Structural Basis of the Immunological Cross-Reactivity between Kiwi and Birch Pollen
Abstract
:1. Introduction
2. Materials and Methods
3. Results
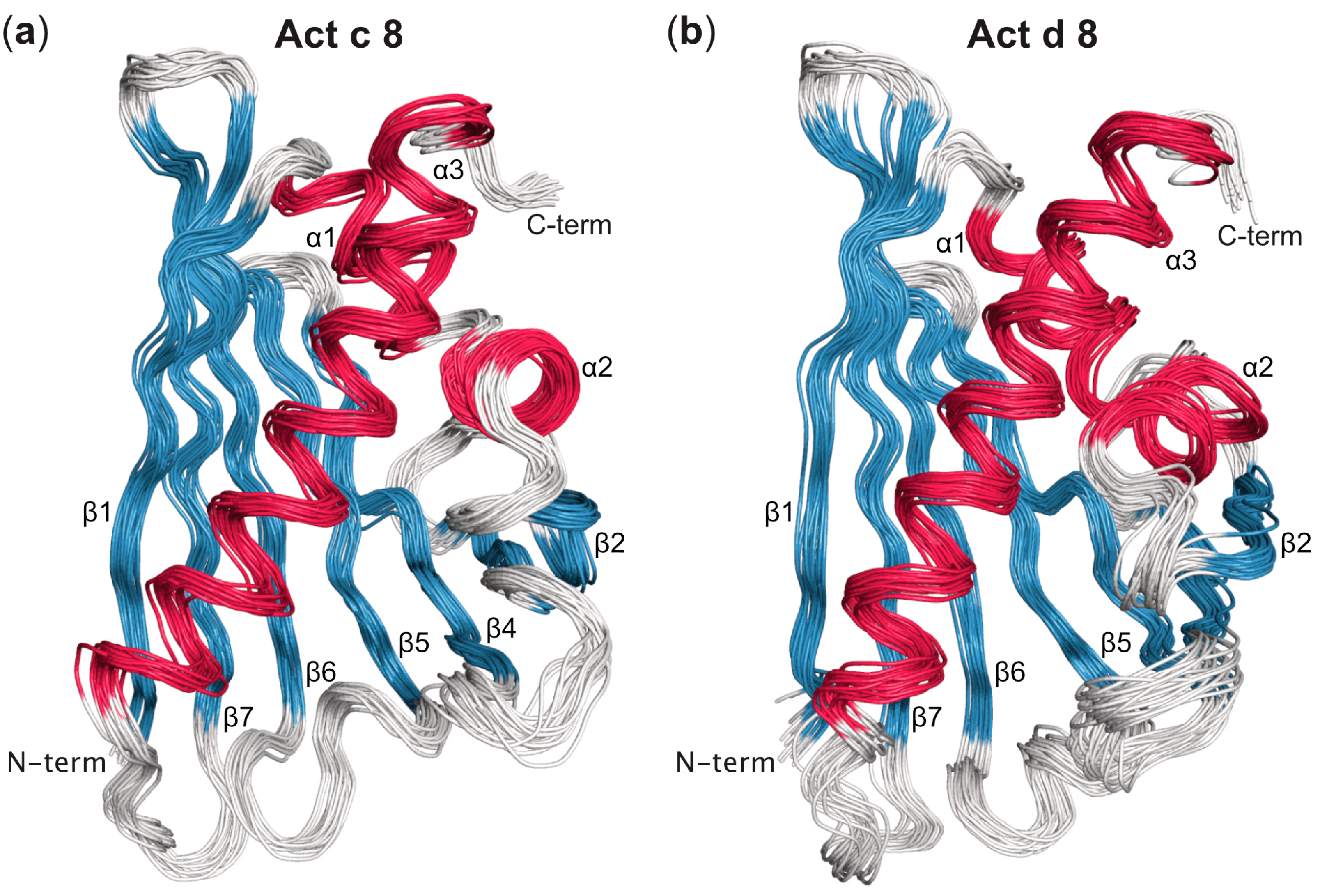
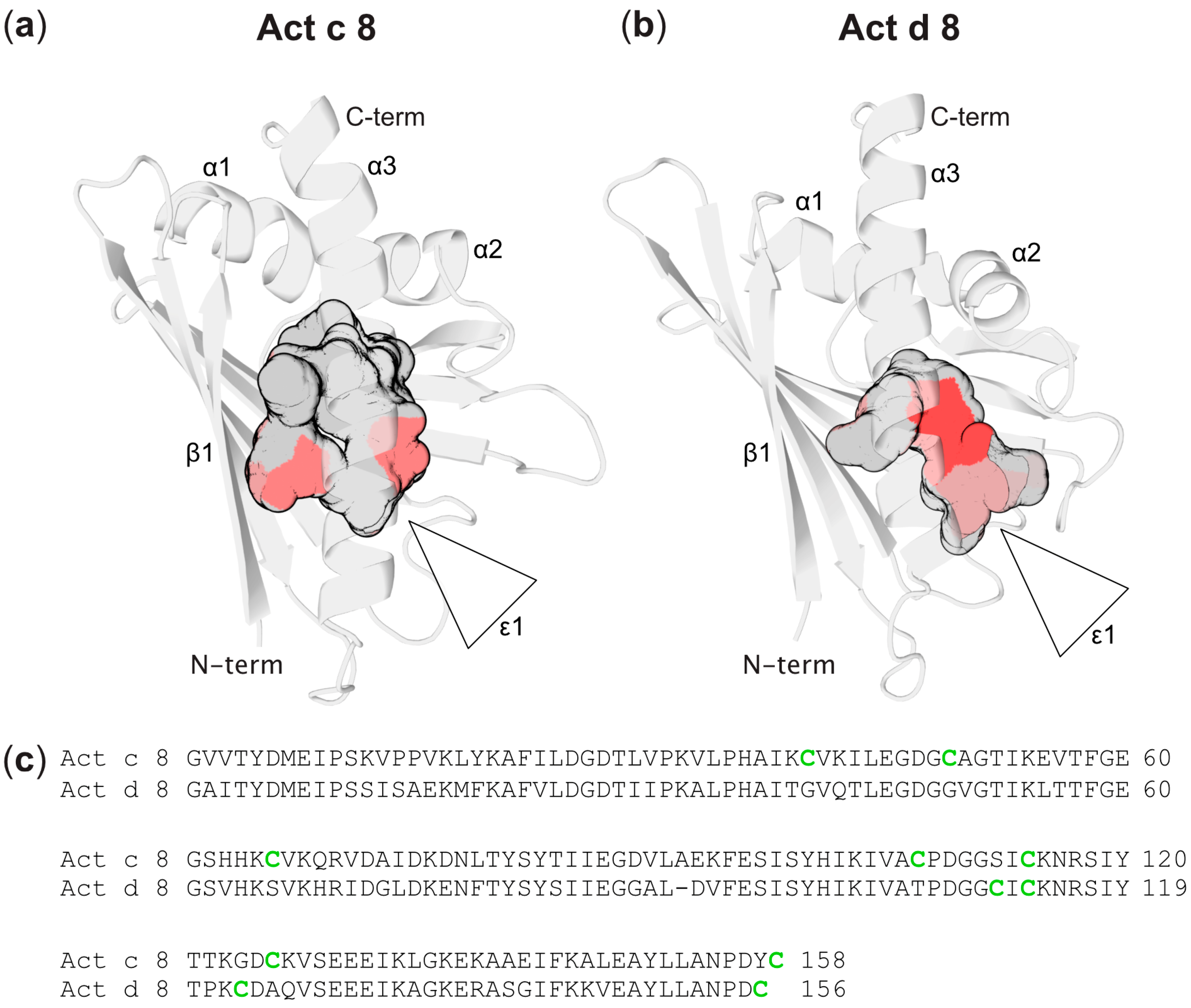
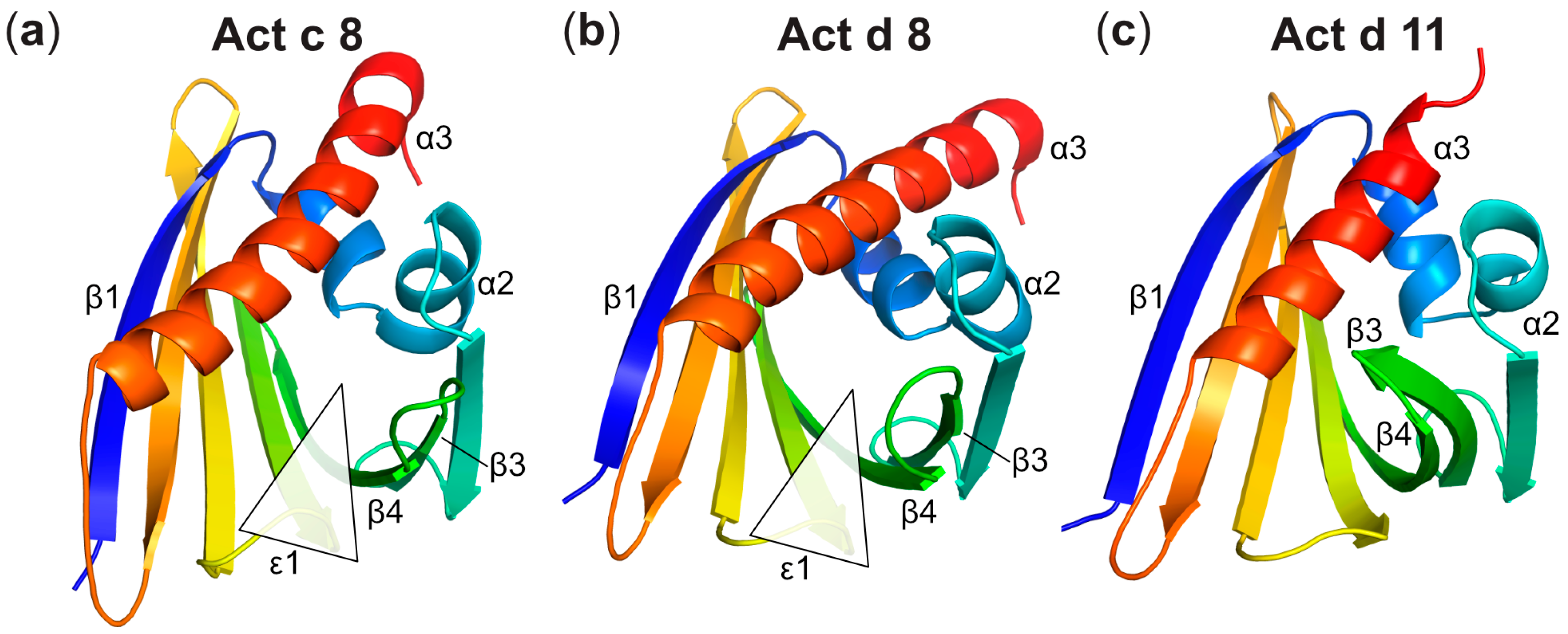
Three-Dimensional Structures of Act c 8 and Act d 8
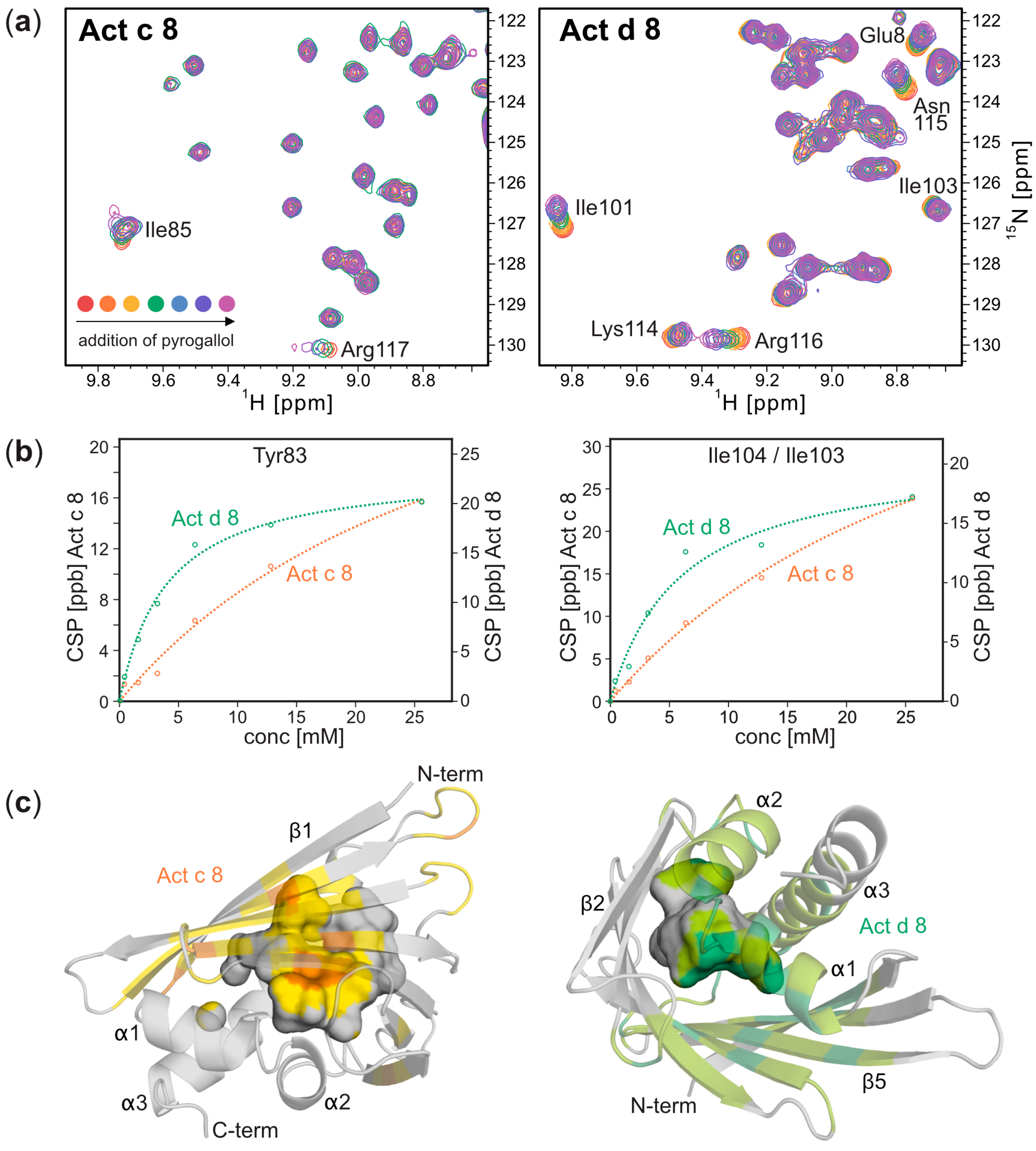
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Drummond, L. The composition and nutritional value of kiwifruit. Adv. Food Nutr. Res. 2013, 68, 33–57. [Google Scholar] [CrossRef] [PubMed]
- Ward, C.; Courtney, D. Kiwifruit: Taking its place in the global fruit bowl. Adv. Food Nutr. Res. 2013, 68, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Lucas, J.S.; Lewis, S.A.; Hourihane, J.O. Kiwi fruit allergy: A review. Pediatr. Allergy Immunol. 2003, 14, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Mempel, M.; Rakoski, J.; Ring, J.; Ollert, M. Severe anaphylaxis to kiwi fruit: Immunologic changes related to successful sublingual allergen immunotherapy. J. Allergy Clin. Immunol. 2003, 111, 1406–1409. [Google Scholar] [CrossRef] [PubMed]
- Radauer, C.; Nandy, A.; Ferreira, F.; Goodman, R.E.; Larsen, J.N.; Lidholm, J.; Pomes, A.; Raulf-Heimsoth, M.; Rozynek, P.; Thomas, W.R.; et al. Update of the WHO/IUIS Allergen Nomenclature Database based on analysis of allergen sequences. Allergy 2014, 69, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Bublin, M. Kiwifruit allergies. Adv. Food Nutr. Res. 2013, 68, 321–340. [Google Scholar] [CrossRef] [PubMed]
- Sirvent, S.; Canto, B.; Gomez, F.; Blanca, N.; Cuesta-Herranz, J.; Canto, G.; Blanca, M.; Rodriguez, R.; Villalba, M.; Palomares, O. Detailed characterization of Act d 12 and Act d 13 from kiwi seeds: Implication in IgE cross-reactivity with peanut and tree nuts. Allergy 2014, 69, 1481–1488. [Google Scholar] [CrossRef]
- Tuppo, L.; Giangrieco, I.; Palazzo, P.; Bernardi, M.L.; Scala, E.; Carratore, V.; Tamburrini, M.; Mari, A.; Ciardiello, M.A. Kiwellin, a modular protein from green and gold kiwi fruits: Evidence of in vivo and in vitro processing and IgE binding. J. Agric. Food Chem. 2008, 56, 3812–3817. [Google Scholar] [CrossRef]
- Bernardi, M.L.; Giangrieco, I.; Camardella, L.; Ferrara, R.; Palazzo, P.; Panico, M.R.; Crescenzo, R.; Carratore, V.; Zennaro, D.; Liso, M.; et al. Allergenic lipid transfer proteins from plant-derived foods do not immunologically and clinically behave homogeneously: The kiwifruit LTP as a model. PLoS ONE 2011, 6, e27856. [Google Scholar] [CrossRef]
- Le, T.M.; Bublin, M.; Breiteneder, H.; Fernandez-Rivas, M.; Asero, R.; Ballmer-Weber, B.; Barreales, L.; Bures, P.; Belohlavkova, S.; de Blay, F.; et al. Kiwifruit allergy across Europe: Clinical manifestation and IgE recognition patterns to kiwifruit allergens. J. Allergy Clin. Immunol. 2013, 131, 164–171. [Google Scholar] [CrossRef]
- Geroldinger-Simic, M.; Zelniker, T.; Aberer, W.; Ebner, C.; Egger, C.; Greiderer, A.; Prem, N.; Lidholm, J.; Ballmer-Weber, B.K.; Vieths, S.; et al. Birch pollen-related food allergy: Clinical aspects and the role of allergen-specific IgE and IgG4 antibodies. J. Allergy Clin. Immunol. 2011, 127, 616–622 e611. [Google Scholar] [CrossRef] [PubMed]
- Breiteneder, H.; Kraft, D. The history and science of the major birch pollen allergen Bet v 1. Biomolecules 2023, 13, 1151. [Google Scholar] [CrossRef] [PubMed]
- Oberhuber, C.; Bulley, S.M.; Ballmer-Weber, B.K.; Bublin, M.; Gaier, S.; DeWitt, A.M.; Briza, P.; Hofstetter, G.; Lidholm, J.; Vieths, S.; et al. Characterization of Bet v 1-related allergens from kiwifruit relevant for patients with combined kiwifruit and birch pollen allergy. Mol. Nutr. Food Res. 2008, 52 (Suppl. S2), S230–S240. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, H.; Michalska, K.; Sikorski, M.; Jaskolski, M. Structural and functional aspects of PR-10 proteins. FEBS J. 2013, 280, 1169–1199. [Google Scholar] [CrossRef] [PubMed]
- Camardella, L.; Carratore, V.; Ciardiello, M.A.; Servillo, L.; Balestrieri, C.; Giovane, A. Kiwi protein inhibitor of pectin methylesterase amino-acid sequence and structural importance of two disulfide bridges. Eur. J. Biochem. 2000, 267, 4561–4565. [Google Scholar] [CrossRef] [PubMed]
- Bublin, M.; Radauer, C.; Knulst, A.; Wagner, S.; Scheiner, O.; Mackie, A.R.; Mills, E.N.; Breiteneder, H. Effects of gastrointestinal digestion and heating on the allergenicity of the kiwi allergens Act d 1, actinidin, and Act d 2, a thaumatin-like protein. Mol. Nutr. Food Res. 2008, 52, 1130–1139. [Google Scholar] [CrossRef] [PubMed]
- Bonavita, A.; Carratore, V.; Ciardiello, M.A.; Giovane, A.; Servillo, L.; D’Avino, R. Influence of pH on the Structure and Function of Kiwi Pectin Methylesterase Inhibitor. J. Agric. Food Chem. 2016, 64, 5866–5876. [Google Scholar] [CrossRef]
- Poojary, M.M.; Hellwig, M.; Henle, T.; Lund, M.N. Covalent bonding between polyphenols and proteins: Synthesis of caffeic acid-cysteine and chlorogenic acid-cysteine adducts and their quantification in dairy beverages. Food Chem. 2023, 403, 134406. [Google Scholar] [CrossRef]
- Unterhauser, J.; Ahammer, L.; Rainer, T.; Eidelpes, R.; Fuhrer, S.; Nothegger, B.; Covaciu, C.E.; Cova, V.; Kamenik, A.S.; Liedl, K.R.; et al. Covalent polyphenol modification of a reactive cysteine in the major apple allergen Mal d 1. Food Chem. 2023, 410, 135374. [Google Scholar] [CrossRef]
- Ahammer, L.; Unterhauser, J.; Eidelpes, R.; Meisenbichler, C.; Nothegger, B.; Covaciu, C.E.; Cova, V.; Kamenik, A.S.; Liedl, K.R.; Breuker, K.; et al. Ascorbylation of a reactive cysteine in the major apple allergen Mal d 1. Foods 2022, 11, 2953. [Google Scholar] [CrossRef]
- Zeindl, R.; Tollinger, M. NMR resonance assignments of the PR-10 allergens Act c 8 and Act d 8 from golden and green kiwifruit. Biomol. NMR Assign. 2021, 15, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Bursal, E.; Gulcin, I. Polyphenol contents and in vitro antioxidant activities of lyophilised aqueous extract of kiwifruit (Actinidia deliciosa). Food Res. Int. 2011, 44, 1482–1489. [Google Scholar] [CrossRef]
- Führer, S.; Kamenik, A.S.; Zeindl, R.; Nothegger, B.; Hofer, F.; Reider, N.; Liedl, K.R.; Tollinger, M. Inverse relation between structural flexibility and IgE reactivity of Cor a 1 hazelnut allergens. Sci. Rep. 2021, 11, 4173. [Google Scholar] [CrossRef] [PubMed]
- Aglas, L.; Soh, W.T.; Kraiem, A.; Wenger, M.; Brandstetter, H.; Ferreira, F. Ligand binding of PR-10 proteins with a particular focus on the Bet v 1 allergen family. Curr. Allergy Asthma Rep. 2020, 20, 25. [Google Scholar] [CrossRef] [PubMed]
- Schanda, P.; Brutscher, B.; Konrat, R.; Tollinger, M. Folding of the KIX domain: Characterization of the equilibrium analog of a folding intermediate using 15N/13C relaxation dispersion and fast 1H/2H amide exchange NMR spectroscopy. J. Mol. Biol. 2008, 380, 726–741. [Google Scholar] [CrossRef] [PubMed]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Vranken, W.F.; Boucher, W.; Stevens, T.J.; Fogh, R.H.; Pajon, A.; Llinas, M.; Ulrich, E.L.; Markley, J.L.; Ionides, J.; Laue, E.D. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 2005, 59, 687–696. [Google Scholar] [CrossRef]
- Shen, Y.; Delaglio, F.; Cornilescu, G.; Bax, A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 2009, 44, 213–223. [Google Scholar] [CrossRef]
- Schwieters, C.D.; Kuszewski, J.J.; Tjandra, N.; Clore, G.M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003, 160, 65–73. [Google Scholar] [CrossRef]
- Case, D.A.; Belfon, K.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E., III; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Giambasu, G.; et al. AMBER 2020; University of California: San Francisco, CA, USA, 2020. [Google Scholar]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Wallnoefer, H.G.; Liedl, K.R.; Fox, T. A challenging system: Free energy prediction for factor Xa. J. Comput. Chem. 2011, 32, 1743–1752. [Google Scholar] [CrossRef] [PubMed]
- Adelman, S.A.; Doll, J.D. Generalized Langevin equation Approach for atom-solid-surface scattering—General formulation for classical scattering off harmonic solids. J. Chem. Phys. 1976, 64, 2375–2388. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; Vangunsteren, W.F.; Dinola, A.; Haak, J.R. Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Miyamoto, S.; Kollman, P.A. Settle—An analytical version of the shake and rattle algorithm for rigid water models. J. Comput. Chem. 1992, 13, 952–962. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald—An N.Log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Tejero, R.; Montelione, G.T. Evaluating protein structures determined by structural genomics consortia. Proteins 2007, 66, 778–795. [Google Scholar] [CrossRef] [PubMed]
- Schrodinger. The PyMOL Molecular Graphics System, Version 2.5.4; Schrodinger: New York, NY, USA, 2022. [Google Scholar]
- Grutsch, S.; Fuchs, J.E.; Freier, R.; Kofler, S.; Bibi, M.; Asam, C.; Wallner, M.; Ferreira, F.; Brandstetter, H.; Liedl, K.R.; et al. Ligand binding modulates the structural dynamics and compactness of the major birch pollen allergen. Biophys. J. 2014, 107, 2972–2981. [Google Scholar] [CrossRef]
- Zeindl, R.; Unterhauser, J.; Röck, M.; Eidelpes, R.; Führer, S.; Tollinger, M. Structural characterization of food allergens by nuclear magnetic resonance spectroscopy. Methods Mol. Biol. 2024, 2717, 159–173. [Google Scholar] [CrossRef]
- Ahammer, L.; Grutsch, S.; Kamenik, A.S.; Liedl, K.R.; Tollinger, M. Structure of the major apple allergen Mal d 1. J. Agric. Food Chem. 2017, 65, 1606–1612. [Google Scholar] [CrossRef]
- Eidelpes, R.; Hofer, F.; Rock, M.; Führer, S.; Kamenik, A.S.; Liedl, K.R.; Tollinger, M. Structure and zeatin binding of the peach allergen Pru p 1. J. Agric. Food Chem. 2021, 69, 8120–8129. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Rajarathnam, K. 13C NMR chemical shifts can predict disulfide bond formation. J. Biomol. NMR 2000, 18, 165–171. [Google Scholar] [CrossRef]
- Chruszcz, M.; Ciardiello, M.A.; Osinski, T.; Majorek, K.A.; Giangrieco, I.; Font, J.; Breiteneder, H.; Thalassinos, K.; Minor, W. Structural and bioinformatic analysis of the kiwifruit allergen Act d 11, a member of the family of ripening-related proteins. Mol. Immunol. 2013, 56, 794–803. [Google Scholar] [CrossRef] [PubMed]
- D’Avino, R.; Bernardi, M.L.; Wallner, M.; Palazzo, P.; Camardella, L.; Tuppo, L.; Alessandri, C.; Breiteneder, H.; Ferreira, F.; Ciardiello, M.A.; et al. Kiwifruit Act d 11 is the first member of the ripening-related protein family identified as an allergen. Allergy 2011, 66, 870–877. [Google Scholar] [CrossRef] [PubMed]
- Führer, S.; Unterhauser, J.; Zeindl, R.; Eidelpes, R.; Fernandez-Quintero, M.L.; Liedl, K.R.; Tollinger, M. The structural flexibility of PR-10 food allergens. Int. J. Mol. Sci. 2022, 23, 8252. [Google Scholar] [CrossRef]
- Miseta, A.; Csutora, P. Relationship between the occurrence of cysteine in proteins and the complexity of organisms. Mol. Biol. Evol. 2000, 17, 1232–1239. [Google Scholar] [CrossRef] [PubMed]
- Baker, E.N. Structure of actinidin, after refinement at 1.7 A resolution. J. Mol. Biol. 1980, 141, 441–484. [Google Scholar] [CrossRef]
- Hamiaux, C.; Maddumage, R.; Middleditch, M.J.; Prakash, R.; Brummell, D.A.; Baker, E.N.; Atkinson, R.G. Crystal structure of kiwellin, a major cell-wall protein from kiwifruit. J. Struct. Biol. 2014, 187, 276–281. [Google Scholar] [CrossRef]
- Offermann, L.R.; Giangrieco, I.; Perdue, M.L.; Zuzzi, S.; Santoro, M.; Tamburrini, M.; Cosgrove, D.J.; Mari, A.; Ciardiello, M.A.; Chruszcz, M. Elusive structural, functional, and immunological features of Act d 5, the green kiwifruit kiwellin. J. Agric. Food Chem. 2015, 63, 6567–6576. [Google Scholar] [CrossRef]
- Di Matteo, A.; Giovane, A.; Raiola, A.; Camardella, L.; Bonivento, D.; De Lorenzo, G.; Cervone, F.; Bellincampi, D.; Tsernoglou, D. Structural basis for the interaction between pectin methylesterase and a specific inhibitor protein. Plant Cell 2005, 17, 849–858. [Google Scholar] [CrossRef]
- O’Malley, A.; Pote, S.; Giangrieco, I.; Tuppo, L.; Gawlicka-Chruszcz, A.; Kowal, K.; Ciardiello, M.A.; Chruszcz, M. Structural characterization of Act c 10.0101 and Pun g 1.0101-allergens from the non-specific lipid transfer protein family. Molecules 2021, 26, 256. [Google Scholar] [CrossRef]
- Wang, L.; Xu, Z.; Yin, W.; Xu, K.; Wang, S.; Shang, Q.; Sa, W.; Liang, J.; Wang, L. Genome-wide analysis of the thaumatin-like gene family in Qingke (Hordeum vulgare L. var. nudum) uncovers candidates involved in plant defense against biotic and abiotic stresses. Front. Plant Sci. 2022, 13, 912296. [Google Scholar] [CrossRef]
- Grutsch, S.; Fuchs, J.E.; Ahammer, L.; Kamenik, A.S.; Liedl, K.R.; Tollinger, M. Conformational flexibility differentiates naturally occurring Bet v 1 isoforms. Int. J. Mol. Sci. 2017, 18, 1192. [Google Scholar] [CrossRef]
- Moschen, T.; Grutsch, S.; Juen, M.A.; Wunderlich, C.H.; Kreutz, C.; Tollinger, M. Measurement of ligand-target residence times by 1H relaxation dispersion NMR spectroscopy. J. Med. Chem. 2016, 59, 10788–10793. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeindl, R.; Franzmann, A.L.; Fernández-Quintero, M.L.; Seidler, C.A.; Hoerschinger, V.J.; Liedl, K.R.; Tollinger, M. Structural Basis of the Immunological Cross-Reactivity between Kiwi and Birch Pollen. Foods 2023, 12, 3939. https://doi.org/10.3390/foods12213939
Zeindl R, Franzmann AL, Fernández-Quintero ML, Seidler CA, Hoerschinger VJ, Liedl KR, Tollinger M. Structural Basis of the Immunological Cross-Reactivity between Kiwi and Birch Pollen. Foods. 2023; 12(21):3939. https://doi.org/10.3390/foods12213939
Chicago/Turabian StyleZeindl, Ricarda, Annika L. Franzmann, Monica L. Fernández-Quintero, Clarissa A. Seidler, Valentin J. Hoerschinger, Klaus R. Liedl, and Martin Tollinger. 2023. "Structural Basis of the Immunological Cross-Reactivity between Kiwi and Birch Pollen" Foods 12, no. 21: 3939. https://doi.org/10.3390/foods12213939
APA StyleZeindl, R., Franzmann, A. L., Fernández-Quintero, M. L., Seidler, C. A., Hoerschinger, V. J., Liedl, K. R., & Tollinger, M. (2023). Structural Basis of the Immunological Cross-Reactivity between Kiwi and Birch Pollen. Foods, 12(21), 3939. https://doi.org/10.3390/foods12213939





