Microbiological and Physico-Chemical Characteristics of Black Tea Kombucha Fermented with a New Zealand Starter Culture
Abstract
1. Introduction
2. Materials and Methods
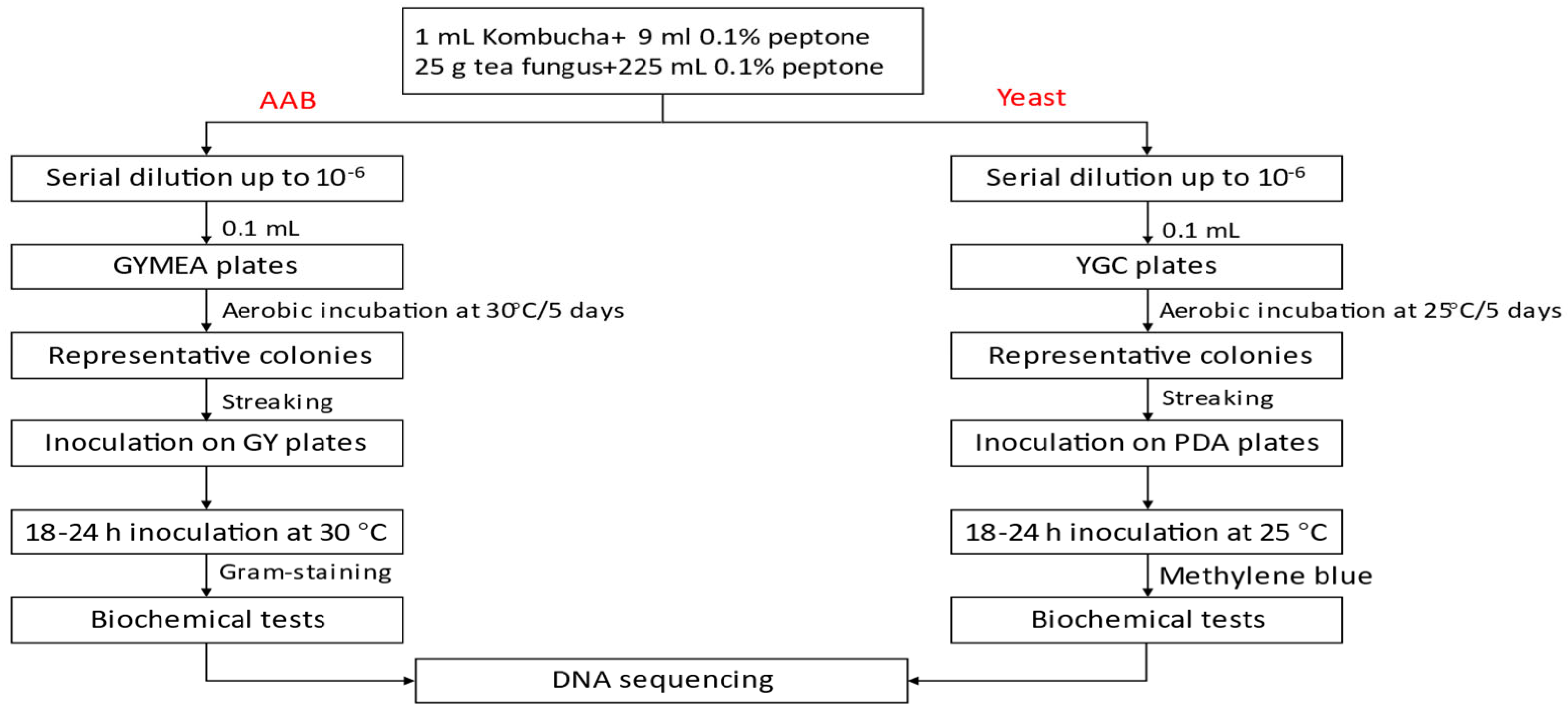
2.1. Preparation of Fermented Kombucha
2.2. Kombucha Fermentation Progress
2.3. Physico-Chemical Characteristics of Kombucha Broth Samples during Fermentation
2.3.1. Determination of pH, Titratable Acidity (TA), and Total Soluble Solids (TSS) of Kombucha Tea Broth during Fermentation
- VNaOH = volume of NaOH (mL)
- MNaOH = molarity of NaOH (M)
- Vsample = volume of sample (mL)
2.3.2. Determination of Wet Yield, Water Holding Capacity (WHC), Moisture Content, and Water Activity of Cellulosic Pellicle at the End of the Fermentation
- W1: weight of empty container (g)
- W2: weight of container and sample before drying (g)
- W3: weight of container and sample after drying (g)
2.4. Isolation and Enumeration of AAB, LAB, and Yeast from Kombucha
2.5. Phenotypic Characterisation of AAB and Yeast Isolated from Kombucha Tea Broth and Cellulosic Pellicle during Fermentation
2.5.1. Phenotypic Characterisation of AAB
2.5.2. Phenotypic Characteristics of Yeast
2.6. Sequence Analysis of Ribosomal RNA Genes
2.7. Statistical Analysis
3. Results and Discussion
3.1. Physico-Chemical Characteristics of Kombucha Samples during Fermentation
Acidity and TSS of Kombucha during Fermentation
3.2. Wet Yield, WHC, Moisture Content, and Water Activity of Cellulosic Pellicle at the End of Fermentation
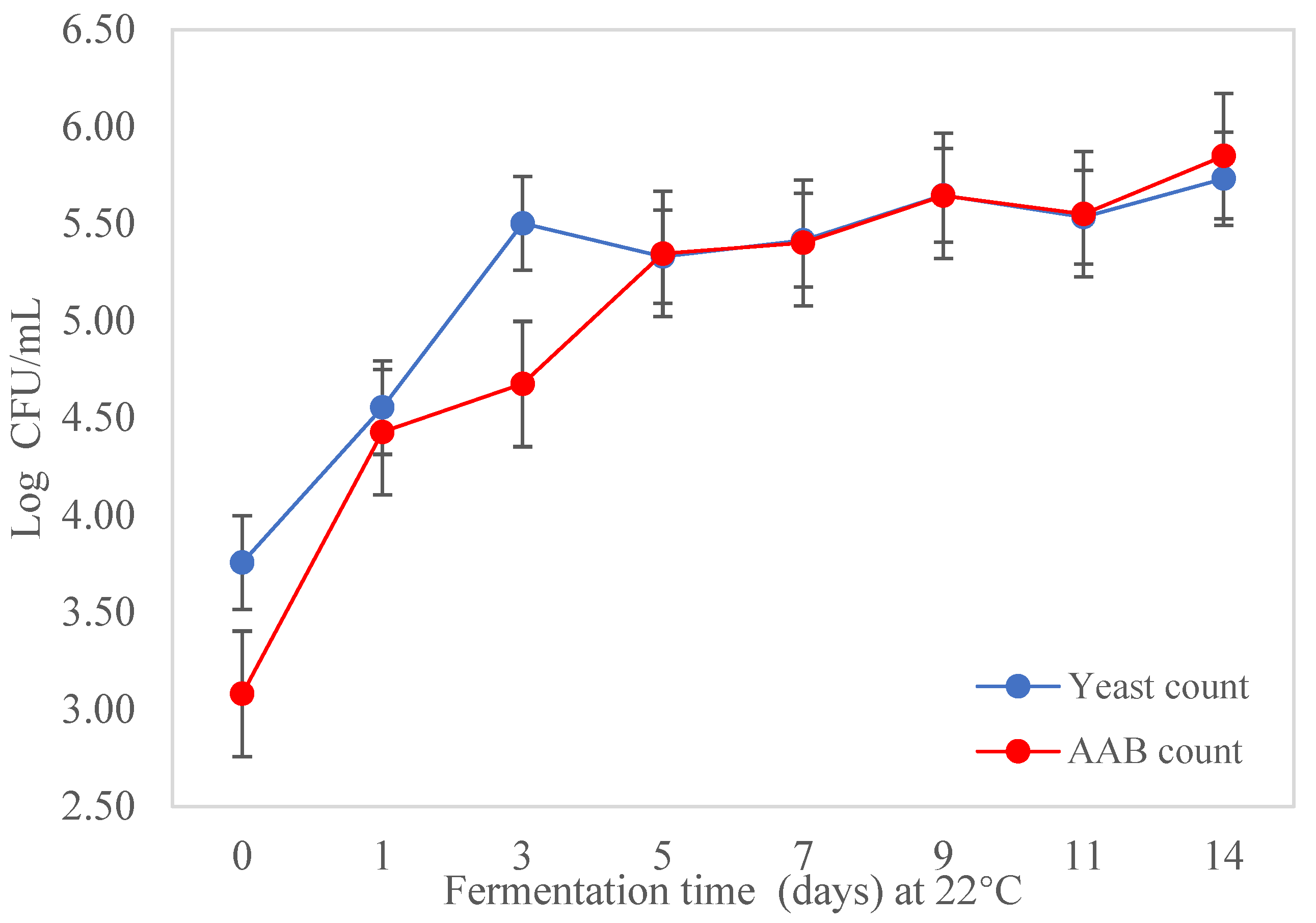
3.3. Isolation and Enumeration of AAB and Yeast during Fermentation of Kombucha
3.4. Phenotypic Characterisation of AAB and Yeast Isolated from the Kombucha Broth and Cellulosic Pellicle
3.4.1. Morphology of AAB and Yeast Isolated from Kombucha Broth and Cellulosic Pellicle
3.4.2. Phenotypic Characteristics of AAB and Yeast Isolated from Kombucha Tea Broth and Pellicle
3.5. Genetic Identification of Representative AAB and Yeast Isolates
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dufresne, C.; Farnworth, E. Tea, Kombucha, and health: A review. Food Res. Int. 2000, 33, 409–421. [Google Scholar] [CrossRef]
- Wang, B.; Rutherfurd-Markwick, K.; Zhang, X.-X.; Mutukumira, A.N. Isolation and characterisation of dominant acetic acid bacteria and yeast isolated from Kombucha samples at point of sale in New Zealand. Curr. Res. Food Sci. 2022, 5, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Neffe-Skocińska, K.; Sionek, B.; Ścibisz, I.; Kołożyn-Krajewska, D. Acid Contents and the Effect of Fermentation Condition of Kombucha Tea Beverages on Physicochemical, Microbiological and Sensory Properties. CyTA-J. Food 2017, 15, 601–607. [Google Scholar] [CrossRef]
- Wang, B.; Rutherfurd-Markwick, K.; Zhang, X.-X.; Mutukumira, A.N. Kombucha: Production and Microbiological Research. Foods 2022, 11, 3456. [Google Scholar] [CrossRef] [PubMed]
- Aung, T.; Eun, J.-B. Impact of time and temperature on the physicochemical, microbiological, and nutraceutical properties of laver kombucha (Porphyra dentata) during fermentation. LWT 2022, 154, 112643. [Google Scholar] [CrossRef]
- Laureys, D.; Britton, S.J.; De Clippeleer, J. Kombucha tea fermentation: A review. J. Am. Soc. Brew. Chem. 2020, 78, 165–174. [Google Scholar] [CrossRef]
- Bishop, P.; Pitts, E.R.; Budner, D.; Thompson-Witrick, K.A. Kombucha: Biochemical and microbiological impacts on the chemical and flavor profile. Food Chem. Adv. 2022, 1, 100025. [Google Scholar] [CrossRef]
- Nyiew, K.Y.; Kwong, P.J.; Yow, Y.Y. An overview of antimicrobial properties of kombucha. Compr. Rev. Food Sci. Food Saf. 2022, 21, 1024–1053. [Google Scholar] [CrossRef]
- Emiljanowicz, K.E.; Malinowska-Pańczyk, E. Kombucha from alternative raw materials—The review. Crit. Rev. Food Sci. Nutri. 2020, 60, 3185–3194. [Google Scholar] [CrossRef]
- Velićanski, A.; Cvetković, D.; Markov, S. Characteristics of Kombucha fermentation on medicinal herbs from Lamiaceae family. Rom. Biotechnol. Lett. 2013, 18, 8034–8042. [Google Scholar]
- Dutta, H.; Paul, S.K. Kombucha drink: Production, quality, and safety aspects. In Production and Management of Beverages; Elsevier: Amsterdam, The Netherlands, 2019; pp. 259–288. [Google Scholar]
- Coelho, R.M.D.; de Almeida, A.L.; do Amaral, R.Q.G.; da Mota, R.N.; de Sousa, P.H.M. Kombucha. Int. J. Gastron. Food Sci. 2020, 22, 100272. [Google Scholar] [CrossRef]
- Savary, O.; Mounier, J.; Thierry, A.; Poirier, E.; Jourdren, J.; Maillard, M.-B.; Penland, M.; Decamps, C.; Coton, E.; Coton, M. Tailor-made microbial consortium for Kombucha fermentation: Microbiota-induced biochemical changes and biofilm formation. Food Res. Int. 2021, 147, 110549. [Google Scholar] [CrossRef] [PubMed]
- Greenwalt, C.J.; Steinkraus, K.H.; Ledford, R.A. Kombucha, the fermented tea: Microbiology, composition, and claimed health effects. J. Food Prot. 2000, 63, 976–981. [Google Scholar] [CrossRef]
- Jayabalan, R.; Malbasa, R.V.; Loncar, E.S.; Vitas, J.S.; Sathishkumar, M. A Review on Kombucha Tea-Microbiology, Composition, Fermentation, Beneficial Effects, Toxicity, and Tea Fungus. Compr. Rev. Food Sci. Food Saf. 2014, 13, 538–550. [Google Scholar] [CrossRef]
- Jayabalan, R.; Chen, P.-N.; Hsieh, Y.-S.; Prabhakaran, K.; Pitchai, P.; Marimuthu, S.; Thangaraj, P.; Swaminathan, K.; Yun, S.E. Effect of solvent fractions of kombucha tea on viability and invasiveness of cancer cells—Characterization of dimethyl 2-(2-hydroxy-2-methoxypropylidine) malonate and vitexin. Indian J. Biotechnol. 2011, 10, 62–75. [Google Scholar]
- Mukadam, T.A.; Punjabi, K.; Deshpande, S.D.; Vaidya, S.P.; Chowdhary, A.S. Isolation and characterization of bacteria and yeast from Kombucha tea. Int. J. Curr. Microbiol. Appl. Sci. 2016, 5, 32–41. [Google Scholar] [CrossRef]
- Nguyen, N.K.; Dong, N.T.; Nguyen, H.T.; Le, P.H. Lactic acid bacteria: Promising supplements for enhancing the biological activities of kombucha. Springerplus 2015, 4, 91. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Jin, W.; Abd El-Aty, A.; Baranenko, D.A.; Gou, X.; Zhang, H.; Geng, J.; Jiang, L.; Chen, D.; Yue, T. Isolation, purification, and structural identification of a new bacteriocin made by Lactobacillus plantarum found in conventional kombucha. Food Control 2020, 110, 106923. [Google Scholar] [CrossRef]
- Diguta, C.F.; Nitoi, G.D.; Matei, F.; Luta, G.; Cornea, C.P. The Biotechnological Potential of Pediococcus spp. Isolated from Kombucha Microbial Consortium. Foods 2020, 9, 1780. [Google Scholar] [CrossRef]
- Yang, J.; Lagishetty, V.; Kurnia, P.; Henning, S.M.; Ahdoot, A.I.; Jacobs, J.P. Microbial and chemical profiles of commercial Kombucha products. Nutrients 2022, 14, 670. [Google Scholar] [CrossRef]
- Bogdan, M.; Justine, S.; Filofteia, D.C.; Petruta, C.C.; Gabriela, L.; Roxana, U.E.; Florentina, M.; Camelia Filofteia, D.; Călina Petruța, C.; Gabriela, L. Lactic acid bacteria strains isolated from Kombucha with potential probiotic effect. Rom. Biotechnol. Lett. 2018, 23, 13592–13598. [Google Scholar]
- Coton, M.; Pawtowski, A.; Taminiau, B.; Burgaud, G.; Deniel, F.; Coulloumme-Labarthe, L.; Fall, A.; Daube, G.; Coton, E. Unraveling microbial ecology of industrial-scale Kombucha fermentations by metabarcoding and culture-based methods. FEMS Microbiol. Ecol. 2017, 93. [Google Scholar] [CrossRef] [PubMed]
- Mayser, P.; Fromme, S.; Leitzmann, C.; Grunder, K. The yeast spectrum of the ‘tea fungus Kombucha’. Mycoses 1995, 38, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Ramadani, A.; Abulreesh, H. Isolation and Identification of Yeast Flora in Local Kombucha Sample: Al Nabtah. Umm Qura Univ. J. App. Sci. 2010, 2, 42–51. [Google Scholar]
- Jayabalan, R.; Malbaśa, R.V.; Sathishkumar, M. Kombucha Tea: Metabolites. In Fungal Metabolites; Springer: Cham, Switzerland, 2017; pp. 965–978. [Google Scholar]
- Teoh, A.L.; Heard, G.; Cox, J. Yeast ecology of Kombucha fermentation. Int. J. Food Microbiol. 2004, 95, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Villarreal-Soto, S.A.; Bouajila, J.; Pace, M.; Leech, J.; Cotter, P.D.; Souchard, J.P.; Taillandier, P.; Beaufort, S. Metabolome-microbiome signatures in the fermented beverage, Kombucha. Int. J. Food Microbiol. 2020, 333, 108778. [Google Scholar] [CrossRef]
- Villarreal-Soto, S.A.; Beaufort, S.; Bouajila, J.; Souchard, J.P.; Taillandier, P. Understanding Kombucha Tea Fermentation: A Review. J. Food Sci. 2018, 83, 580–588. [Google Scholar] [CrossRef]
- Czaja, W.K.; Young, D.J.; Kawecki, M.; Brown, R.M. The future prospects of microbial cellulose in biomedical applications. Biomacromolecules 2007, 8, 1–12. [Google Scholar] [CrossRef]
- Oliveira, Í.A.C.L.D.; Rolim, V.A.d.O.; Gaspar, R.P.L.; Rossini, D.Q.; de Souza, R.; Bogsan, C.S.B. The Technological Perspectives of Kombucha and Its Implications for Production. Fermentation 2022, 8, 185. [Google Scholar] [CrossRef]
- Waisundara, V.Y. Usage of Kombucha ‘Tea Fungus’ for Enhancement of Functional Properties of Herbal Beverages. In Frontiers and New Trends in the Science of Fermented Food and Beverages; IntechOpen: London, UK, 2018. [Google Scholar]
- Amarasinghe, H.; Weerakkody, N.S.; Waisundara, V.Y. Evaluation of physicochemical properties and antioxidant activities of kombucha “Tea Fungus” during extended periods of fermentation. Food Sci. Nutr. 2018, 6, 659–665. [Google Scholar] [CrossRef]
- Aung, T.; Eun, J.B. Production and characterization of a novel beverage from laver (Porphyra dentata) through fermentation with kombucha consortium. Food Chem. 2021, 350, 129274. [Google Scholar] [CrossRef] [PubMed]
- Avcioglu, N.H.; Birben, M.; Bilkay, I.S. Optimization and physicochemical characterization of enhanced microbial cellulose production with a new Kombucha consortium. Process Biochem. 2021, 108, 60–68. [Google Scholar] [CrossRef]
- Du Toit, W.J.; Lambrechts, M.G. The enumeration and identification of acetic acid bacteria from South African red wine fermentations. Int. J. Food Microbiol. 2002, 74, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Reiner, K. Catalase Test Protocol. Am. Soc. Micorbiol. 2010, 1–6. [Google Scholar]
- Shields, P.; Cathcart, L. Oxidase Test Protocol; American Society for Microbiology: Washington DC, USA, 2010; pp. 1–10. [Google Scholar]
- Gomes, R.J.; Borges, M.F.; Rosa, M.F.; Castro-Gomez, R.J.H.; Spinosa, W.A. Acetic Acid Bacteria in the Food Industry: Systematics, Characteristics and Applications. Food Technol. Biotechnol. 2018, 56, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Gullo, M.; Giudici, P. Acetic acid bacteria in traditional balsamic vinegar: Phenotypic traits relevant for starter cultures selection. Int. J. Food Microbiol. 2008, 125, 46–53. [Google Scholar] [CrossRef]
- Arifuzzaman, M.; Hasan, M.Z.; Rahman, S.B.; Pramanik, M.K. Isolation and Characterization of Acetobacter and Gluconobacter spp. from Sugarcane and Rotten Fruits. Res. Revs. Biosci. 2014, 8, 359–365. [Google Scholar]
- Lavasani, P.S.; Motevaseli, E.; Shirzad, M.; Modarressi, M.H. Isolation and identification of Komagataeibacter xylinus from Iranian traditional vinegars and molecular analyses. Iran. J. Microbiol. 2017, 9, 338–347. [Google Scholar]
- Semjonovs, P.; Ruklisha, M.; Paegle, L.; Saka, M.; Treimane, R.; Skute, M.; Rozenberga, L.; Vikele, L.; Sabovics, M.; Cleenwerck, I. Cellulose synthesis by Komagataeibacter rhaeticus strain P 1463 isolated from Kombucha. Appl. Microbiol. Biotechnol. 2017, 101, 1003–1012. [Google Scholar] [CrossRef]
- Asai, T.; Iizuka, H.; Komagata, K. The flagellation and taxonomy of genera Gluconobacter and Acetobacter with reference to the existence of intermediate strains. J. Gen. Appl. Microbiol. 1964, 10, 95–126. [Google Scholar] [CrossRef]
- Swings, J. Phenotypic Identification of Acetic Acid Bacteria. Identification Methods in Applied and Environmental Microbiology (The Society for Applied Bacteriology Technical Series No. 29). 1992, pp. 103–110. Available online: http://hdL.handle.net/1854/LU-222097 (accessed on 30 May 2023).
- Matthews, C. On the Staining of Yeast Cells by Methylene Blue, etc. J. Inst. Brew. 1914, 20, 488–496. [Google Scholar] [CrossRef]
- Painting, K.; Kirsop, B. A quick method for estimating the percentage of viable cells in a yeast population, using methylene blue staining. World J. Microbiol. Biotechnol. 1990, 6, 346–347. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Fell, J.W.; Boekhout, T.; Robert, V. Methods for isolation, phenotypic characterization and maintenance of yeasts. In The Yeasts; Elsevier: Amsterdam, The Netherlands, 2011; pp. 87–110. [Google Scholar]
- Kurtzman, C.P.; Boekhout, T.; Robert, V.; Fell, J.W.; Deak, T. Methods to identify yeasts. In Yeasts Food; Woodhead Publishing: Sawston, UK, 2003; pp. 69–121. [Google Scholar] [CrossRef]
- Yuan, Y.; Feng, F.; Chen, L.; Yao, Q.; Chen, K. Directional Isolation of ethanol-tolerant Acetic Acid Bacteria from Industrial Fermented Vinegar. Eur. Food Res. Technol. 2013, 236, 573–578. [Google Scholar] [CrossRef]
- Matei, B.; Diguță, C.F.; Popa, O.; Cornea, C.P.; Matei, F. Molecular identification of yeast isolated from different kombucha sources. Ann. Univ. Dunarea Jos Galati. Fascicle VI-Food Technol. 2018, 42, 17–25. [Google Scholar]
- Reller, L.B.; Weinstein, M.P.; Petti, C.A. Detection and identification of microorganisms by gene amplification and sequencing. Clinic. Infect. Dis. 2007, 44, 1108–1114. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A Taxonomically United Database of 16S rRNA Gene Sequences and Whole-Genome Assemblies. Int. J. Systematic Evol. Microbiol. 2017, 67, 1613. [Google Scholar] [CrossRef]
- Chu, S.-C.; Chen, C. Effects of origins and fermentation time on the antioxidant activities of kombucha. Food Chem. 2006, 98, 502–507. [Google Scholar] [CrossRef]
- Nummer, B.A. Kombucha brewing under the Food and Drug Administration model Food Code: Risk analysis and processing guidance. J. Environ. Health 2013, 76, 8–11. [Google Scholar]
- de Miranda, J.F.; Ruiz, L.F.; Silva, C.B.; Uekane, T.M.; Silva, K.A.; Gonzalez, A.G.M.; Fernandes, F.F.; Lima, A.R. Kombucha: A review of substrates, regulations, composition, and biological properties. J. Food Sci. 2022, 87, 503–527. [Google Scholar] [CrossRef]
- Jacek, P.; da Silva, F.A.S.; Dourado, F.; Bielecki, S.; Gama, M. Optimization and characterization of bacterial nanocellulose produced by Komagataeibacter rhaeticus K3. Carbohydr. Polym. Technol. Appl. 2021, 2, 100022. [Google Scholar] [CrossRef]
- Wu, J.-M.; Liu, R.-H. Cost-effective production of bacterial cellulose in static cultures using distillery wastewater. J. Biosci. Bioeng. 2013, 115, 284–290. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Lu, S.; Yang, Y. Production of bacterial cellulose from byproduct of citrus juice processing (citrus pulp) by Gluconacetobacter hansenii. Cellulose 2018, 25, 6977–6988. [Google Scholar] [CrossRef]
- Yamada, Y.; Yukphan, P. Genera and Species in Acetic Acid Bacteria. Int. J. Food Microbiol. 2008, 125, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Mamlouk, D.; Gullo, M. Acetic Acid Bacteria: Physiology and Carbon Sources Oxidation. Indian J. Microbiol. 2013, 53, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Raspor, P.; Goranovic, D. Biotechnological Applications of Acetic Acid Bacteria. Crit. Rev. Biotechnol. 2008, 28, 101–124. [Google Scholar] [CrossRef]
- Drysdale, G.; Fleet, G. Acetic Acid Bacteria in Winemaking: A Review. Am. J. Enol. Vitic. 1988, 39, 143–154. [Google Scholar] [CrossRef]
- Dellaglio, F.; Cleenwerck, I.; Felis, G.E.; Engelbeen, K.; Janssens, D.; Marzotto, M. Description of Gluconacetobacter swingsii sp. nov. and Gluconacetobacter rhaeticus sp. nov., isolated from Italian apple fruit. Int. J. Syst. Evolut. Microbiol. 2005, 55, 2365–2370. [Google Scholar] [CrossRef]
- Yamada, Y.; Yukphan, P.; Vu, H.T.L.; Muramatsu, Y.; Ochaikul, D.; Tanasupawat, S.; Nakagawa, Y. Description of Komagataeibacter gen. nov., with proposals of new combinations (Acetobacteraceae). J. Gen. Appl. Microbiol. 2012, 58, 397–404. [Google Scholar] [CrossRef]
- Machado, R.T.; Gutierrez, J.; Tercjak, A.; Trovatti, E.; Uahib, F.G.; de Padua Moreno, G.; Nascimento, A.P.; Berreta, A.A.; Ribeiro, S.J.; Barud, H.S. Komagataeibacter rhaeticus as an alternative bacteria for cellulose production. Carbohydr. Polym. 2016, 152, 841–849. [Google Scholar] [CrossRef]
- Thorat, M.N.; Dastager, S.G. High yield production of cellulose by a Komagataeibacter rhaeticus PG2 strain isolated from pomegranate as a new host. RSC Adv. 2018, 8, 29797–29805. [Google Scholar] [CrossRef]
- Gupte, Y.; Kulkarni, A.; Raut, B.; Sarkar, P.; Choudhury, R.; Chawande, A.; Kumar, G.R.K.; Bhadra, B.; Satapathy, A.; Das, G.; et al. Characterization of nanocellulose production by strains of Komagataeibacter sp. isolated from organic waste and Kombucha. Carbohydr. Polym. 2021, 266, 118176. [Google Scholar] [CrossRef] [PubMed]
- Gopu, G.; Govindan, S. Production of bacterial cellulose from Komagataeibacter saccharivorans strain BC1 isolated from rotten green grapes. Prep. Biochem. Biotechnol. 2018, 48, 842–852. [Google Scholar] [CrossRef] [PubMed]
- Steels, H.; Bond, C.J.; Collins, M.D.; Roberts, I.N.; Stratford, M.; James, S.A. Zygosaccharomyces lentus sp. nov., a new member of the yeast genus Zygosaccharomyces Barker. Int. J. Syst. Evolut. Microbiol. 1999, 49, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Hopfe, S.; Flemming, K.; Lehmann, F.; Mockel, R.; Kutschke, S.; Pollmann, K. Leaching of rare earth elements from fluorescent powder using the tea fungus Kombucha. Waste Manag. 2017, 62, 211–221. [Google Scholar] [CrossRef]
- Marsh, A.J.; O’Sullivan, O.; Hill, C.; Ross, R.P.; Cotter, P.D. Sequence-based analysis of the bacterial and fungal compositions of multiple kombucha (tea fungus) samples. Food Microbiol. 2014, 38, 171–178. [Google Scholar] [CrossRef]
- Steels, H.; James, S.; Roberts, I.; Stratford, M. Zygosaccharomyces lentus: A significant new osmophilic, preservative-resistant spoilage yeast, capable of growth at low temperature. J. Appl. Microbiol. 1999, 87, 520–527. [Google Scholar] [CrossRef]
- De Filippis, F.; Troise, A.D.; Vitaglione, P.; Ercolini, D. Different temperatures select distinctive acetic acid bacteria species and promotes organic acids production during Kombucha tea fermentation. Food Microbiol. 2018, 73, 11–16. [Google Scholar] [CrossRef]
- Chakravorty, S.; Bhattacharya, S.; Chatzinotas, A.; Chakraborty, W.; Bhattacharya, D.; Gachhui, R. Kombucha tea fermentation: Microbial and biochemical dynamics. Int. J. Food Microbiol. 2016, 220, 63–72. [Google Scholar] [CrossRef]
- Moyer, C.L.; Dobbs, F.C.; Karl, D.M. Estimation of diversity and community structure through restriction fragment length polymorphism distribution analysis of bacterial 16S rRNA genes from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl. Environ. Microbiol. 1994, 60, 871–879. [Google Scholar] [CrossRef]
- Song, Z.; Cao, Y.; Zhang, Y.; Zhang, Z.; Shi, X.; Zhang, W.; Wen, P. Effects of storage methods on the microbial community and quality of Sichuan smoked bacon. LWT 2022, 158, 113115. [Google Scholar] [CrossRef]
- Filippousi, R.; Tsouko, E.; Mordini, K.; Ladakis, D.; Koutinas, A.A.; Aggelis, G.; Papanikolaou, S. Sustainable arabitol production by a newly isolated Debaryomyces prosopidis strain cultivated on biodiesel-derived glycerol. Carbon Resour. Convers. 2022, 5, 92–99. [Google Scholar] [CrossRef]



| Characteristics | Cellulosic Pellicle |
|---|---|
| Wet yield (%) | 27.75 ± 0.08 |
| WHC (g water/g dry pellicle) | 17.82 ± 0.16 |
| Moisture content (%) | 94.68 ± 0.05 |
| Water activity at 21.45 °C | 0.996 ± 0.002 |
| Colony Group | Description of Appearance |
|---|---|
| Group I | Circular and slightly umbonate in the centre, white colour, smooth surface, and entire margin |
| Group II | Circular, brownish cream colour, flat and smooth surface |
| Test | Group I | Group II | Group III | Group IV | Group V | Group VI |
|---|---|---|---|---|---|---|
| Oxidase | - | - | - | - | - | - |
| Catalase | + | + | + | + | + | + |
| Growth at different temperature | ||||||
| Growth at 25 °C | + | + | + | + | + | + |
| Growth at 30 °C | + | + | + | + | + | + |
| Growth at 37 °C | + | + | + | + | + | + |
| Growth at different pH | ||||||
| pH 2 | - | - | - | - | - | - |
| pH 3 | + | + | + | + | + | + |
| Cellulose formation | + | + | + | + | + | + |
| Growth without acetic acid | + | + | + | + | + | + |
| Oxidation of acetate | + | + | + | + | + | + |
| Oxidation of lactate | + | + | + | + | + | + |
| Alcoholic tolerance (v/v) | ||||||
| 2% | + | + | + | + | + | + |
| 4% | + | - | - | + | + | + |
| 6% | - | - | - | + | + | - |
| 8% | - | - | - | + | - | - |
| 10% | - | - | - | + | - | - |
| Acid produced from: | ||||||
| D-glucose | + | + | + | + | + | + |
| Sucrose | - | - | - | - | - | - |
| Lactose | - | - | - | - | - | - |
| Trehalose | - | - | - | - | - | - |
| Oxidation of ethanol | - | + | - | + | - | - |
| Oxidation of ethanol to water and CO2 | - | + | - | + | - | - |
| Ketogenesis of glycerol to DHA | + | + | + | + | + | + |
| Tests | Group I | Group II |
|---|---|---|
| Growth in broth at 37 °C | - | w |
| Growth in broth at 30 °C | - | + |
| Growth in broth at 25 °C | + | + |
| Growth in medium containing 0.01% cycloheximide | + | + |
| Growth in medium containing 0.1% cycloheximide | w | + |
| Growth in medium containing 1% glacial acetic acid | + | w |
| Growth in medium containing 5% glucose and 10% NaCl | - | w |
| Growth in broth at pH 2 | + | + |
| Growth in broth at pH 3 | + | + |
| Acid produced from: | ||
| D-glucose | + | + |
| Sucrose | + | + |
| Trehalose | - | + |
| Lactose | - | - |
| Representative Strains | Top Hit Taxon | Type Strain | GenBank Accession | % Similarity | Variation |
|---|---|---|---|---|---|
| AAB | |||||
| TFT1AAB26 | Komagataibacter rhaeticus | DST GL02 | AY180961 | 99.92 | 1/1292 |
| Yeast | |||||
| GI: D3T3Y9 | Zygosaccharomyces lentus | CBS 8574 | NR_156001.1 | 99.84 | 1/634 |
| GII: TFT1Y39 | Debaryomyces prosopidis | JCM 9913 | NR_077067.1 | 100.00 | 0/600 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, B.; Rutherfurd-Markwick, K.; Naren, N.; Zhang, X.-X.; Mutukumira, A.N. Microbiological and Physico-Chemical Characteristics of Black Tea Kombucha Fermented with a New Zealand Starter Culture. Foods 2023, 12, 2314. https://doi.org/10.3390/foods12122314
Wang B, Rutherfurd-Markwick K, Naren N, Zhang X-X, Mutukumira AN. Microbiological and Physico-Chemical Characteristics of Black Tea Kombucha Fermented with a New Zealand Starter Culture. Foods. 2023; 12(12):2314. https://doi.org/10.3390/foods12122314
Chicago/Turabian StyleWang, Boying, Kay Rutherfurd-Markwick, Naran Naren, Xue-Xian Zhang, and Anthony N. Mutukumira. 2023. "Microbiological and Physico-Chemical Characteristics of Black Tea Kombucha Fermented with a New Zealand Starter Culture" Foods 12, no. 12: 2314. https://doi.org/10.3390/foods12122314
APA StyleWang, B., Rutherfurd-Markwick, K., Naren, N., Zhang, X.-X., & Mutukumira, A. N. (2023). Microbiological and Physico-Chemical Characteristics of Black Tea Kombucha Fermented with a New Zealand Starter Culture. Foods, 12(12), 2314. https://doi.org/10.3390/foods12122314







