 infections, there has been a renewed interest in phage research. This is evident in the accumulated evidence suggesting that phage application is an effective method of biocontrol of foodborne bacterial pathogens on fresh produce and other foods. Phages has been used as a natural antimicrobial method to reduce E. coli O157:H7 from the food supply. Phage-based medications have been approved by the Food and Drug Administration (FDA) in certain countries, such as the United States, Canada, and Israel for the treatment of E. coli strains, including the use of various phage cocktails. Numerous studies have also produced novel phages and showed their effectiveness in inhibiting the growth of E. coli O157:H7 in food products without changing the organoleptic properties. They have also been proven to reduce E. coli O157:H7 in vivo through the use of mice. Ongoing research involves the construction of phage libraries, thereby enabling the use of phage cocktails to inhibit phage mutant strains, increase host range, and increase infectivity. Nevertheless, the European Union remains concerned about the utilization of these products due to the limited availability of safety data and comprehensive research on the potential consequences of phage discharge into the environment. One of the most important aspects of creating One Health approaches to lowering health risks for people, agricultural systems, and natural ecosystems is the effective use of “safe” phages as a biocontrol agent in combating pathogens like E. coli O157:H7 and resolving the issue of antibiotic resistance in preharvest livestock environments. In order to advance the development of novel antibacterial strategies, there is a need to conduct further investigation into the phage properties that make them safe and suitable as a biocontrol and the underlying mechanism through which phages can effectively inhibit the expression of host genes. The utilization of advanced technologies like WGS to carefully scrutinize and characterize phage genomes presents a solution to this concern. This, in turn, will improve both patient care and infection control strategies, ultimately leading to a decrease in the incidence of severe E. coli O157:H7 infections. This review highlights the renewed hope provided by modern technologies in combating E. coli O157:H7 infections, reducing the persistence and spread of antimicrobial resistance, and an enhanced potential in improving livestock production. These further provide an indication of increased food security and safety, and thus a significant contribution towards achieving the goals of the highly integrated “One health” approach.
Author Contributions
Conceptualization, B.O.O. and C.N.A.; methodology, B.O.O., H.A.N., P.K.M. and C.N.A.; validation, B.O.O., I.D.P., H.A.N., P.K.M. and C.N.A.; investigation, B.O.O. writing—original draft preparation, B.O.O.; writing—review and editing, B.O.O., D.J.A., T.O.A., I.D.P., H.A.N., P.K.M. and C.N.A.; supervision, I.D.P., H.A.N., P.K.M. and C.N.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Research Foundation (NRF), grant number 121802.
Data Availability Statement
Not applicable.
Acknowledgments
The authors would like to acknowledge the Faculty of Natural and Agricultural Sciences and the department of Microbiology, School of Biological Sciences, North-West University, South Africa for their support.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Vila, J.; Sáez-López, E.; Johnson, J.R.; Römling, U.; Dobrindt, U.; Cantón, R.; Giske, C.; Naas, T.; Carattoli, A.; Martínez-Medina, M. Escherichia coli: An old friend with new tidings. FEMS Microbiol. Rev. 2016, 40, 437–463. [Google Scholar] [CrossRef]
- Farrokh, C.; Jordan, K.; Auvray, F.; Glass, K.; Oppegaard, H.; Raynaud, S.; Thevenot, D.; Condron, R.; De Reu, K.; Govaris, A. Review of Shiga-toxin-producing Escherichia coli (STEC) and their significance in dairy production. Int. J. Food. Microbiol. 2013, 162, 190–212. [Google Scholar] [CrossRef]
- Ombarak, R.A.; Hinenoya, A.; Awasthi, S.P.; Iguchi, A.; Shima, A.; Elbagory, A.-R.M.; Yamasaki, S. Prevalence and pathogenic potential of Escherichia coli isolates from raw milk and raw milk cheese in Egypt. Int. J. Food Microbiol. 2016, 221, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Akindolire, M.A.; Ateba, C.N. Use of pulsed field gel electrophoresis genetic typing for tracing contamination with virulent Escherichia coli O157: H7 in beef-cattle producing farms. Gene Rep. 2018, 13, 59–65. [Google Scholar] [CrossRef]
- Sugiyama, A.; Iwade, Y.; Akachi, S.; Nakano, Y.; Matsuno, Y.; Yano, T.; Yamauchi, A.; Nakayama, O.; Sakai, H.; Yamamoto, K.-i. An outbreak of Shigatoxin-producing Escherichia coli O157: H7 in a nursery school in Mie Prefecture. Jpn. J. Infect. Dis. 2005, 58, 398. [Google Scholar]
- Shen, J.; Rump, L.; Ju, W.; Shao, J.; Zhao, S.; Brown, E.; Meng, J. Virulence characterization of non-O157 Shiga toxin-producing Escherichia coli isolates from food, humans and animals. Food Microb. 2015, 50, 20–27. [Google Scholar] [CrossRef]
- Ateba, C.N.; Mbewe, M. Detection of Escherichia coli O157: H7 virulence genes in isolates from beef, pork, water, human and animal species in the northwest province, South Africa: Public health implications. Res. Microbiol. 2011, 162, 240–248. [Google Scholar] [CrossRef]
- Ateba, C.N.; Mbewe, M. Genotypic characterization of Escherichia coli O157: H7 isolates from different sources in the north-west province, South Africa, using enterobacterial repetitive intergenic consensus PCR analysis. Int. J. Mol. Sci. 2014, 15, 9735–9747. [Google Scholar] [CrossRef] [PubMed]
- Law, D. Virulence factors of Escherichia coli O157 and other Shiga toxin-producing E. coli. J. Appl. Microb. 2000, 88, 729–745. [Google Scholar] [CrossRef]
- Rivera-Betancourt, M.; Shackelford, S.D.; Arthur, T.M.; Westmoreland, K.E.; Bellinger, G.; Rossman, M.; Reagan, J.O.; Koohmaraie, M. Prevalence of Escherichia coli O157: H7, Listeria monocytogenes, and Salmonella in two geographically distant commercial beef processing plants in the United States. J. Food Protect. 2004, 67, 295–302. [Google Scholar] [CrossRef]
- Shu, X.; Singh, M.; Karampudi, N.B.R.; Bridges, D.F.; Kitazumi, A.; Wu, V.C.; De los Reyes, B.G. Responses of Escherichia coli and Listeria monocytogenes to ozone treatment on non-host tomato: Efficacy of intervention and evidence of induced acclimation. PLoS ONE 2021, 16, e0256324. [Google Scholar] [CrossRef]
- Liu, D.; Lawrence, M.L.; Ainsworth, A.J.; Austin, F.W. Comparative assessment of acid, alkali and salt tolerance in Listeria monocytogenes virulent and avirulent strains. FEMS Microb. Lett. 2005, 243, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Bridges, D.F.; Rane, B.; Wu, V.C. The effectiveness of closed-circulation gaseous chlorine dioxide or ozone treatment against bacterial pathogens on produce. Food Cont. 2018, 91, 261–267. [Google Scholar] [CrossRef]
- WHO. WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: http://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/en/ (accessed on 8 August 2023).
- Gould, L.H.; Mody, R.K.; Ong, K.L.; Clogher, P.; Cronquist, A.B.; Garman, K.N.; Lathrop, S.; Medus, C.; Spina, N.L.; Webb, T.H. Increased recognition of non-O157 Shiga toxin–producing Escherichia coli infections in the United States during 2000–2010: Epidemiologic features and comparison with E. coli O157 infections. Foodborne Pathog. Dis. 2013, 10, 453–460. [Google Scholar] [CrossRef]
- Karlsson, M.E.; Uhlig, E.; Håkansson, Å.; Alsanius, B.W. Seed inoculation with antagonistic bacteria limits occurrence of E. coli O157: H7gfp+ on baby spinach leaves. BMC Microbiol. 2022, 22, 131. [Google Scholar] [CrossRef] [PubMed]
- Ben, Y.; Fu, C.; Hu, M.; Liu, L.; Wong, M.H.; Zheng, C. Human health risk assessment of antibiotic resistance associated with antibiotic residues in the environment: A review. Environ. Res. 2019, 169, 483–493. [Google Scholar] [CrossRef]
- Griffin, P. Escherichia coli O157: H7 and other enterohemorrhagic Escherichia coli. Infect. Gas. Tract. 1995, 169, 739–761. [Google Scholar]
- Ammar, A.M.; Abd El-Hamid, M.I.; El-Malt, R.M.; Azab, D.S.; Albogami, S.; Al-Sanea, M.M.; Soliman, W.E.; Ghoneim, M.M.; Bendary, M.M. Molecular detection of fluoroquinolone resistance among multidrug-, extensively drug-, and pan-drug-resistant Campylobacter species in Egypt. Antibiotics 2021, 10, 1342. [Google Scholar] [CrossRef]
- Huchin, C.; Briceño, M.A.; Mendoza, T.; Martínez, A.P.; Ramírez, M.A.; Torres, J.C. Prevalence and drug-resistance patterns of Enterotoxigenic Escherichia coli and Shigella species among children with diarrhea in Merida City, Mexico. J. Biosci. Med. 2017, 6, 22–33. [Google Scholar]
- Arbab, S.; Ullah, H.; Wang, W.; Zhang, J. Antimicrobial drug resistance against Escherichia coli and its harmful effect on animal health. Vet. Med. Sci. 2022, 8, 1780–1786. [Google Scholar] [CrossRef] [PubMed]
- Gambushe, S.M.; Zishiri, O.T.; El Zowalaty, M.E. Review of Escherichia coli O157: H7 prevalence, pathogenicity, heavy metal and antimicrobial resistance, African perspective. Infect. Drug Resist. 2022, 15, 4645–4673. [Google Scholar] [CrossRef] [PubMed]
- Rivas, L.; Coffey, B.; McAuliffe, O.; McDonnell, M.J.; Burgess, C.M.; Coffey, A.; Ross, R.P.; Duffy, G. In vivo and ex vivo evaluations of bacteriophages e11/2 and e4/1c for use in the control of Escherichia coli O157: H7. Appl. Environ. Microbiol. 2010, 76, 7210–7216. [Google Scholar] [CrossRef] [PubMed]
- Kannan, G.; Mahapatra, A.K.; Degala, H.L. Preharvest management and postharvest intervention strategies to reduce Escherichia coli contamination in goat meat: A review. Animals 2021, 11, 2943. [Google Scholar] [CrossRef]
- Jeong, K.C.; Kang, M.Y.; Kang, J.; Baumler, D.J.; Kaspar, C.W. Reduction of Escherichia coli O157: H7 shedding in cattle by addition of chitosan microparticles to feed. Appl. Environ. Microbiol. 2011, 77, 2611–2616. [Google Scholar] [CrossRef]
- Anderson, N.M. Recent advances in low moisture food pasteurization. Curr. Opin. Food Sci. 2019, 29, 109–115. [Google Scholar] [CrossRef]
- Acuff, G. Chemical decontamination strategies for meat. In Improving the Safety of Fresh Meat; Sofos, J., Ed.; Woodhead Publishing: Sawston, Cambridge, UK, 2005; pp. 350–363. [Google Scholar]
- Gil, M.I.; Selma, M.V.; López-Gálvez, F.; Allende, A. Fresh-cut product sanitation and wash water disinfection: Problems and solutions. Int. J. Food Microbial. 2009, 134, 37–45. [Google Scholar] [CrossRef]
- Harada, L.K.; Silva, E.C.; Campos, W.F.; Del Fiol, F.S.; Vila, M.; Dąbrowska, K.; Krylov, V.N.; Balcão, V.M. Biotechnological applications of bacteriophages: State of the art. Microbiol. Res. 2018, 212, 38–58. [Google Scholar] [CrossRef]
- Tan, T.; Chan, K.; Lee, L. Application of bacteriophage in biocontrol of major foodborne bacterial pathogens. J. Mol. Biol. Mol. Imaging 2014, 1, 4658187. [Google Scholar]
- Moye, Z.D.; Woolston, J.; Sulakvelidze, A. Bacteriophage applications for food production and processing. Viruses 2018, 10, 205. [Google Scholar] [CrossRef]
- Zinno, P.; Devirgiliis, C.; Ercolini, D.; Ongeng, D.; Mauriello, G. Bacteriophage P22 to challenge Salmonella in foods. Int. J. Food Microbiol. 2014, 191, 69–74. [Google Scholar] [CrossRef]
- Pinto, G.; Minnich, S.A.; Hovde, C.J.; Oliveira, H.; Smidt, H.; Almeida, C.; Azeredo, J. The interactions of bacteriophage Ace and Shiga toxin-producing Escherichia coli during biocontrol. FEMS Microbiol. Ecol. 2021, 97, fiab105. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, T.; Yu, M.; Chen, Y.-L.; Jin, M. The life cycle transitions of temperate phages: Regulating factors and potential ecological implications. Viruses 2022, 14, 1904. [Google Scholar] [CrossRef] [PubMed]
- Law, D. The history and evolution of Escherichia coli O157 and other Shiga toxin-producing E. coli. World J. Microbiol. 2000, 16, 701–709. [Google Scholar] [CrossRef]
- Wick, L.M.; Qi, W.; Lacher, D.W.; Whittam, T.S. Evolution of genomic content in the stepwise emergence of Escherichia coli O157: H7. J. Bacteriol. 2005, 187, 1783–1791. [Google Scholar] [CrossRef]
- Feng, P.; Lampel, K.A.; Karch, H.; Whittam, T.S. Genotypic and phenotypic changes in the emergence of Escherichia coli O157: H7. J. Infect. Dis. 1998, 177, 1750–1753. [Google Scholar] [CrossRef] [PubMed]
- Idland, L.; Bø-Granquist, E.G.; Aspholm, M.; Lindbäck, T. The Ability of Shiga Toxin-Producing Escherichia coli to Grow in Raw Cow’s Milk Stored at Low Temperatures. Foods 2022, 11, 3411. [Google Scholar] [CrossRef] [PubMed]
- Al-Aalim, A.M.A. Immune Responses Induced by Standard and Extracted Lipopolysaccharide (LPS) from Escherichia coli. Ph.D. Thesis, University of Basrah, Basrah, Iraq, 2022. [Google Scholar]
- Gebisa, E.S.; Gerasu, M.A.; Leggese, D.T. A Review on Virulence Factors of Escherichia coli. Vet. Anim. Sci. 2019, 7, 83–93. [Google Scholar] [CrossRef]
- Karch, H.; Bielaszewska, M. Sorbitol-fermenting Shiga toxin-producing Escherichia coli O157: H−strains: Epidemiology, phenotypic and molecular characteristics, and microbiological diagnosis. J. Clin. Micobiol. 2001, 39, 2043–2049. [Google Scholar] [CrossRef]
- Monday, S.R.; Minnich, S.A.; Feng, P.C. A 12-base-pair deletion in the flagellar master control gene flhC causes nonmotility of the pathogenic German sorbitol-fermenting Escherichia coli O157: H−strains. J. Bacteriol. 2004, 186, 2319–2327. [Google Scholar] [CrossRef]
- Lim, J.Y.; Yoon, J.W.; Hovde, C.J. A brief overview of Escherichia coli O157: H7 and its plasmid. J. Microbiol. Biotechnol. 2010, 20, 5. [Google Scholar] [CrossRef]
- Stanton, E. The Impact of Mobile Genetic Elements as Drivers of Genome Diversification in Bovine Escherichia coli O157: H7; The University of Wisconsin-Madison: Madison, WI, USA, 2020. [Google Scholar]
- Nguyen, T.T.H.; Kikuchi, T.; Tokunaga, T.; Iyoda, S.; Iguchi, A. Diversity of the tellurite resistance gene operon in Escherichia coli. Front. Microbiol. 2021, 12, 681175. [Google Scholar] [CrossRef]
- Lee, M.-S.; Tesh, V.L. Roles of Shiga toxins in immunopathology. J. Toxins 2019, 11, 212. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Rubio, L.; Haarmann, N.; Schwidder, M.; Muniesa, M.; Schmidt, H. Bacteriophages of Shiga toxin-producing Escherichia coli and their contribution to pathogenicity. J. Pathog. 2021, 10, 404. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Bolton, F.; Fiefield, D.; Lamb, P.; Woloschin, E.; Smith, N.; McCann, R. An outbreak of E. coli O157 associated with a swimming pool: An unusual vehicle of transmission. Epidemiol. Infect. 2007, 135, 989–992. [Google Scholar] [CrossRef]
- Ababu, A.; Endashaw, D.; Fesseha, H. Isolation and antimicrobial susceptibility profile of Escherichia coli O157: H7 from raw milk of dairy cattle in Holeta district, Central Ethiopia. Int. J. Microbiol. 2020, 2020, 6626488. [Google Scholar] [CrossRef]
- Smith, J.L.; Fratamico, P.M.; Gunther, N.W., IV. Shiga toxin-producing Escherichia coli. Adv. Appl. Microbiol. 2014, 86, 145–197. [Google Scholar] [CrossRef] [PubMed]
- Montso, P.K.; Mlambo, V.; Ateba, C.N. The first isolation and molecular characterization of Shiga Toxin-producing virulent multi-drug resistant atypical enteropathogenic Escherichia coli O177 serogroup from South African Cattle. Front. Cell. Infect. Microbiol. 2019, 9, 333. [Google Scholar] [CrossRef]
- Nicola, S.; Cocetta, G.; Ferrante, A.; Ertani, A. Fresh-cut produce quality: Implications for postharvest. In Postharvest Handling; Elsevier: Amsterdam, The Netherlands, 2022; pp. 187–250. [Google Scholar]
- Zhao, X.; Sun, Y.; Ma, Y.; Xu, Y.; Guan, H.; Wang, D. Research advances on the contamination of vegetables by Enterohemorrhagic Escherichia coli: Pathways, processes and interaction. Crit. Rev. Food Sci. Nutr. 2022, 1–15. [Google Scholar] [CrossRef]
- Kim, D.-K.; Shin, M.; Kim, H.-S.; Kang, D.-H. Inactivation efficacy of combination treatment of blue light-emitting diodes (LEDs) and riboflavin to control E. coli O157: H7 and S. typhimurium in apple juice. Innov. Food Sci. Emerg. Technol. 2022, 78, 103014. [Google Scholar] [CrossRef]
- Angelopoulou, M.; Petrou, P.; Misiakos, K.; Raptis, I.; Kakabakos, S. Simultaneous Detection of Salmonella typhimurium and Escherichia coli O157: H7 in Drinking Water and Milk with Mach–Zehnder Interferometers Monolithically Integrated on Silicon Chips. Biosensors 2022, 12, 507. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, H.J.; Knox, J.P.; Boraston, A.B. Advances in understanding the molecular basis of plant cell wall polysaccharide recognition by carbohydrate-binding modules. Curr. Opin. Struc. Biol. 2013, 23, 669–677. [Google Scholar] [CrossRef]
- Xie, Y.; Zhu, L.; Lyu, G.; Lu, L.; Ma, J.; Ma, J. Persistence of E. coli O157: H7 in urban recreational waters from Spring and Autumn: A comparison analysis. Environ. Sci. Pollut. Res. 2022, 29, 39088–39101. [Google Scholar] [CrossRef] [PubMed]
- Iwu, C.D.; du Plessis, E.; Korsten, L.; Okoh, A.I. Antibiogram imprints of E. coli O157: H7 recovered from irrigation water and agricultural soil samples collected from two district municipalities in South Africa. Int. J. Environ. Stud. 2021, 78, 940–953. [Google Scholar] [CrossRef]
- Odo, S.E.; Uchechukwu, C.F.; Ezemadu, U.R. Foodborne Diseases and Intoxication in Nigeria: Prevalence of Escherichia coli 0157: H7, Salmonella, Shigella and Staphylococcus aureus. J. Adv. Microbiol. 2021, 20, 84–94. [Google Scholar] [CrossRef]
- Lopes, F.A.; Davies-Colley, R.; Piazi, J.; Silveira, J.S.; Leite, A.C.; Lopes, N.I.A. Challenges for contact recreation in a tropical urban lake: Assessment by a water quality index. Environ. Dev. Sustain. 2020, 22, 5409–5423. [Google Scholar] [CrossRef]
- Chávez-Díaz, L.V.; Gutiérrez-Cacciabue, D.; Poma, H.R.; Rajal, V.B. Sediments quality must be considered when evaluating freshwater aquatic environments used for recreational activities. Int. J. Hyg. Environ. Health 2020, 223, 159–170. [Google Scholar] [CrossRef]
- Ihekweazu, C.; Barlow, M.; Roberts, S.; Christensen, H.; Guttridge, B.; Lewis, D.A.; Painter, S. Outbreak of E. coli O157 infection in the south west of the UK: Risks from streams crossing seaside beaches. Eurosurveillance 2006, 11, 5–6. [Google Scholar] [CrossRef]
- Akindolire, M.A. Molecular Characterisation of Escherichia coli O157: H7 Specific Bacteriophages from Cattle Faeces. Ph.D. Thesis, North-West University (South Africa), Mahikeng, South Africa, 2019. [Google Scholar]
- Seabela, M.D.L. Assessment of Food Safety Hazards among Day Care Centres in Mbombela, Republic of South Africa. Master’s Dissertation, University of Johannesburg, Johannesburg, South Africa, 2020. [Google Scholar]
- Barkley, J.; Viveiros, B.; Michael Gosciminski, M.; Utpala Bandy, M. Preventing foodborne and enteric illnesses among at-risk populations in the United States and Rhode Island. Rhode Isl. Med. J. 2016, 99, 25. [Google Scholar]
- Mesele, F.; Abunna, F. Escherichia coli O157: H7 in foods of animal origin and its food safety implications. Adv. Biol. Res. 2019, 13, 134–145. [Google Scholar]
- Rangel, J.M.; Sparling, P.H.; Crowe, C.; Griffin, P.M.; Swerdlow, D.L. Epidemiology of Escherichia coli O157: H7 outbreaks, united states, 1982–2002. Emerg. Infect. Dis. 2005, 11, 603. [Google Scholar] [CrossRef]
- Lupindua, A.M. Epidemiology of Shiga toxin-producing Escherichia coli O157: H7 in Africa in review. S. Afr. J. Infect. Dis. 2018, 33, 24–30. [Google Scholar] [CrossRef]
- Browning, N.; Botha, J.; Sacho, H.; Moore, P. Escherichia coli O157: H7 haemorrhagic colitis. Report of the first South African case. S. Afr. J. Surg. 1990, 28, 28–29. [Google Scholar] [PubMed]
- Germani, Y.; Soro, B.; Vohito, M.; Morel, O.; Morvan, J. Enterohaemorrhagic Escherichia coli in Central African Republic. Lancet 1997, 349, 1670. [Google Scholar] [CrossRef] [PubMed]
- Raji, M.; Minga, U.; Machang’u, R. Prevalence and characterization of verotocytoxin producing Escherichia coli O157 from diarrhoea patients in Morogoro, Tanzania. Tanzan. J. Health Res. 2008, 10, 151–158. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ghali-Mohammed, I.; Odetokun, I.A.; Raufu, I.A.; Alhaji, N.B.; Adetunji, V.O. Prevalence of Escherichia coli O157 isolated from marketed raw cow milk in Kwara State, Nigeria. Sci. Afr. 2023, 19, e01469. [Google Scholar] [CrossRef]
- Mandomando, I.M.; Macete, E.V.; Ruiz, J.; Sanz, S.; Abacassamo, F.; Valles, X.; Sacarlal, J.; Navia, M.M.; Vila, J.; Alonso, P.L. Etiology of diarrhea in children younger than 5 years of age admitted in a rural hospital of southern Mozambique. Am. J. Trop. Med. Hyg. 2007, 76, 522–527. [Google Scholar] [CrossRef]
- Gwavava, C.; Chihota, V.; Gangaidzo, I.; Gumbo, T. Dysentery in patients infected with human immunodeficiency virus in Zimbabwe: An emerging role for Schistosoma mansoni and Escherichia coli O157? Ann. Trop. Med. Parasitol. 2001, 95, 509–513. [Google Scholar] [CrossRef]
- Croxen, M.A.; Law, R.J.; Scholz, R.; Keeney, K.M.; Wlodarska, M.; Finlay, B.B. Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 2013, 26, 822–880. [Google Scholar] [CrossRef]
- Jores, J.; Rumer, L.; Wieler, L.H. Impact of the locus of enterocyte effacement pathogenicity island on the evolution of pathogenic Escherichia coli. Int. J. Med. Microbiol. 2004, 294, 103–113. [Google Scholar] [CrossRef]
- Farfan, M.J.; Torres, A.G. Molecular mechanisms that mediate colonization of Shiga toxin-producing Escherichia coli strains. Infect. Immun. 2012, 80, 903–913. [Google Scholar] [CrossRef]
- Bielaszewska, M.; Aldick, T.; Bauwens, A.; Karch, H. Hemolysin of enterohemorrhagic Escherichia coli: Structure, transport, biological activity and putative role in virulence. Int. J. Med. Microbiol. 2014, 304, 521–529. [Google Scholar] [CrossRef]
- Pinto, G.; Almeida, C.; Azeredo, J. Bacteriophages to control Shiga toxin-producing E. coli—Safety and regulatory challenges. Crit. Rev. Biotechnol. 2020, 40, 1081–1097. [Google Scholar] [CrossRef]
- WHO. The Fight against Antimicrobial Resistance Is Closely Linked to the Sustainable Development Goals. Available online: https://apps.who.int/iris/bitstream/handle/10665/337519/WHO-EURO-2020-1634-41385-56394-eng.pdf (accessed on 26 December 2022).
- Ackermann, H.-W.; Martin, M.; Vieu, J.-F.; Nicolle, P. Félix d’Hérelle: His life and work and the foundation of a bacteriophage reference center. ASM News 1982, 48, 347–348. [Google Scholar]
- Aswani, V.H.; Shukla, S.K. An Early History of Phage Therapy in the United States: Is it Time to Reconsider? J. Clin. Med. Res. 2021, 19, 82–89. [Google Scholar] [CrossRef]
- Clokie, M.R.; Millard, A.D.; Letarov, A.V.; Heaphy, S. Phages in nature. Bacteriophage 2011, 1, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Fernández, L.; Rodríguez, A.; García, P. Phage or foe: An insight into the impact of viral predation on microbial communities. ISME J. 2018, 12, 1171–1179. [Google Scholar] [CrossRef]
- Melo, L.D.; Oliveira, H.; Santos, S.B.; Sillankorva, S.; Azeredo, J. Phages against infectious diseases. In Bioprospecting; Springer: Berlin/Heidelberg, Germany, 2017; pp. 269–294. [Google Scholar]
- d’Herelle, F. An invisible microbe that is antagonistic to the dysentery Bacillus. CR Acad. Sci. 1917, 165, 373–375. [Google Scholar]
- Strathdee, S.A.; Hatfull, G.F.; Mutalik, V.K.; Schooley, R.T. Phage therapy: From biological mechanisms to future directions. Cell 2023, 186, 17–31. [Google Scholar] [CrossRef]
- Wittebole, X.; De Roock, S.; Opal, S.M. A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence 2014, 5, 226–235. [Google Scholar] [CrossRef]
- Wernicki, A.; Nowaczek, A.; Urban-Chmiel, R. Bacteriophage therapy to combat bacterial infections in poultry. Virol. J. 2017, 14, 179. [Google Scholar] [CrossRef]
- Sillankorva, S.M.; Oliveira, H.; Azeredo, J. Bacteriophages and their role in food safety. Int. J. Microbiol. 2012, 2012, 863945. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Gómez, C.; Blanco-Picazo, P.; Brown-Jaque, M.; Quirós, P.; Rodríguez-Rubio, L.; Cerdà-Cuellar, M.; Muniesa, M. Infectious phage particles packaging antibiotic resistance genes found in meat products and chicken feces. Sci. Rep. 2019, 9, 13281. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.W.; Rukayadi, Y.; Hasan, H.; Abdul-Mutalib, N.-A.; Jambari, N.N.; Hara, H.; Thung, T.Y.; Lee, E.; Radu, S. Isolation and characterization of six Vibrio parahaemolyticus lytic bacteriophages from seafood samples. Front. Microbiol. 2021, 12, 616548. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.; Bandara, N.; Shin, E.; Ryu, S.; Kim, K.-p. Prevalence of Bacillus cereus bacteriophages in fermented foods and characterization of phage JBP901. Res. Microbiol. 2011, 162, 791–797. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Lin, H.; Ji, X.; Yan, G.; Lei, L.; Han, W.; Gu, J.; Huang, J. Therapeutic applications of lytic phages in human medicine. Microb. Pathog. 2020, 142, 104048. [Google Scholar] [CrossRef]
- Brovko, L.Y.; Anany, H.; Griffiths, M.W. Bacteriophages for detection and control of bacterial pathogens in food and food-processing environment. Adv. Food Nutr. Res. 2012, 67, 241–288. [Google Scholar] [CrossRef]
- Bandara, N.; Jo, J.; Ryu, S.; Kim, K.-P. Bacteriophages BCP1-1 and BCP8-2 require divalent cations for efficient control of Bacillus cereus in fermented foods. Food Microbiol. 2012, 31, 9–16. [Google Scholar] [CrossRef]
- Imran, A.; Shehzadi, U.; Islam, F.; Afzaal, M.; Ali, R.; Ali, Y.A.; Chauhan, A.; Biswas, S.; Khurshid, S.; Usman, I. Bacteriophages and food safety: An updated overview. Food Sci. Nutr. 2023, 11, 3621–3630. [Google Scholar] [CrossRef]
- O’Sullivan, L.; Bolton, D.; McAuliffe, O.; Coffey, A. Bacteriophages in food applications: From foe to friend. Annu. Rev. Food Sci. Technol. 2019, 10, 151–172. [Google Scholar] [CrossRef] [PubMed]
- Bonilla, N.; Rojas, M.I.; Cruz, G.N.F.; Hung, S.-H.; Rohwer, F.; Barr, J.J. Phage on tap–a quick and efficient protocol for the preparation of bacteriophage laboratory stocks. PeerJ 2016, 4, e2261. [Google Scholar] [CrossRef] [PubMed]
- Pirnay, J.-P.; Verbeken, G.; Ceyssens, P.-J.; Huys, I.; De Vos, D.; Ameloot, C.; Fauconnier, A. The magistral phage. Viruses 2018, 10, 64. [Google Scholar] [CrossRef] [PubMed]
- Rehman, S.; Ali, Z.; Khan, M.; Bostan, N.; Naseem, S. The dawn of phage therapy. Rev. Med. Virol. 2019, 29, e2041. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.B.; Vallad, G.E.; Iriarte, F.B.; Obradović, A.; Wernsing, M.H.; Jackson, L.E.; Balogh, B.; Hong, J.C.; Momol, M.T. Considerations for using bacteriophages for plant disease control. Bacteriophage 2012, 2, e23857. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Vivas, J.; Elexpuru-Zabaleta, M.; Samano, M.L.; Barrera, A.P.; Forbes-Hernández, T.Y.; Giampieri, F.; Battino, M. Phages and enzybiotics in food biopreservation. Molecules 2021, 26, 5138. [Google Scholar] [CrossRef] [PubMed]
- Dion, M.B.; Oechslin, F.; Moineau, S. Phage diversity, genomics and phylogeny. Nat. Rev. Microbiol. 2020, 18, 125–138. [Google Scholar] [CrossRef]
- Qu, Q.; Chen, T.; He, P.; Geng, H.; Zeng, P.; Luan, G. Isolation and characterization of a novel lytic bacteriophage vB_Efm_LG62 infecting Enterococcus faecium. Virus Genes 2023, 59, 763–774. [Google Scholar] [CrossRef]
- Bulannga, R.B.; Schmidt, S. Two predators, one prey—The interaction between bacteriophage, bacterivorous ciliates, and Escherichia coli. Microb. Ecol. 2023, 86, 1620–1631. [Google Scholar] [CrossRef]
- Montso, P.K.; Mlambo, V.; Ateba, C.N. Characterization of lytic bacteriophages infecting multidrug-resistant shiga toxigenic atypical Escherichia coli O177 strains isolated from cattle feces. Front. Public Health 2019, 7, 355. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.J.; Lefkowitz, E.J.; King, A.M.; Harrach, B.; Harrison, R.L.; Knowles, N.J.; Kropinski, A.M.; Krupovic, M.; Kuhn, J.H.; Mushegian, A.R. 50 years of the International Committee on Taxonomy of Viruses: Progress and prospects. Arch. Virol. 2017, 162, 1441–1446. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.; Shkoporov, A.N.; Lood, C.; Millard, A.D.; Dutilh, B.E.; Alfenas-Zerbini, P.; van Zyl, L.J.; Aziz, R.K.; Oksanen, H.M.; Poranen, M.M. Abolishment of morphology-based taxa and change to binomial species names: 2022 taxonomy update of the ICTV bacterial viruses subcommittee. Arch. Virol. 2023, 168, 74. [Google Scholar] [CrossRef] [PubMed]
- Navez, M.; Antoine, C.; Laforêt, F.; Goya-Jorge, E.; Douny, C.; Scippo, M.-L.; Vermeersch, M.; Duprez, J.-N.; Daube, G.; Mainil, J. In Vitro Effect on Piglet Gut Microbiota and In Vivo Assessment of Newly Isolated Bacteriophages against F18 Enterotoxigenic Escherichia coli (ETEC). Viruses 2023, 15, 1053. [Google Scholar] [CrossRef] [PubMed]
- Śliwka, P.; Weber-Dąbrowska, B.; Żaczek, M.; Kuźmińska-Bajor, M.; Dusza, I.; Skaradzińska, A. Characterization and comparative genomic analysis of three virulent E. coli bacteriophages with the potential to reduce antibiotic-resistant bacteria in the environment. Int. J. Mol. Sci. 2023, 24, 5696. [Google Scholar] [CrossRef]
- Mallmann, W.; Hemstreet, C. Isolation of an inhibitory substance from plants. Agric. Res. 1924, 28, 599–602. [Google Scholar]
- Buttimer, C.; McAuliffe, O.; Ross, R.P.; Hill, C.; O’Mahony, J.; Coffey, A. Bacteriophages and bacterial plant diseases. Front. Microbiol. 2017, 8, 34. [Google Scholar] [CrossRef]
- Das, M.; Bhowmick, T.S.; Ahern, S.J.; Young, R.; Gonzalez, C.F. Control of Pierce’s disease by phage. PLoS ONE 2015, 10, e0128902. [Google Scholar] [CrossRef] [PubMed]
- Dalmasso, M.; Strain, R.; Neve, H.; Franz, C.M.; Cousin, F.J.; Ross, R.P.; Hill, C. Three new Escherichia coli phages from the human gut show promising potential for phage therapy. PLoS ONE 2016, 11, e0156773. [Google Scholar] [CrossRef]
- Viazis, S.; Akhtar, M.; Feirtag, J.; Diez-Gonzalez, F. Reduction of Escherichia coli O157: H7 viability on leafy green vegetables by treatment with a bacteriophage mixture and trans-cinnamaldehyde. Food Microbiol. 2011, 28, 149–157. [Google Scholar] [CrossRef]
- Snyder, A.B.; Perry, J.J.; Yousef, A.E. Developing and optimizing bacteriophage treatment to control enterohemorrhagic Escherichia coli on fresh produce. Int. J. Food Microbiol. 2016, 236, 90–97. [Google Scholar] [CrossRef]
- Coffey, B.; Rivas, L.; Duffy, G.; Coffey, A.; Ross, R.P.; McAuliffe, O. Assessment of Escherichia coli O157: H7-specific bacteriophages e11/2 and e4/1c in model broth and hide environments. Int. J. Food Microbiol. 2011, 147, 188–194. [Google Scholar] [CrossRef]
- Viazis, S.; Akhtar, M.; Feirtag, J.; Diez-Gonzalez, F. Reduction of Escherichia coli O157: H7 viability on hard surfaces by treatment with a bacteriophage mixture. Int. J. Food Microbiol. 2011, 145, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Anany, H.; Chen, W.; Pelton, R.; Griffiths, M. Biocontrol of Listeria monocytogenes and Escherichia coli O157: H7 in meat by using phages immobilized on modified cellulose membranes. Appl. Environ. Microbiol. 2011, 77, 6379–6387. [Google Scholar] [CrossRef] [PubMed]
- Tomat, D.; Casabonne, C.; Aquili, V.; Balagué, C.; Quiberoni, A. Evaluation of a novel cocktail of six lytic bacteriophages against Shiga toxin-producing Escherichia coli in broth, milk and meat. Food Microbiol. 2018, 76, 434–442. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, S.; Roberts, C.; Handy, E.; Sharma, M. Lytic bacteriophages reduce Escherichia coli O157: H7 on fresh cut lettuce introduced through cross-contamination. Bacteriophage 2013, 3, e24323. [Google Scholar] [CrossRef] [PubMed]
- Hudson, J.; Billington, C.; Premaratne, A.; On, S.L. Inactivation of Escherichia coli O157: H7 using ultraviolet light-treated bacteriophages. Food Sci. Technol. Int. 2016, 22, 3–9. [Google Scholar] [CrossRef]
- Liu, H.; Niu, Y.; Meng, R.; Wang, J.; Li, J.; Johnson, R.; McAllister, T.; Stanford, K. Control of Escherichia coli O157 on beef at 37, 22 and 4 C by T5-, T1-, T4-and O1-like bacteriophages. Food Microbiol. 2015, 51, 69–73. [Google Scholar] [CrossRef]
- Le, T.S.; Southgate, P.C.; O’Connor, W.; Poole, S.; Kurtböke, D.I. Bacteriophages as biological control agents of enteric bacteria contaminating edible oysters. Curr. Microbiol. 2018, 75, 611–619. [Google Scholar] [CrossRef]
- Cui, H.; Bai, M.; Yuan, L.; Surendhiran, D.; Lin, L. Sequential effect of phages and cold nitrogen plasma against Escherichia coli O157: H7 biofilms on different vegetables. Int. J. Food Microbiol. 2018, 268, 1–9. [Google Scholar] [CrossRef]
- Patel, J.; Sharma, M.; Millner, P.; Calaway, T.; Singh, M. Inactivation of Escherichia coli O157: H7 attached to spinach harvester blade using bacteriophage. Foodborne Pathog. Dis. 2011, 8, 541–546. [Google Scholar] [CrossRef]
- Ramirez, K.; Cazarez-Montoya, C.; Lopez-Moreno, H.S.; Castro-del Campo, N. Bacteriophage cocktail for biocontrol of Escherichia coli O157: H7: Stability and potential allergenicity study. PLoS ONE 2018, 13, e0195023. [Google Scholar] [CrossRef]
- Aslam, S.; Schooley, R.T. What’s old is new again: Bacteriophage therapy in the 21st century. Antimicrob. Agents Chemother. 2019, 64, e01987-19. [Google Scholar] [CrossRef]
- Hesse, S.; Adhya, S. Phage therapy in the twenty-first century: Facing the decline of the antibiotic era; is it finally time for the age of the phage? Annu. Rev. Microbiol. 2019, 73, 155–174. [Google Scholar] [CrossRef] [PubMed]
- Nale, J.Y.; Redgwell, T.A.; Millard, A.; Clokie, M.R. Efficacy of an optimised bacteriophage cocktail to clear Clostridium difficile in a batch fermentation model. Antibiotics 2018, 7, 13. [Google Scholar] [CrossRef] [PubMed]
- Melo, L.D.; Pires, D.P.; Monteiro, R.; Azeredo, J. Phage therapy of infectious biofilms: Challenges and strategies. In Phage Therapy: A Practical Approach; Springer: Berlin/Heidelberg, Germany, 2019; pp. 295–313. [Google Scholar]
- Muthuvelu, K.S.; Ethiraj, B.; Pramnik, S.; Raj, N.K.; Venkataraman, S.; Rajendran, D.S.; Bharathi, P.; Palanisamy, E.; Narayanan, A.S.; Vaidyanathan, V.K. Biopreservative technologies of food: An alternative to chemical preservation and recent developments. Food Sci. Biotechnol. 2023, 32, 1337–1350. [Google Scholar] [CrossRef] [PubMed]
- Shuwen, H.; Kefeng, D. Intestinal phages interact with bacteria and are involved in human diseases. Gut Microbes 2022, 14, 2113717. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lv, P.; Shi, D.; Zhao, H.; Yuan, X.; Jin, X.; Wang, X. A cocktail of three virulent phages controls multidrug-resistant Salmonella enteritidis infection in poultry. Front. Microbiol. 2022, 13, 940525. [Google Scholar] [CrossRef]
- Bruttin, A.; Brüssow, H. Human volunteers receiving Escherichia coli phage T4 orally: A safety test of phage therapy. Antimicrob. Agents Chemother. 2005, 49, 2874–2878. [Google Scholar] [CrossRef]
- Sarker, S.A.; Sultana, S.; Reuteler, G.; Moine, D.; Descombes, P.; Charton, F.; Bourdin, G.; McCallin, S.; Ngom-Bru, C.; Neville, T. Oral phage therapy of acute bacterial diarrhea with two coliphage preparations: A randomized trial in children from Bangladesh. EBioMedicine 2016, 4, 124–137. [Google Scholar] [CrossRef]
- Intralytix. EcoShieldTM PX. Available online: https://www.intralytix.com/index.php/product/11 (accessed on 10 August 2023).
- Wójcicki, M.; Błażejak, S.; Gientka, I.; Brzezicka, K. The concept of using bacteriophages to improve the microbiological quality of minimally processed foods. Acta Sci.Pol. Technol. Aliment. 2019, 18, 373–383. [Google Scholar] [CrossRef]
- FDA. GRAS Notice Inventory. Available online: https://www.fda.gov/food/generally-recognized-safe-gras/gras-notice-inventory (accessed on 10 August 2023).
- Górski, A.; Międzybrodzki, R.; Łobocka, M.; Głowacka-Rutkowska, A.; Bednarek, A.; Borysowski, J.; Jończyk-Matysiak, E.; Łusiak-Szelachowska, M.; Weber-Dąbrowska, B.; Bagińska, N. Phage therapy: What have we learned? Viruses 2018, 10, 288. [Google Scholar] [CrossRef]
- Brüssow, H. Hurdles for phage therapy to become a reality—An editorial comment. Viruses 2019, 11, 557. [Google Scholar] [CrossRef]
- Dennehy, J.J.; Abedon, S.T. Adsorption: Phage acquisition of bacteria. In Bacteriophages: Biology, Technology, Therapy; Springer: Cham, Switzerland, 2021; pp. 93–117. [Google Scholar]
- Lopez, M.E.S.; Gontijo, M.T.P.; Cardoso, R.R.; Batalha, L.S.; Eller, M.R.; Bazzolli, D.M.S.; Vidigal, P.M.P.; Mendonça, R.C.S. Complete genome analysis of Tequatrovirus ufvareg1, a Tequatrovirus species inhibiting Escherichia coli O157: H7. Front. Cell. Infect. Microbiol. 2023, 13, 560. [Google Scholar] [CrossRef]
- Zhu, W.; Ding, Y.; Huang, C.; Wang, J.; Wang, J.; Wang, X. Genomic characterization of a novel bacteriophage STP55 revealed its prominent capacity in disrupting the dual-species biofilm formed by Salmonella Typhimurium and Escherichia coli O157: H7 strains. Arch. Microbiol. 2022, 204, 597. [Google Scholar] [CrossRef] [PubMed]
- Osman, A.-H.; Kotey, F.C.; Odoom, A.; Darkwah, S.; Yeboah, R.K.; Dayie, N.T.; Donkor, E.S. The Potential of Bacteriophage-Antibiotic Combination Therapy in Treating Infections with Multidrug-Resistant Bacteria. Antibiotics 2023, 12, 1329. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Huang, H.-H.; Ma, L.Z.; Masuda, Y.; Honjoh, K.-i.; Miyamoto, T. Inactivation of mixed Escherichia coli O157: H7 biofilms on lettuce by bacteriophage in combination with slightly acidic hypochlorous water (SAHW) and mild heat treatment. Food Microbiol. 2022, 104, 104010. [Google Scholar] [CrossRef]
- Górski, A.; Międzybrodzki, R.; Węgrzyn, G.; Jończyk-Matysiak, E.; Borysowski, J.; Weber-Dąbrowska, B. Phage therapy: Current status and perspectives. Med. Res. Rev. 2020, 40, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Rohde, C.; Resch, G.; Pirnay, J.-P.; Blasdel, B.G.; Debarbieux, L.; Gelman, D.; Górski, A.; Hazan, R.; Huys, I.; Kakabadze, E. Expert opinion on three phage therapy related topics: Bacterial phage resistance, phage training and prophages in bacterial production strains. Viruses 2018, 10, 178. [Google Scholar] [CrossRef]
- Simmons, E.L.; Drescher, K.; Nadell, C.D.; Bucci, V. Phage mobility is a core determinant of phage–bacteria coexistence in biofilms. ISME J. 2018, 12, 531–543. [Google Scholar] [CrossRef] [PubMed]
- Rostøl, J.T.; Marraffini, L. (Ph) ighting phages: How bacteria resist their parasites. Cell Host Microbe 2019, 25, 184–194. [Google Scholar] [CrossRef]
- Tomat, D.; Quiberoni, A.; Mercanti, D.; Balagué, C. Hard surfaces decontamination of enteropathogenic and Shiga toxin-producing Escherichia coli using bacteriophages. Food Res. Int. 2014, 57, 123–129. [Google Scholar] [CrossRef]
- Fu, Q.; Li, S.; Wang, Z.; Shan, W.; Ma, J.; Cheng, Y.; Wang, H.; Yan, Y.; Sun, J. H-NS Mutation-Mediated CRISPR-Cas activation inhibits phage release and toxin production of Escherichia coli Stx2 phage lysogen. Front. Microbiol. 2017, 8, 652. [Google Scholar] [CrossRef]
- Ershova, A.; Rusinov, I.; Spirin, S.; Karyagina, A.; Alexeevski, A. Role of restriction-modification systems in prokaryotic evolution and ecology. Biochemistry 2015, 80, 1373–1386. [Google Scholar] [CrossRef] [PubMed]
- Lopatina, A.; Tal, N.; Sorek, R. Abortive infection: Bacterial suicide as an antiviral immune strategy. Annu. Rev. Vir. 2020, 7, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Orsi, R.H.; Luo, H.; Ge, C.; Zhang, G.; Baker, R.C.; Stevenson, A.; Wiedmann, M. Assessment and comparison of molecular subtyping and characterization methods for Salmonella. Front. Microbiol. 2019, 10, 1591. [Google Scholar] [CrossRef]
- Lee, C.; Choi, I.Y.; Park, D.H.; Park, M.-K. Isolation and characterization of a novel Escherichia coli O157: H7-specific phage as a biocontrol agent. J. Environ. Health Sci. Eng. 2020, 18, 189–199. [Google Scholar] [CrossRef]
- Liao, Y.-T.; Sun, X.; Quintela, I.A.; Bridges, D.F.; Liu, F.; Zhang, Y.; Salvador, A.; Wu, V.C. Discovery of Shiga toxin-producing Escherichia coli (STEC)-specific bacteriophages from non-fecal composts using genomic characterization. Front. Microbiol. 2019, 10, 627. [Google Scholar] [CrossRef] [PubMed]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef]
- Tanji, Y.; Shimada, T.; Fukudomi, H.; Miyanaga, K.; Nakai, Y.; Unno, H. Therapeutic use of phage cocktail for controlling Escherichia coli O157: H7 in gastrointestinal tract of mice. J. Biosci. Bioeng. 2005, 100, 280–287. [Google Scholar] [CrossRef]
- Sheng, H.; Knecht, H.J.; Kudva, I.T.; Hovde, C.J. Application of bacteriophages to control intestinal Escherichia coli O157: H7 levels in ruminants. Appl. Environ. Microbiol. 2006, 72, 5359–5366. [Google Scholar] [CrossRef]
- Stanford, K.; McAllister, T.; Niu, Y.; Stephens, T.; Mazzocco, A.; Waddell, T.; Johnson, R. Oral delivery systems for encapsulated bacteriophages targeted at Escherichia coli O157: H7 in feedlot cattle. J. Food Protect. 2010, 73, 1304–1312. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).