Effects of ESA_00986 Gene on Adhesion/Invasion and Virulence of Cronobacter sakazakii and Its Molecular Mechanism
Abstract
1. Introduction
2. Experimental Materials and Procedures
2.1. Strains and Plasmids
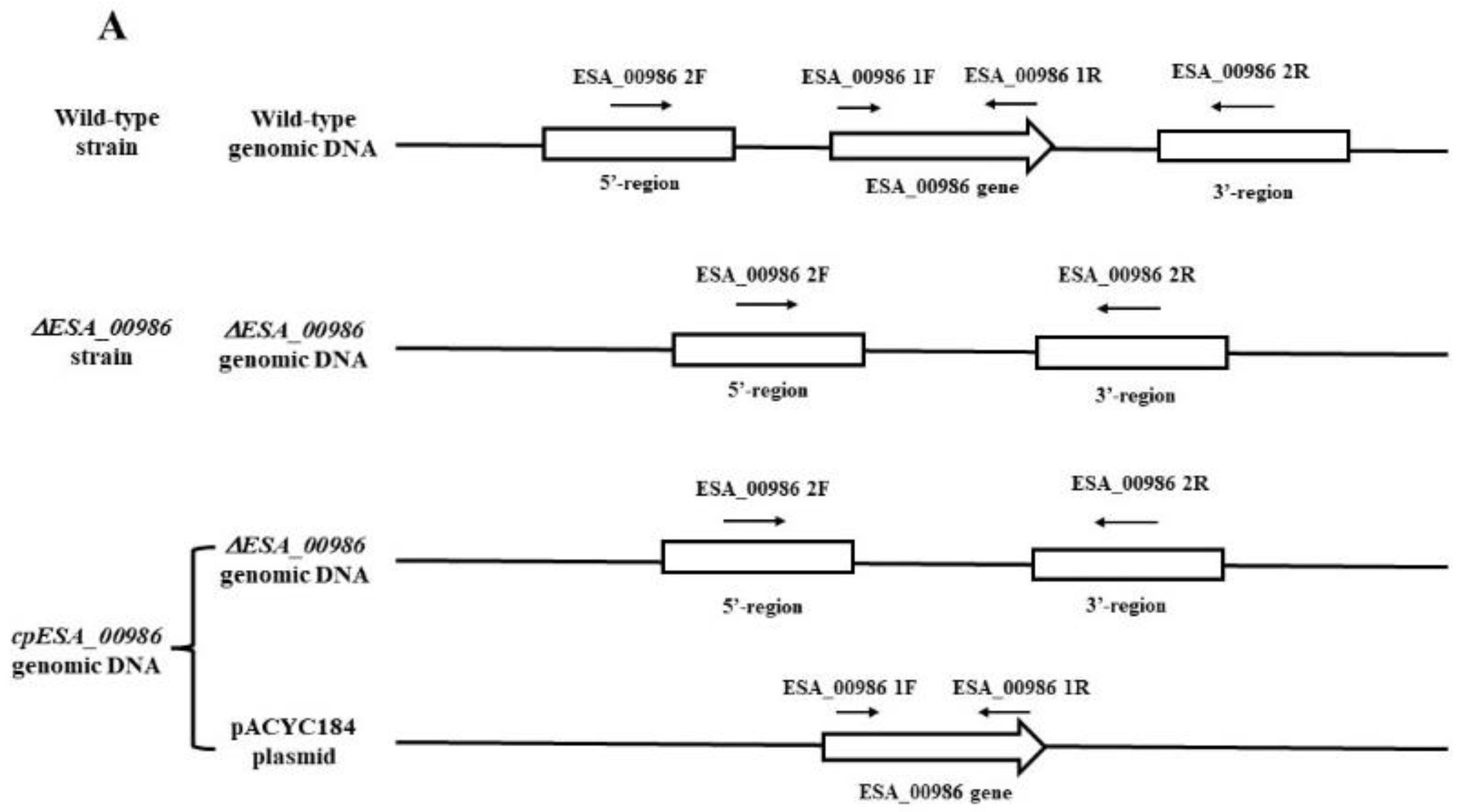
2.2. Construction of ESA_00986 Mutant
2.3. Complementation
2.4. Growth Curve Analysis
2.5. Motility Analysis
2.6. Hydrophobicity Analysis
2.7. Outer Membrane Permeability Analysis
2.8. Biofilm Formation Ability Analysis
2.9. Adhesion/Invasion Capacity Analysis
2.10. Rat Virulence Assay
2.11. Quantification of Bacteria in Tissues
2.12. Serum Inflammatory Levels Analysis
2.13. Gene Expression Analysis by qRT-PCR
2.14. Histopathological Analysis
2.15. Statistical Analysis
3. Results
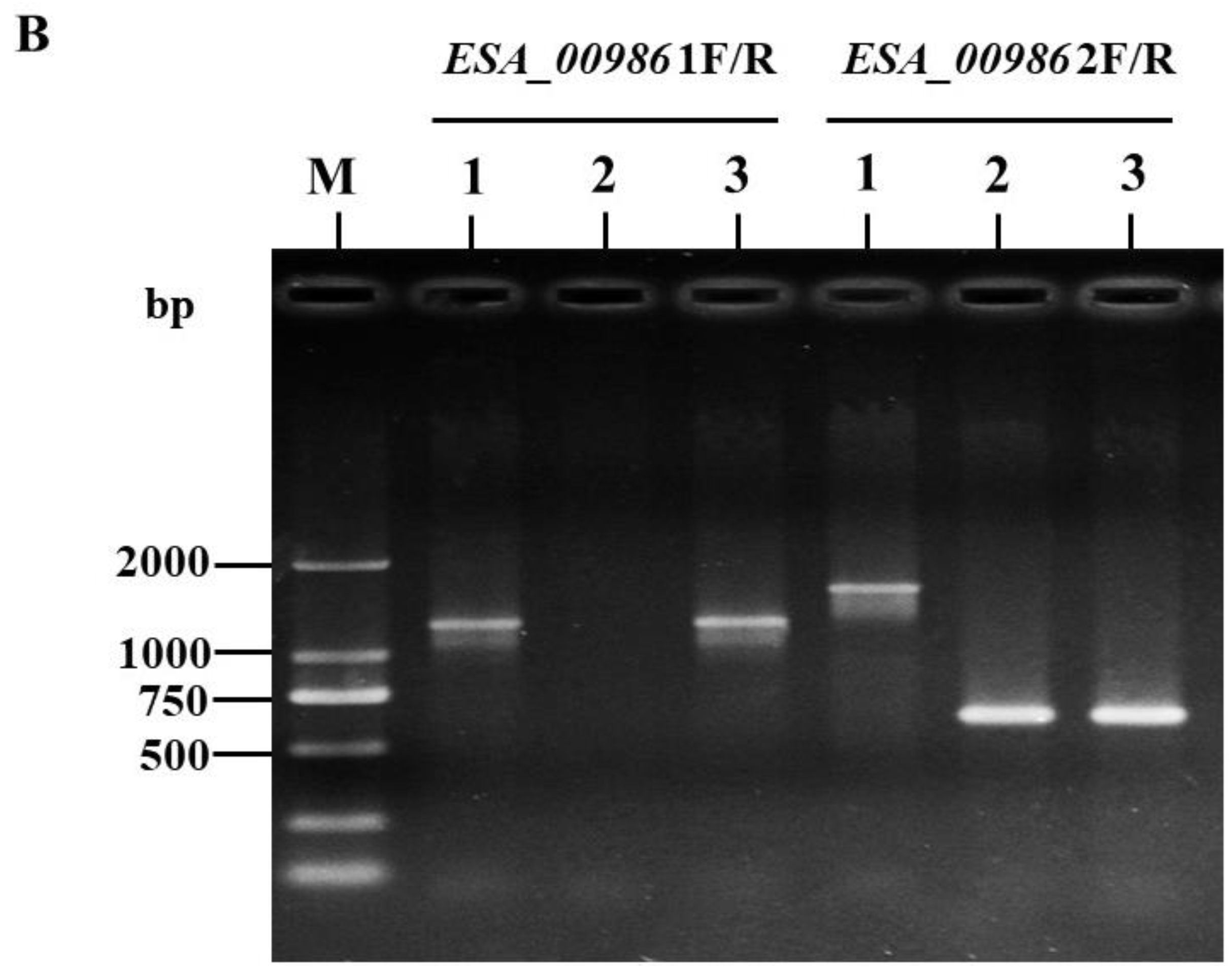
3.1. Construction and Validation of ΔESA_00986 and Complementary Strains
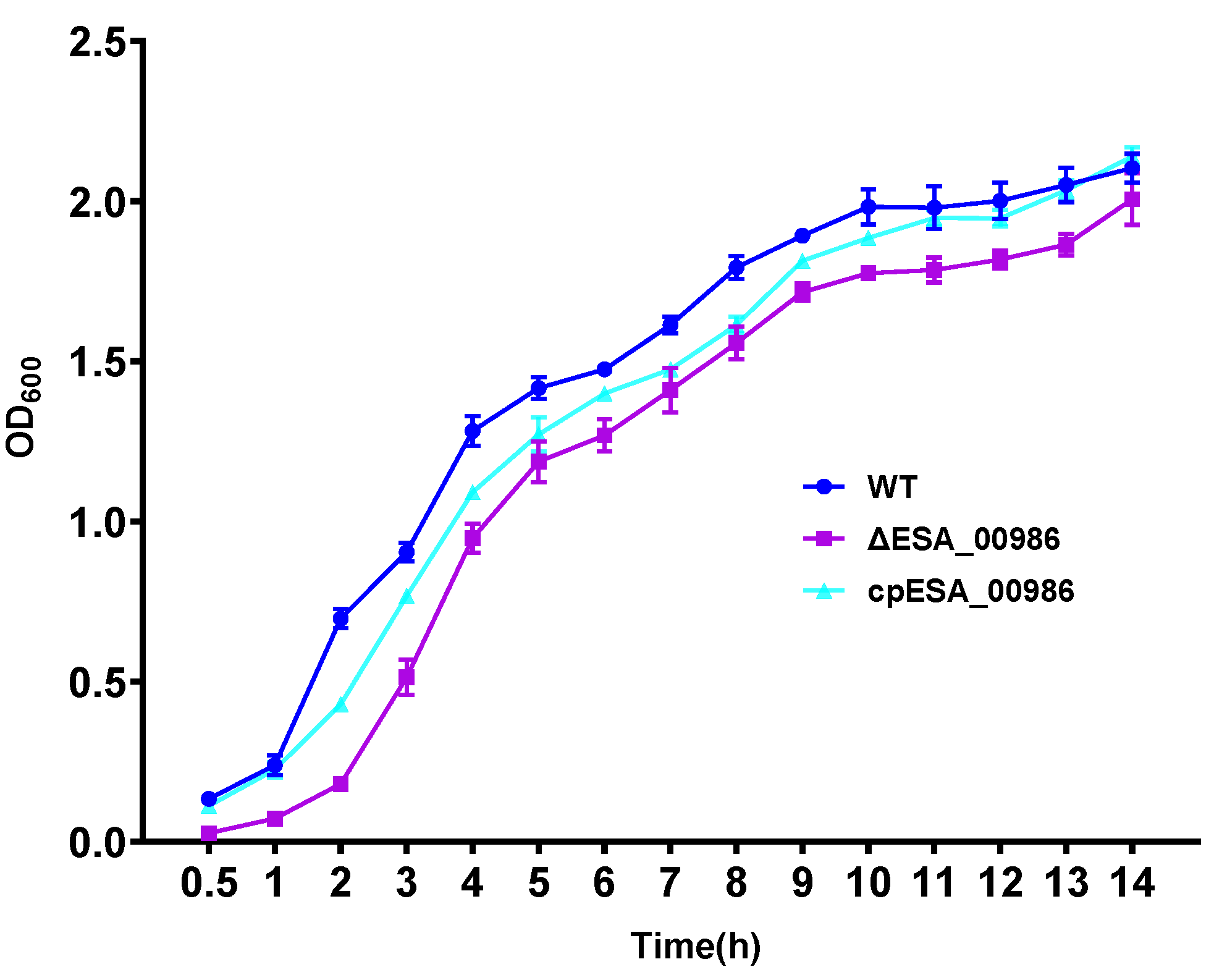
3.2. Effects of ESA_00986 Gene on Growth
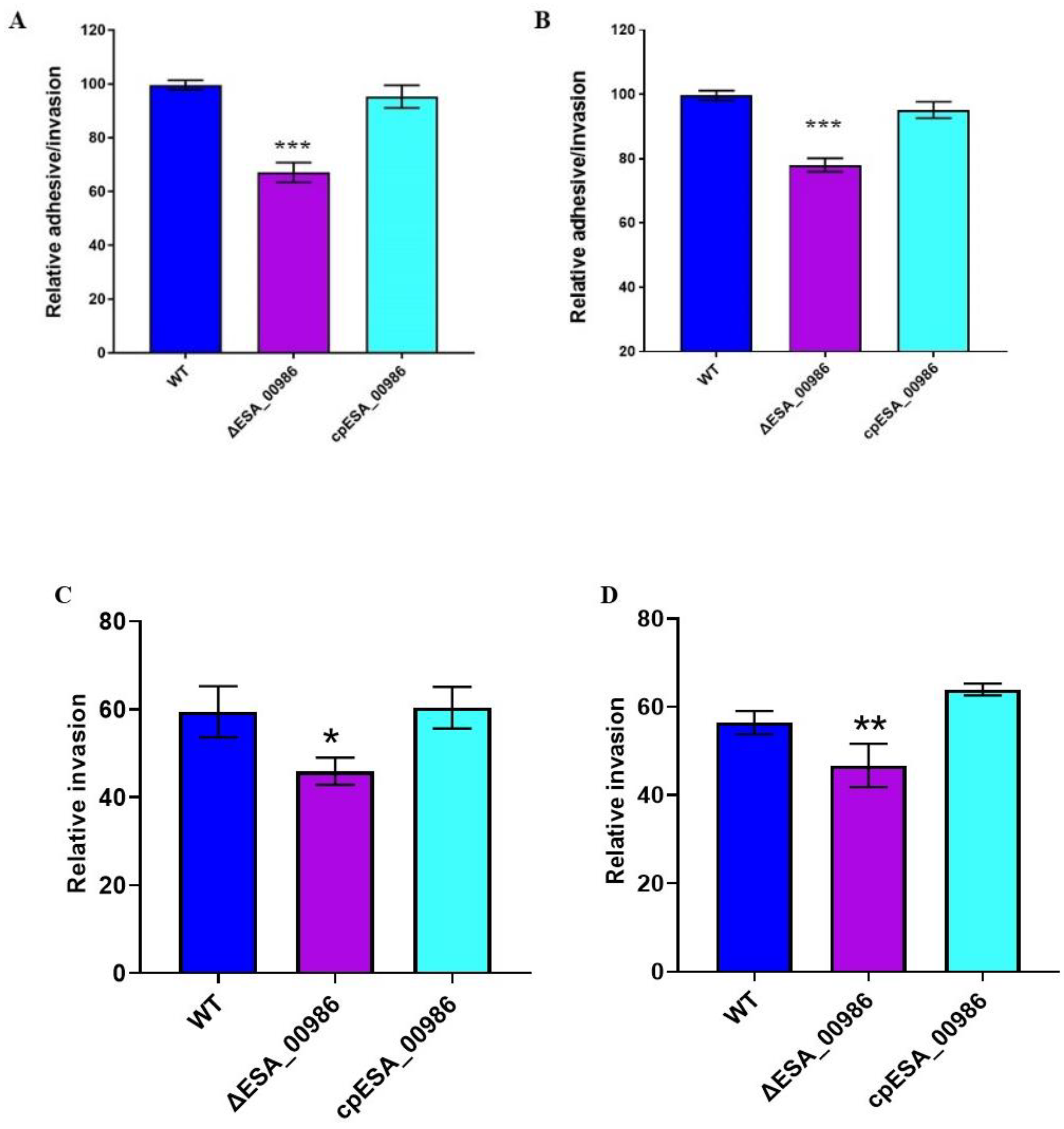
3.3. Effects of ESA_00986 Gene on Cell Adhesion/Invasion
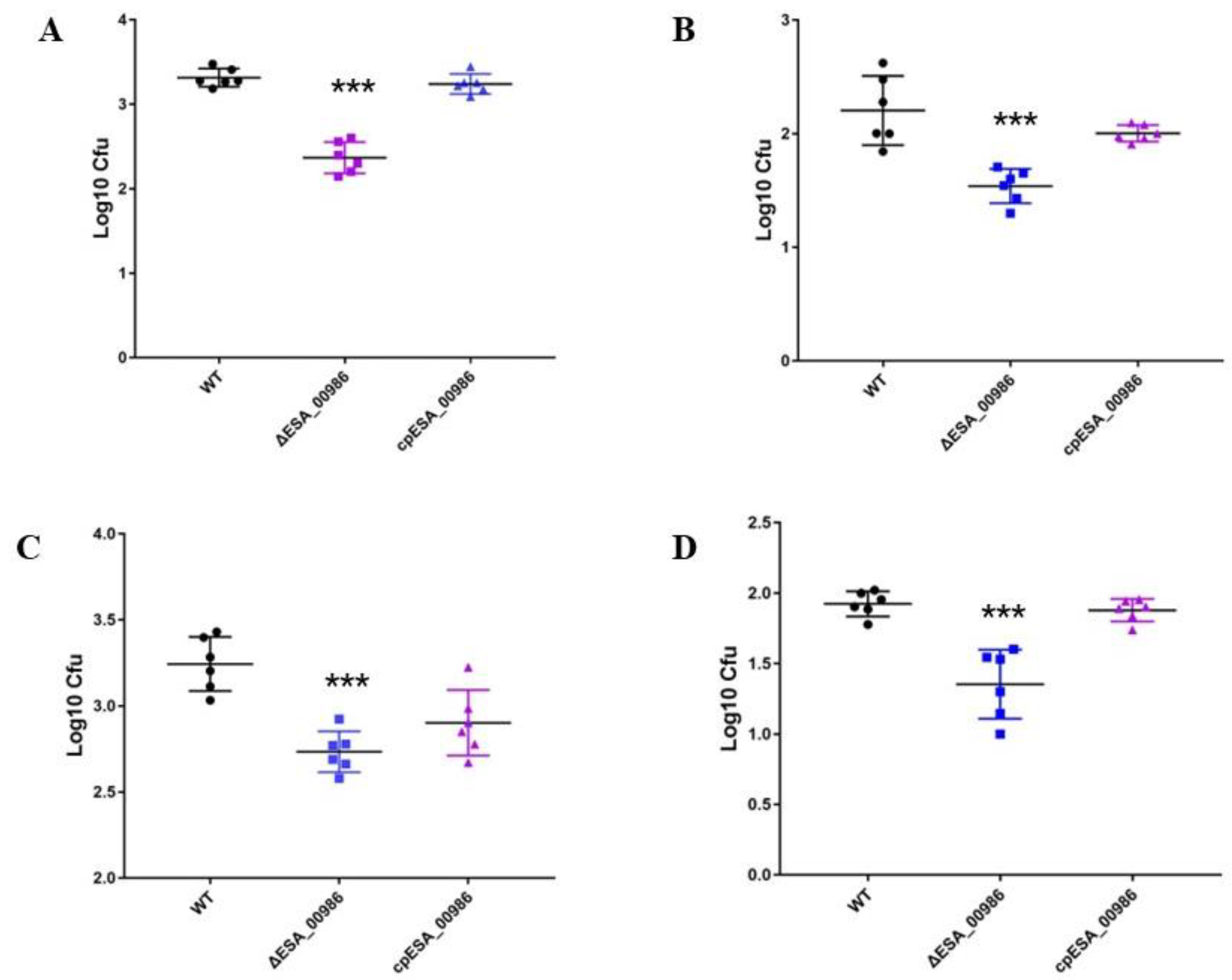
3.4. Effects of ESA_00986 Gene on Virulence of C. sakazakii In Vivo
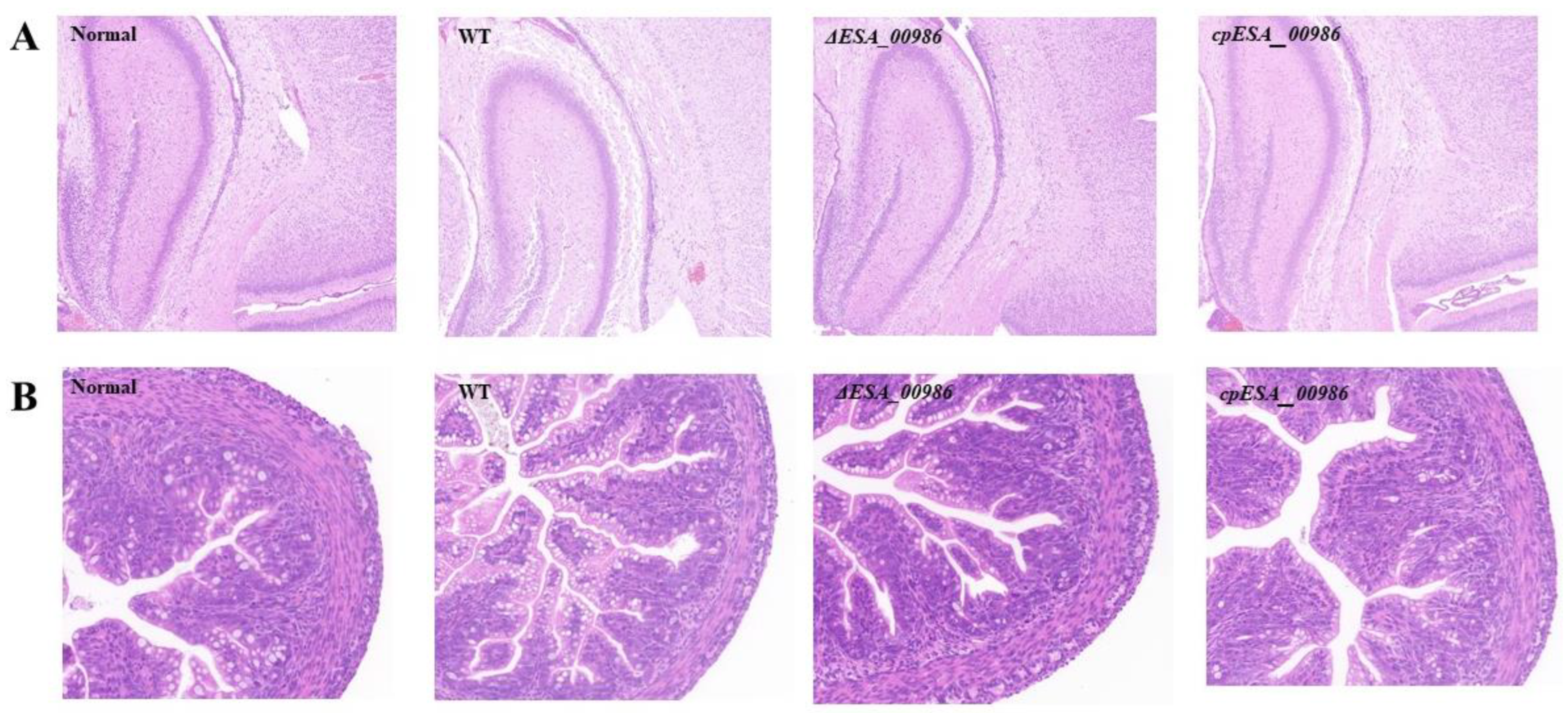
3.5. Pathological Analysis
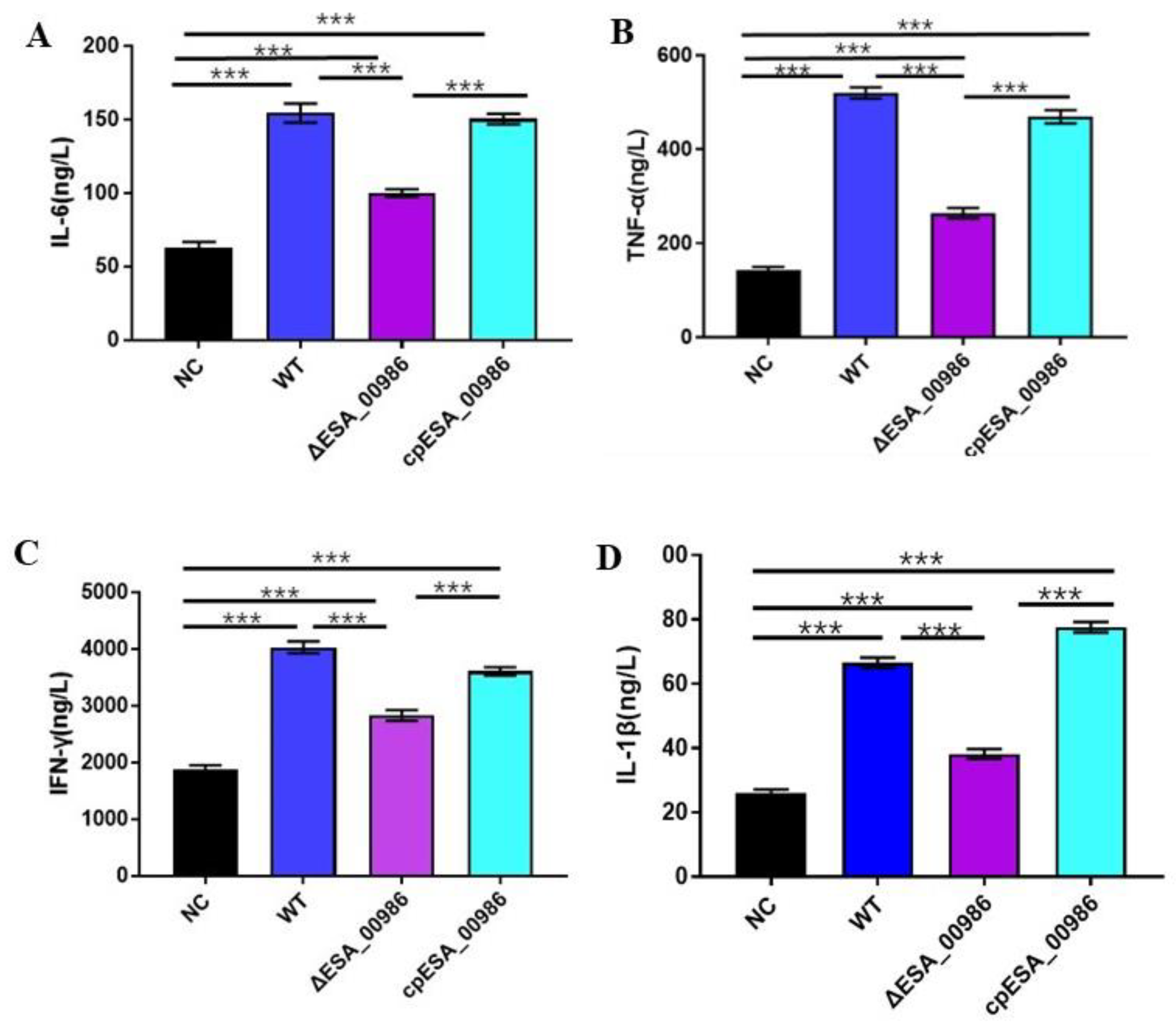
3.6. Effect of ESA_00986 on the Serum Inflammatory Cytokines
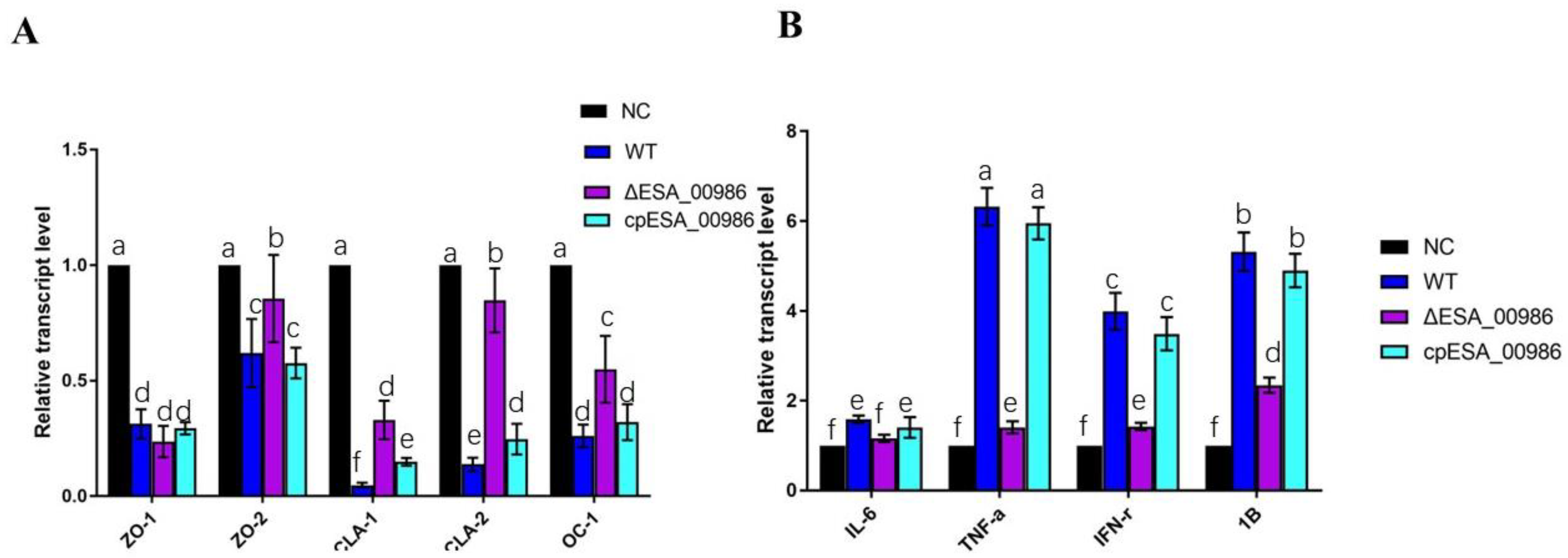
3.7. Effect of ESA_00986 on the Expression of Genes Involved in Inflammation and Intestinal Integrity
3.8. Effects of ESA_00986 Gene on the Motility of C. sakazakii
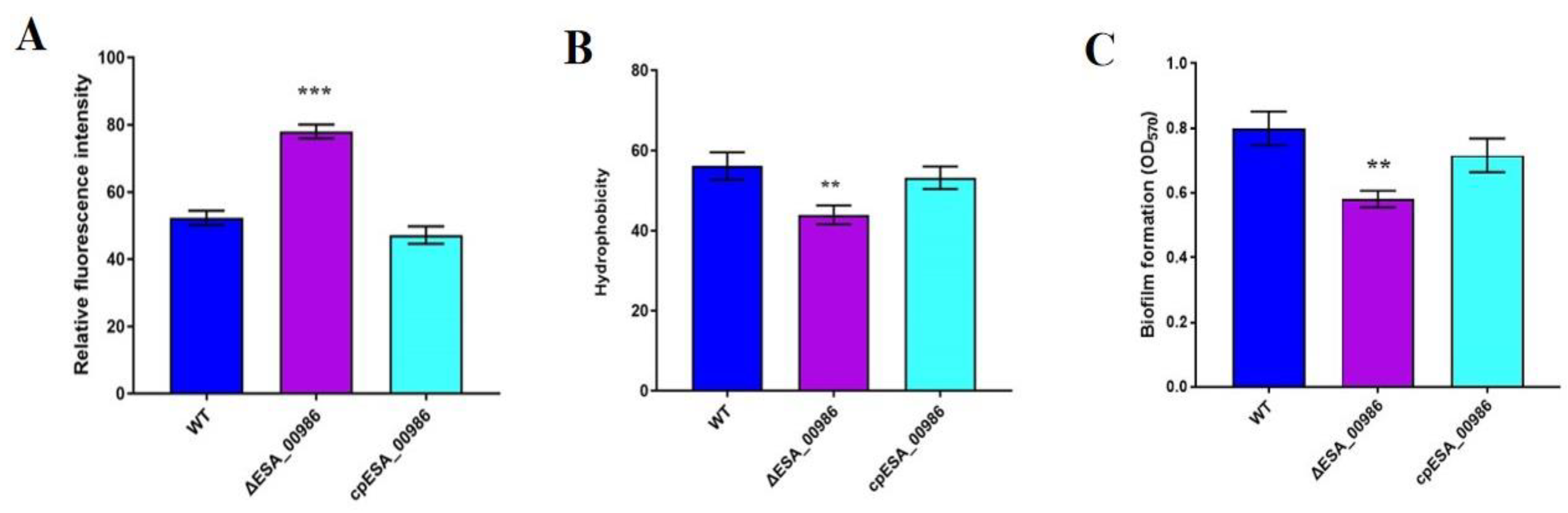
3.9. Effects of ESA_00986 Gene on Physiological Properties of Bacterial Surface
3.10. Effects of ESA_00986 Gene on Biofilm Synthesis
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Feeney, A.; Kropp, K.A.; O’connor, R.; Sleator, R.D. Cronobacter sakazakii: Stress survival and virulence potential in an opportunistic foodborne pathogen. Gut Microbes 2014, 5, 711–718. [Google Scholar] [CrossRef] [PubMed]
- Iversen, C.; Lane, M.; Forsythe, S. The growth profile, thermotolerance and biofilm formation of Enterobacter sakazakii grown in infant formula milk. Lett. Appl. Microbiol. 2004, 38, 378–382. [Google Scholar] [CrossRef] [PubMed]
- Kucerova, E.; Clifton, S.W.; Xia, X.-Q.; Long, F.; Porwollik, S.; Fulton, L.; Fronick, C.; Minx, P.; Kyung, K.; Warren, W.; et al. Genome sequence of Cronobacter sakazakii BAA-894 and comparative genomic hybridization analysis with other Cronobacter species. PLoS ONE 2010, 5, e9556. [Google Scholar] [CrossRef]
- Jing, C.E.; Du, X.J.; Li, P.; Wang, S. Transcriptome analysis of Cronobacter sakazakii ATCC BAA-894 after interaction with human intestinal epithelial cell line HCT-8. Appl. Microbiol. Biotechnol. 2016, 100, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Fakruddin; Rahaman, M.; Ahmed, M.M.; Hoque, M. Stress tolerant virulent strains of Cronobacter sakazakii from food. Biol. Res. 2014, 47, 63. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Condell, O.; Power, K.; Butler, F.; Tall, B.; Fanning, S. Cronobacter species (formerly known as Enterobacter sakazakii) in powdered infant formula: A review of our current understanding of the biology of this bacterium. J. Appl. Microbiol. 2012, 113, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, G.G.; Calarga, A.P.; Teodoro, J.R.; Queiroz, M.M.; Astudillo-Trujillo, C.A.; Levy, C.E.; Brocchi, M.; Kabuki, D.Y. Isolation, comparison of identification methods and antibiotic resistance of Cronobacter spp. in infant foods. Food Res. Int. 2020, 137, 109643. [Google Scholar] [CrossRef]
- Yan, Q.; Wang, J.; Gangiredla, J.; Cao, Y.; Martins, M.; Gopinath, G.R.; Stephan, R.; Lampel, K.; Tall, B.D.; Fanning, S. Comparative Genotypic and Phenotypic Analysis of Cronobacter species Cultured from Four Powdered Infant Formula Production Facilities: Indication of Pathoadaptation along the Food Chain. Appl. Environ. Microbiol. 2015, 81, 4388–4402. [Google Scholar] [CrossRef]
- Jaradat, Z.W.; Al Mousa, W.; Elbetieha, A.; Al Nabulsi, A.; Tall, B.D. Cronobacter spp.—Opportunistic food-borne pathogens. A review of their virulence and environmental-adaptive traits. J. Med. Microbiol. 2014, 63 Pt 8, 1023–1037. [Google Scholar] [CrossRef]
- Giri, C.P.; Shima, K.; Tall, B.D.; Curtis, S.; Sathyamoorthy, V.; Hanisch, B.; Kim, K.S.; Kopecko, D.J. Cronobacter spp. (previously Enterobacter sakazakii) invade and translocate across both cultured human intestinal epithelial cells and human brain microvascular endothelial cells. Microb. Pathog. 2012, 52, 140–147. [Google Scholar] [CrossRef]
- Franco, A.A.; Hu, L.; Grim, C.J.; Gopinath, G.; Sathyamoorthy, V.; Jarvis, K.G.; Lee, C.; Sadowski, J.; Kim, J.; Kothary, M.H.; et al. Characterization of putative virulence genes on the related RepFIB plasmids harbored by Cronobacter spp. Appl. Environ. Microbiol. 2011, 77, 3255–3267. [Google Scholar] [CrossRef]
- Hartmann, I.; Carranza, P.; Lehner, A.; Stephan, R.; Eberl, L.; Riedel, K. Genes involved in Cronobacter sakazakii biofilm formation. Appl. Environ. Microbiol. 2010, 76, 2251–2261. [Google Scholar] [CrossRef] [PubMed]
- Townsend, S.; Barron, J.C.; Loc-Carrillo, C.; Forsythe, S. The presence of endotoxin in powdered infant formula milk and the influence of endotoxin and Enterobacter sakazakii on bacterial translocation in the infant rat. Food Microbiol. 2007, 24, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Han, Z.; Li, P.; Zhang, H.; Du, X.; Wang, S. Outer Membrane Protein F Is Involved in Biofilm Formation, Virulence and Antibiotic Resistance in Cronobacter sakazakii. Microorganisms 2021, 9, 2338. [Google Scholar] [CrossRef] [PubMed]
- Nair, M.K.M.; Venkitanarayanan, K.; Silbart, L.K.; Kim, K.S.; Awadallah, M.A.; Ahmed, H.A.; Merwad, A.M.; Elez, R.M.A.; Saleh, K.M.; Li, Z.; et al. Outer membrane protein A (OmpA) of Cronobacter sakazakii binds fibronectin and contributes to invasion of human brain microvascular endothelial cells. Foodborne Pathog. Dis. 2009, 6, 495–501. [Google Scholar] [CrossRef]
- Singh, N.; Goel, G.; Raghav, M. Insights into virulence factors determining the pathogenicity of Cronobacter sakazakii. Virulence 2015, 6, 433–440. [Google Scholar] [CrossRef]
- Kim, K.; Kim, K.P.; Choi, J.; Lim, J.A.; Lee, J.; Hwang, S.; Ryu, S. Outer membrane proteins A (OmpA) and X (OmpX) are essential for basolateral invasion of Cronobacter sakazakii. Appl. Environ. Microbiol. 2010, 76, 5188–5198. [Google Scholar] [CrossRef]
- Chandrapala, D.; Kim, K.; Choi, Y.; Senevirathne, A.; Kang, D.-H.; Ryu, S.; Kim, K.-P. Putative Inv is essential for basolateral invasion of Caco-2 cells and acts synergistically with OmpA to affect in vitro and in vivo virulence of Cronobacter sakazakii ATCC 29544. Infect. Immun. 2014, 82, 1755–1765. [Google Scholar] [CrossRef]
- Li, P.; Zong, W.; Zhang, Z.; Lv, W.; Ji, X.; Zhu, D.; Du, X.; Wang, S. Effects and molecular mechanism of flagellar gene flgK on the motility, adhesion/invasion, and desiccation resistance of Cronobacter sakazakii. Food Res. Int. 2023, 164, 112418. [Google Scholar] [CrossRef]
- Choi, Y.; Kim, K.P.; Kim, K.; Choi, J.; Shin, H.; Kang, D.H.; Ryu, S. Possible roles of LysR-type transcriptional regulator (LTTR) homolog as a global regulator in Cronobacter sakazakii ATCC 29544. Int. J. Med. Microbiol. 2012, 302, 270–275. [Google Scholar] [CrossRef]
- Reis, R.S.; Horn, F. Enteropathogenic Escherichia coli, Samonella, Shigella and Yersinia: Cellular aspects of host-bacteria interactions in enteric diseases. Gut Pathog. 2010, 2, 8. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Frey, E.A.; Pfuetzner, R.A.; Creagh, A.L.; Knoechel, D.G.; Haynes, C.A.; Finlay, B.B.; Strynadka, N.C.J. Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex. Nature 2000, 405, 1073–1077. [Google Scholar] [CrossRef] [PubMed]
- Hamburger, Z.A.; Brown, M.S.; Isberg, R.R.; Bjorkman, P.J. Crystal structure of invasin: A bacterial integrin-binding protein. Science 1999, 286, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Jerse, A.E.; Yu, J.; Tall, B.; Kaper, J.B. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. USA 1990, 87, 7839–7843. [Google Scholar] [CrossRef]
- Isberg, R.R.; Leong, J.M. Multiple beta 1 chain integrins are receptors for invasin, a protein that promotes bacterial penetration into mammalian cells. Cell 1990, 60, 861–871. [Google Scholar] [CrossRef]
- Sadana, P.; Mönnich, M.; Unverzagt, C.; Scrima, A. Structure of the Y. pseudotuberculosis adhesin InvasinE. Protein Sci. 2017, 26, 1182–1195. [Google Scholar] [CrossRef]
- Bodelón, G.; Palomino, C.; Fernández, L. Immunoglobulin domains in Escherichia coli and other enterobacteria: From pathogenesis to applications in antibody technologies. FEMS Microbiol. Rev. 2013, 37, 204–250. [Google Scholar] [CrossRef]
- Seo, K.S.; Kim, J.W.; Park, J.Y.; Viall, A.K.; Rohde, H.N.; Schnider, D.R.; Lim, S.Y.; Hong, J.B.; Hinnebusch, B.J.; O’Loughlin, J.L.; et al. Role of a new intimin/invasin-like protein in Yersinia pestis virulence. Infect. Immun. 2012, 80, 3559–3569. [Google Scholar] [CrossRef]
- Clark, M.A.; Hirst, B.H.; Jepson, M.A. Inoculum composition and Salmonella pathogenicity island 1 regulate M-cell invasion and epithelial destruction by Salmonella typhimurium. Infect. Immun. 1998, 66, 724–731. [Google Scholar] [CrossRef]
- Ji, X.; Lu, P.; Xue, J.; Zhao, N.; Zhang, Y.; Dong, L.; Zhang, X.; Li, P.; Hu, Y.; Wang, J.; et al. The lipoprotein NlpD in Cronobacter sakazakii responds to acid stress and regulates macrophage resistance and virulence by maintaining membrane integrity. Virulence 2021, 12, 415–429. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, D.; Lian, S.; Gu, X.; Hou, Q.; Xia, P.; Zhu, G. Mechanism of nitrite transporter NirC in motility, biofilm formation, and adhesion of avian pathogenic Escherichia coli. Arch. Microbiol. 2021, 203, 4221–4231. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.H.; Kornberg, A. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2000, 97, 4885–4890. [Google Scholar] [CrossRef] [PubMed]
- Andriantsoanirina, V.; Teolis, A.-C.; Xin, L.X.; Butel, M.J.; Aires, J. Bifidobacterium longum and Bifidobacterium breve isolates from preterm and full term neonates: Comparison of cell surface properties. Anaerobe 2014, 28, 212–215. [Google Scholar] [CrossRef]
- Gao, J.X.; Li, P.; Du, X.J.; Han, Z.H.; Xue, R.; Liang, B.; Wang, S. A Negative Regulator of Cellulose Biosynthesis, bcsR, Affects Biofilm Formation, and Adhesion/Invasion Ability of Cronobacter sakazakii. Front. Microbiol. 2017, 8, 1839. [Google Scholar] [CrossRef] [PubMed]
- Ganan, M.; Campos, G.; Muñoz, R.; Carrascosa, A.; de Pascual-Teresa, S.; Martinez-Rodriguez, A. Effect of growth phase on the adherence to and invasion of Caco-2 epithelial cells by Campylobacter. Int. J. Food Microbiol. 2010, 140, 14–18. [Google Scholar] [CrossRef]
- Pizarro-Cerdá, J.; Cossart, P. Bacterial Adhesion and Entry into Host Cells. Cell 2006, 124, 715–727. [Google Scholar] [CrossRef]
- Ribet, D.; Cossart, P. How bacterial pathogens colonize their hosts and invade deeper tissues. Microbes Infect. 2015, 17, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Pilatti, L.; de Paiva, J.B.; Rojas, T.C.G.; Leite, J.L.; Conceição, R.A.; Nakazato, G.; da Silveira, W.D. The virulence factor ychO has a pleiotropic action in an Avian Pathogenic Escherichia coli (APEC) strain. BMC Microbiol. 2016, 16, 35. [Google Scholar] [CrossRef]
- Siljan, W.W.; Holter, J.C.; Nymo, S.H.; Husebye, E.; Ueland, T.; Aukrust, P.; Mollnes, T.E.; Heggelund, L. Cytokine responses, microbial aetiology and short-term outcome in community-acquired pneumonia. Eur. J. Clin. Investg. 2018, 48, e12865. [Google Scholar] [CrossRef]
- Esmon, C.T. The impact of the inflammatory response on coagulation. Thromb. Res. 2004, 114, 321–327. [Google Scholar] [CrossRef]
- Papasian, C.J.; Silverstein, R.; Gao, J.J.; Bamberger, D.M.; Morrison, D.C. Anomalous role of tumor necrosis factor alpha in experimental enterococcal infection. Infect. Immun. 2002, 70, 6628–6637. [Google Scholar] [CrossRef] [PubMed]
- Leendertse, M.; Heikens, E.; Wijnands, L.M.; van Luit-Asbroek, M.; Teske, G.J.D.; Roelofs, J.J.; Bonten, M.J.M.; van der Poll, T.; Willems, R.J.L. Enterococcal surface protein transiently aggravates Enterococcus faecium-induced urinary tract infection in mice. J. Infect. Dis. 2009, 200, 1162–1165. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Shankar, N. Surface protein Esp enhances pro-inflammatory cytokine expression through NF-κB activation during enterococcal infection. Innate Immun. 2016, 22, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Watkins, R.L.; Pallister, K.B.; Voyich, J.M. The SaeR/S gene regulatory system induces a pro-inflammatory cytokine response during Staphylococcus aureus infection. PLoS ONE 2011, 6, e19939. [Google Scholar] [CrossRef] [PubMed]
- Okumura, R.; Takeda, K. Roles of intestinal epithelial cells in the maintenance of gut homeostasis. Exp. Mol. Med. 2017, 49, e338. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.Y.; Yang, W.X.; Hu, Y.J. The role of epithelial tight junctions involved in pathogen infections. Mol. Biol. Rep. 2014, 41, 6591–6610. [Google Scholar] [CrossRef]
- Awad, W.A.; Hess, C.; Hess, M. Enteric Pathogens and Their Toxin-Induced Disruption of the Intestinal Barrier through Alteration of Tight Junctions in Chickens. Toxins 2017, 9, 60. [Google Scholar] [CrossRef]
- Zheng, M.; Sun, S.; Zhou, J.; Liu, M. Virulence factors impair epithelial junctions during bacterial infection. J. Clin. Lab. Anal. 2021, 35, e23627. [Google Scholar] [CrossRef]
- Hatayama, S.; Shimohata, T.; Amano, S.; Kido, J.; Nguyen, A.Q.; Sato, Y.; Kanda, Y.; Tentaku, A.; Fukushima, S.; Nakahashi, M.; et al. Cellular Tight Junctions Prevent Effective Campylobacter jejuni Invasion and Inflammatory Barrier Disruption Promoting Bacterial Invasion from Lateral Membrane in Polarized Intestinal Epithelial Cells. Front. Cell Infect. Microbiol. 2018, 8, 15. [Google Scholar] [CrossRef]
- Chen, M.L.; Ge, Z.; Fox, J.G.; Schauer, D.B. Disruption of tight junctions and induction of proinflammatory cytokine responses in colonic epithelial cells by Campylobacter jejuni. Infect. Immun. 2006, 74, 6581–6589. [Google Scholar] [CrossRef]
- Thanabalasuriar, A.; Koutsouris, A.; Weflen, A.; Mimee, M.; Hecht, G.; Gruenheid, S. The bacterial virulence factor NleA is required for the disruption of intestinal tight junctions by enteropathogenic Escherichia coli. Cell Microbiol. 2010, 12, 31–41. [Google Scholar] [CrossRef]
- Zhang, Y.-G.; Wu, S.; Xia, Y.; Sun, J. Salmonella infection upregulates the leaky protein claudin-2 in intestinal epithelial cells. PLoS ONE 2013, 8, e58606. [Google Scholar] [CrossRef] [PubMed]
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Rafiq, M.; Kamil, M.A. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 2018, 81, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Guerra, M.E.S.; Destro, G.; Vieira, B.; Lima, A.S.; Ferraz, L.F.C.; Hakansson, A.P.; Darrieux, M.; Converso, T.R. Klebsiella pneumoniae Biofilms and Their Role in Disease Pathogenesis. Front. Cell Infect. Microbiol. 2022, 12, 877995. [Google Scholar] [CrossRef] [PubMed]
- Del Pozo, J.L. Biofilm-related disease. Expert Rev. Anti Infect. Ther. 2018, 16, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Naziri, Z.; Kilegolan, J.A.; Moezzi, M.S.; Derakhshandeh, A. Biofilm formation by uropathogenic Escherichia coli: A complicating factor for treatment and recurrence of urinary tract infections. J. Hosp. Infect. 2021, 117, 9–16. [Google Scholar] [CrossRef]
- Zamani, H.; Salehzadeh, A. Biofilm formation in uropathogenic Escherichia coli: Association with adhesion factor genes. Turk. J. Med. Sci. 2018, 48, 162–167. [Google Scholar] [CrossRef]
- Piperaki, E.T.; Syrogiannopoulos, G.A.; Tzouvelekis, L.S.; Daikos, G.L. Klebsiella pneumoniae: Virulence, Biofilm and Antimicrobial Resistance. Pediatr. Infect. Dis. J. 2017, 36, 1002–1005. [Google Scholar] [CrossRef]
- Lehner, A.; Riedel, K.; Eberl, L.; Breeuwer, P.; Diep, B.; Stephan, R. Biofilm formation, extracellular polysaccharide production, and cell-to-cell signaling in various Enterobacter sakazakii strains: Aspects promoting environmental persistence. J. Food Prot. 2005, 68, 2287–2294. [Google Scholar] [CrossRef]
- Kim, H.; Ryu, J.-H.; Beuchat, L.R. Attachment of and biofilm formation by Enterobacter sakazakii on stainless steel and enteral feeding tubes. Appl. Environ. Microbiol. 2006, 72, 5846–5856. [Google Scholar] [CrossRef]


| Strain or Plasmid | Genotype or Characteristics | Source |
|---|---|---|
| Cronobacter sakazakii | ||
| ATCC BAA-894 | WT | ATCC |
| ΔESA_00986 | ΔESA_00986::ampr | This study |
| cpESA_00986 | ΔESA_00986 with pACYC184-ESA_00986 | This study |
| E. coli | ||
| S17 lambda pir | Strain for construction harboring lambda pir | 29 |
| Plasmids | ||
| pCVD442 | Suicide plasmid for deletion: ampr | 29 |
| pCVD442-Q-H | pCVD442 with homologous arms | This study |
| pACYC184 | p15A ori Cmr Tetr | |
| pACYC184-ESA_00986 | pACYC184 with ESA_00986 | This study |
| Primer | Gene Amplified | Primer Sequences (5′-3′) | Amplification Size (bp) |
|---|---|---|---|
| pCVD442 F | Suicide plasmid for markerless deletion | CAATAACCCTGATAAATGCTTCAA | 6345 |
| pCVD442 R | CTCATGAGCGGATACATATTTG | ||
| ESA_00986 QF | Upstream of ESA_00986 gene | TGATAAATGCTTCAACGTCAGCGTCACCTGGAACG | 828 |
| ESA_00986 QR | GTGGTAGTTCAGGCGACTATTTTCCTGACGGAAACAGACG | ||
| ESA_00986 HF | Downstream of ESA_00986 gene | ATAGTCGCCTGAACTACCACAAATGG | 780 |
| ESA_00986 HR | GCGGATACATATTTGCCAGCCCGTCAGTCGTAATG | ||
| ESA_00986 1F | ESA_00986 gene sequence | TACATGGCAACTTATTCTGTATTTAC | 1166 |
| ESA_00986 1R | TGACCATTTGTGGTAGTTCAGG | ||
| ESA_00986 2F | Both ends of the ESA_00986 gene sequence | GCTCTGTCTCGGTGATGTAG | 1620(WT)/600(ΔESA_00986) |
| ESA_00986 2R | TGATCCTTCAATCCCAGTAA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, Y.; Li, P.; Zhu, D.; Zhao, C.; Jiao, J.; Ji, X.; Du, X. Effects of ESA_00986 Gene on Adhesion/Invasion and Virulence of Cronobacter sakazakii and Its Molecular Mechanism. Foods 2023, 12, 2572. https://doi.org/10.3390/foods12132572
Fan Y, Li P, Zhu D, Zhao C, Jiao J, Ji X, Du X. Effects of ESA_00986 Gene on Adhesion/Invasion and Virulence of Cronobacter sakazakii and Its Molecular Mechanism. Foods. 2023; 12(13):2572. https://doi.org/10.3390/foods12132572
Chicago/Turabian StyleFan, Yufei, Ping Li, Dongdong Zhu, Chumin Zhao, Jingbo Jiao, Xuemeng Ji, and Xinjun Du. 2023. "Effects of ESA_00986 Gene on Adhesion/Invasion and Virulence of Cronobacter sakazakii and Its Molecular Mechanism" Foods 12, no. 13: 2572. https://doi.org/10.3390/foods12132572
APA StyleFan, Y., Li, P., Zhu, D., Zhao, C., Jiao, J., Ji, X., & Du, X. (2023). Effects of ESA_00986 Gene on Adhesion/Invasion and Virulence of Cronobacter sakazakii and Its Molecular Mechanism. Foods, 12(13), 2572. https://doi.org/10.3390/foods12132572






