Survey of Colistin Resistance in Commensal Bacteria from Penaeus vannamei Farms in China
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples and Bacterial Isolation
2.2. Colistin Resistance Screening
2.3. 16S rRNA Gene Analyses
2.4. Electroporation Transformation
2.5. Antimicrobial Susceptibility Testing
2.6. Multilocus Sequence Typing (MLST)
2.7. Whole Genome Sequencing (WGS) and Analysis
2.8. Phylogenetic Analysis
3. Results
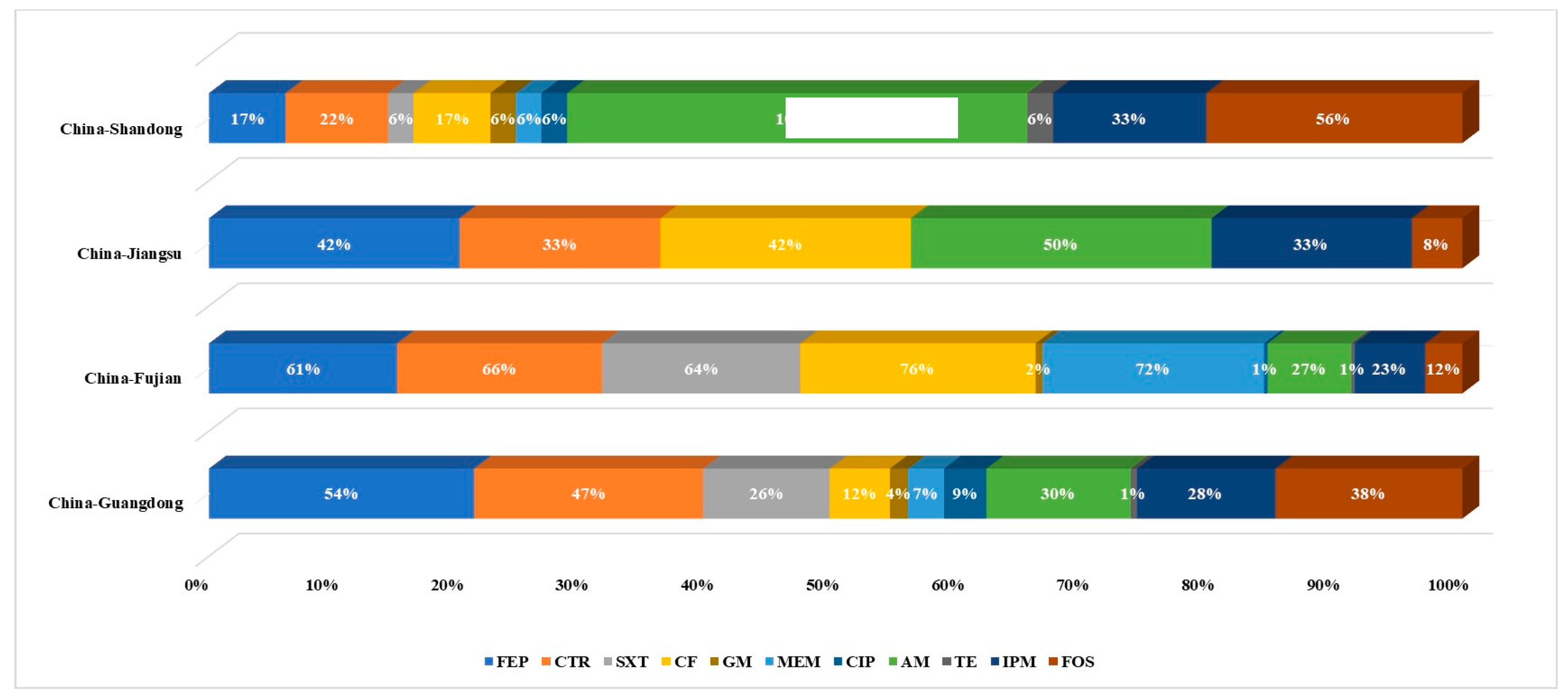
3.1. Prevalence of Colistin-Resistant Isolates from Four Chinese Provinces
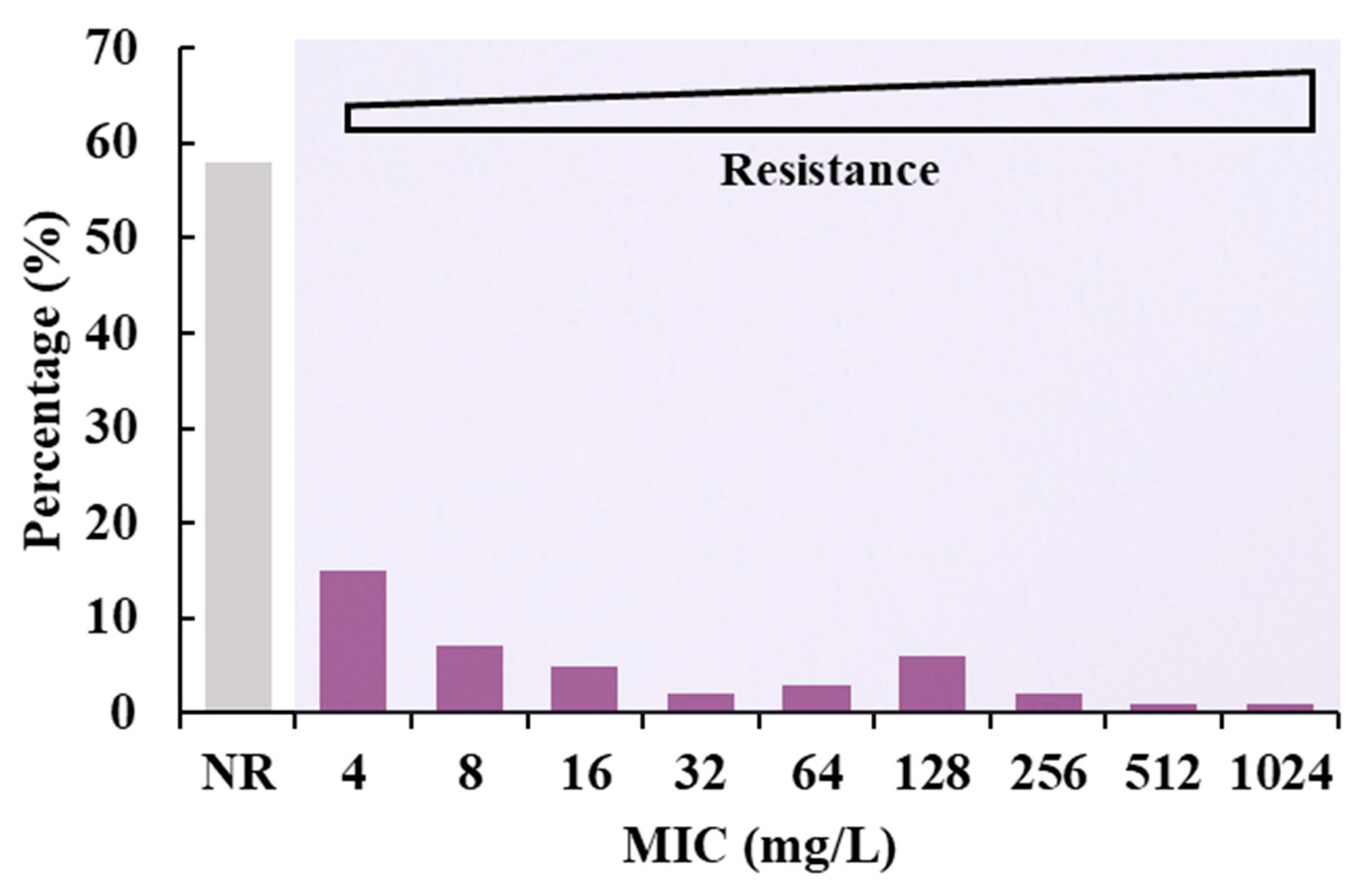
3.2. Determination of MIC Value of Electron Transformants
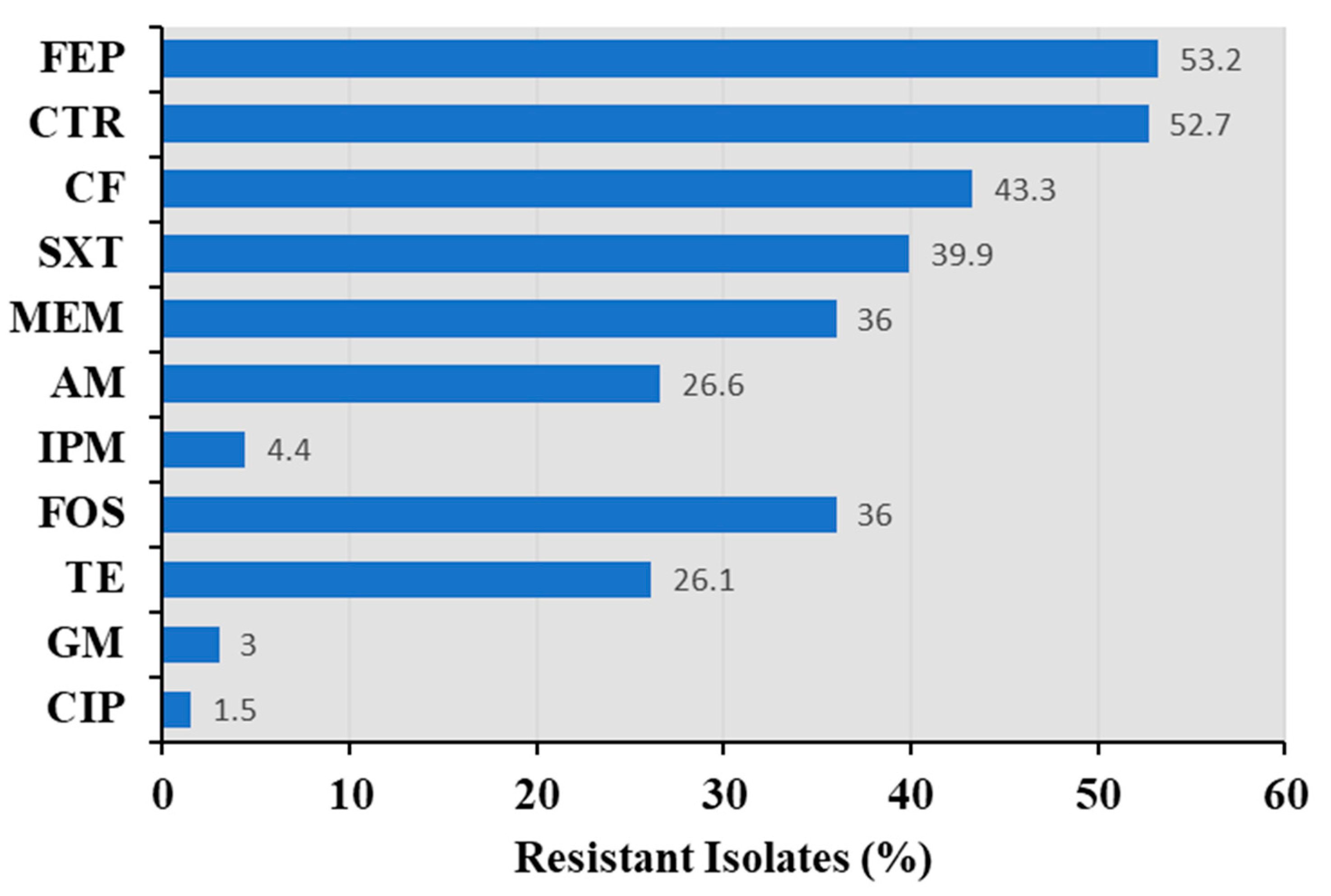
3.3. Antimicrobial Susceptibility of Bacillus spp.
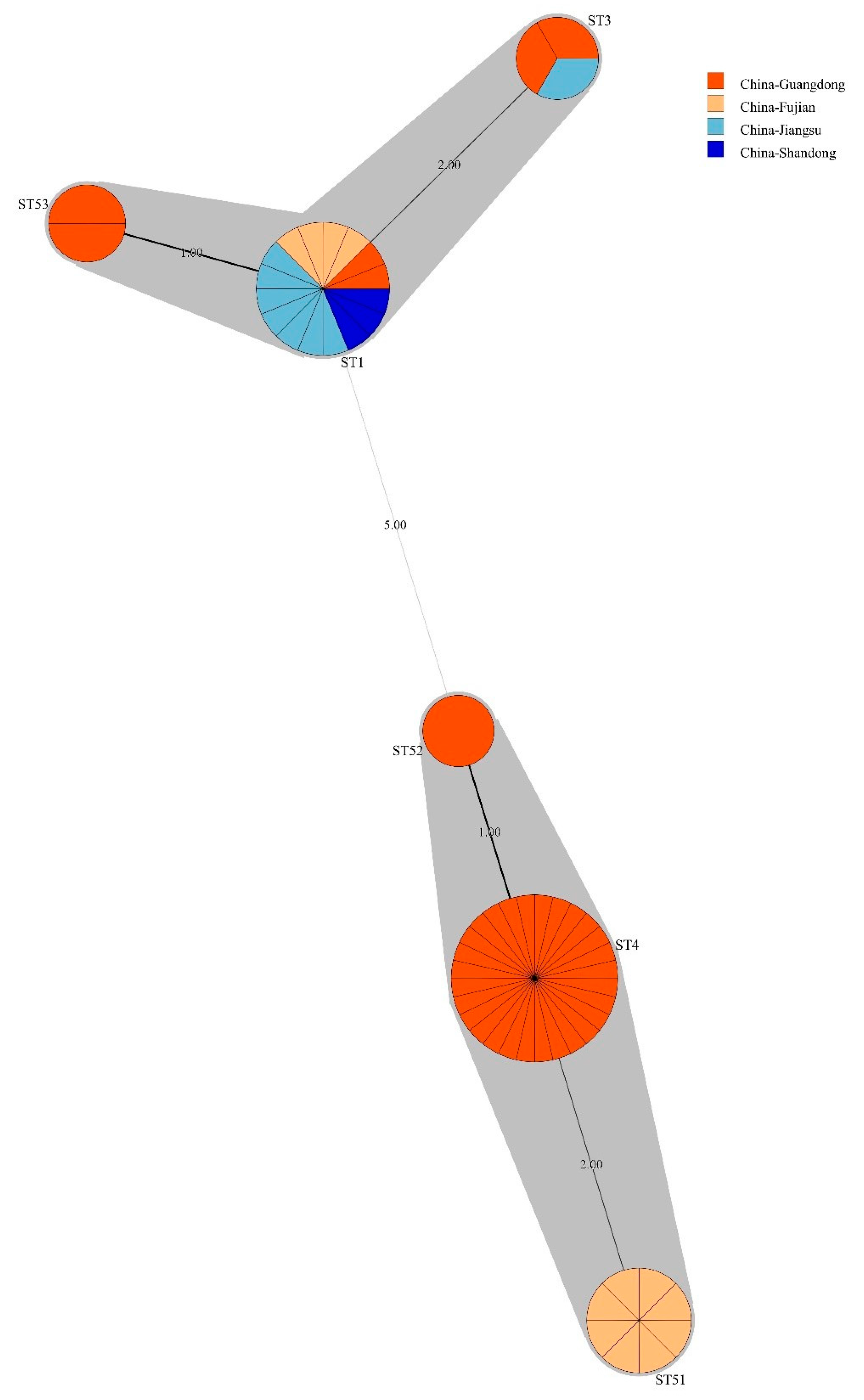
3.4. Bacillus licheniformis ST Diversity
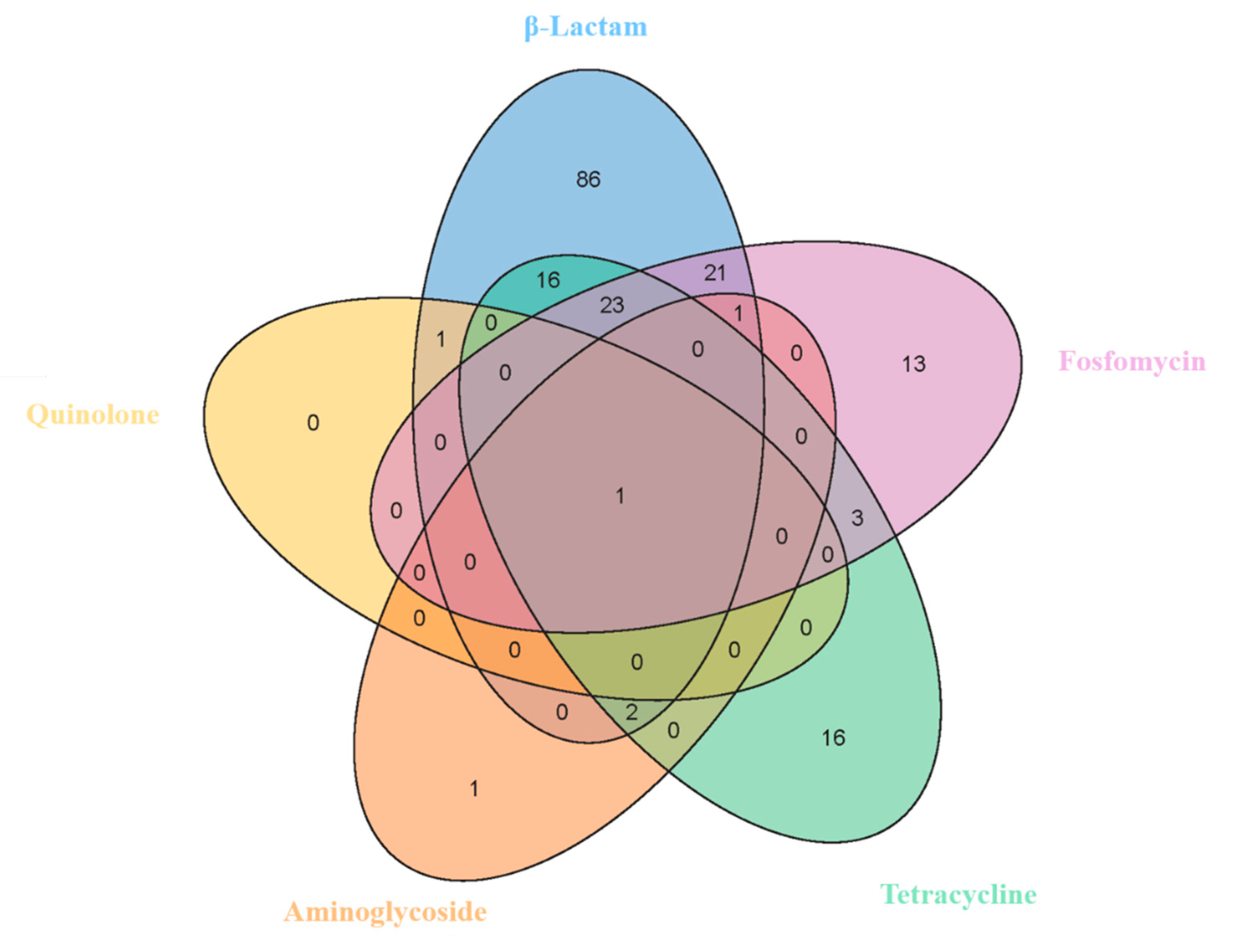
3.5. Antibiotic Resistance Genes of Bacillus licheniformis
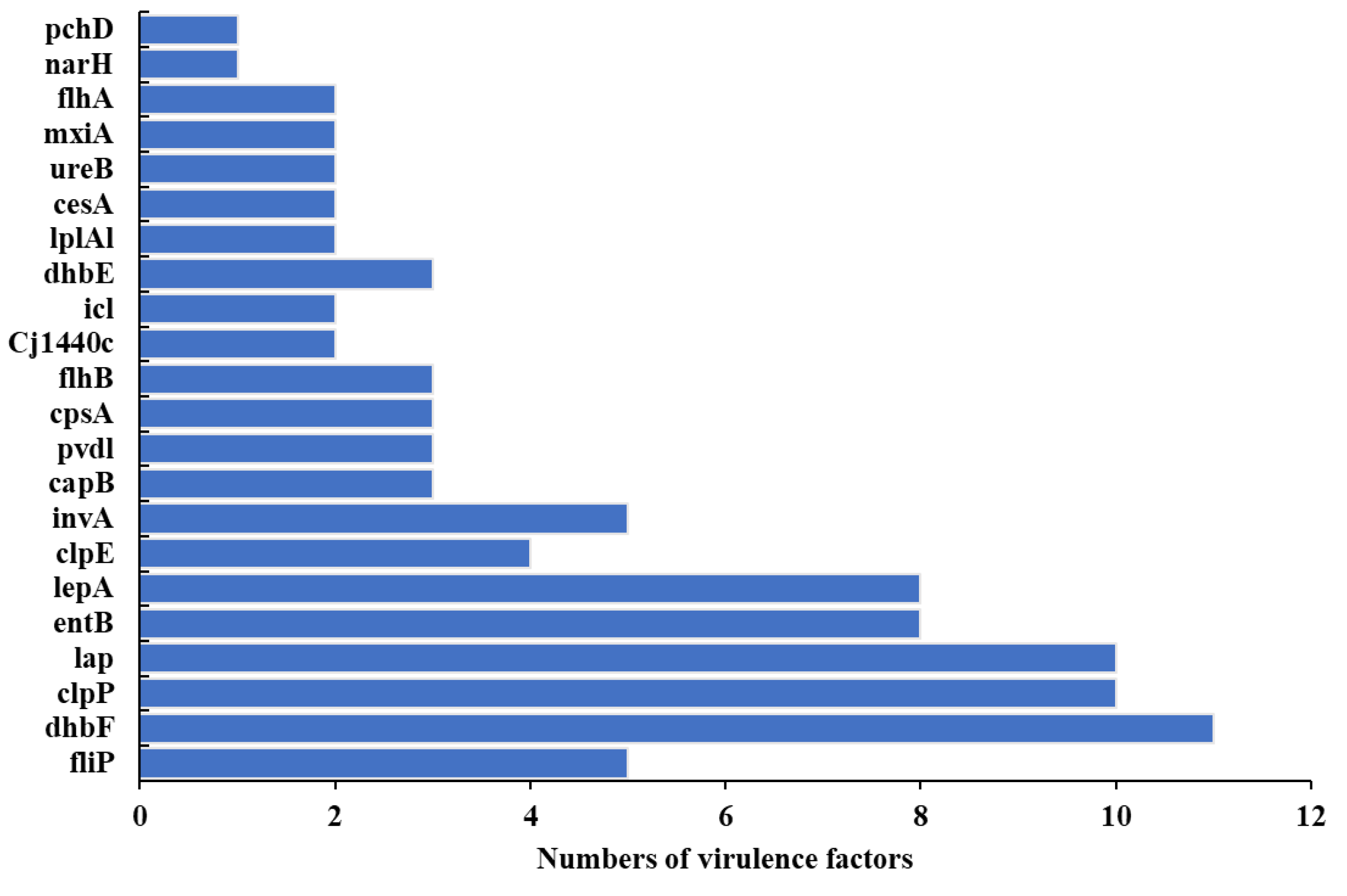
3.6. Pathogenicity, Virulomes, and Mobile Genetic Elements
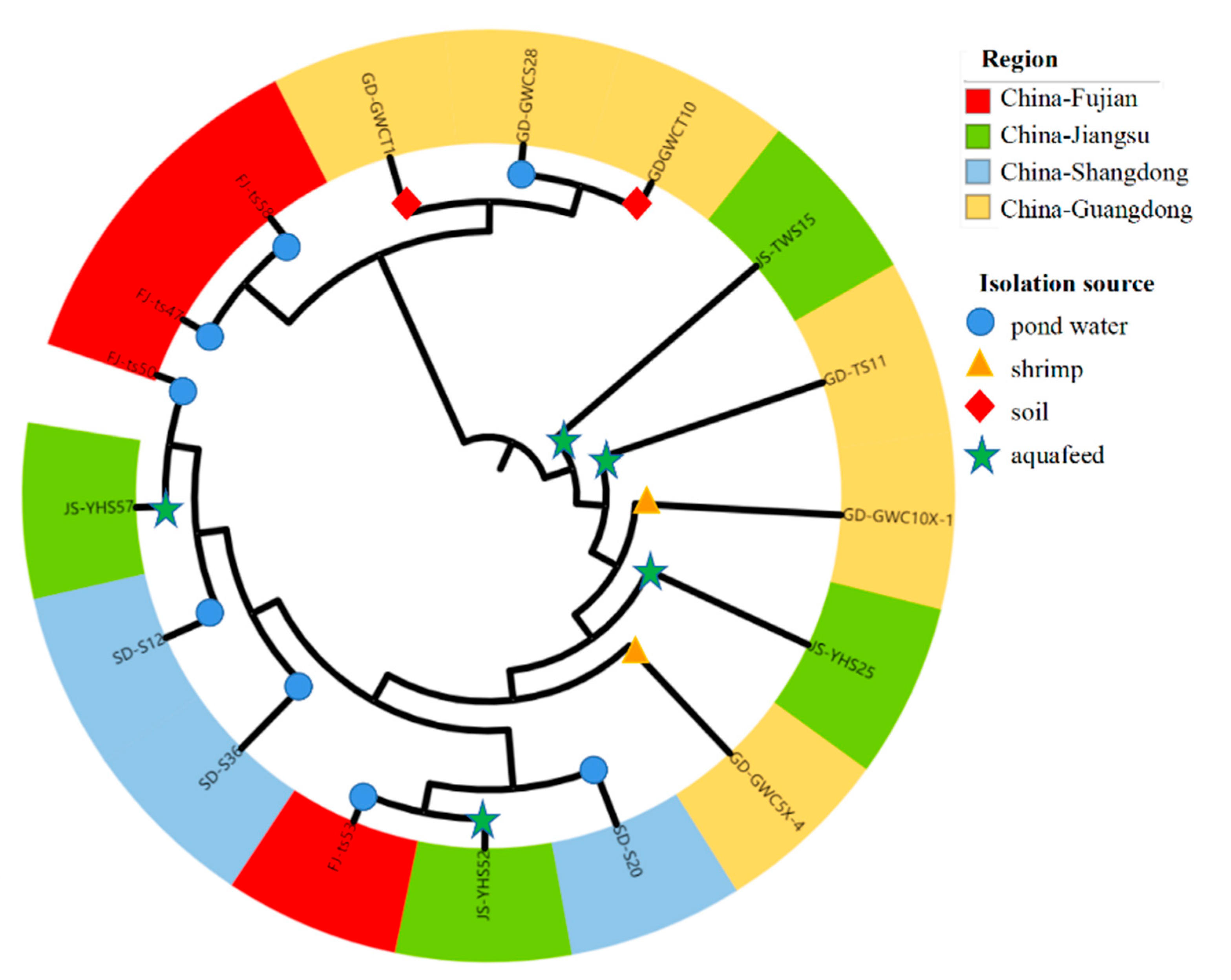
3.7. Phylogenetic Lineages of Bacillus licheniformis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Anokyewaa, M.A.; Amoah, K.; Li, Y.; Lu, Y.; Kuebutornye, F.K.A.; Asiedu, B.; Seidu, I. Prevalence of virulence genes and antibiotic susceptibility of Bacillus used in commercial aquaculture probiotics in China. Aquac. Rep. 2021, 21, 100784. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Blue, S.; Singh, V.; Saubolle, M.A. Bacillus licheniformis bacteremia: Five cases associated with indwelling central venous catheters. Clin. Infect. Dis. 1995, 20, 629–633. [Google Scholar] [CrossRef]
- Boinapally, K.; Jiang, X. Comparing antibiotic resistance in commensal and pathogenic bacteria isolated from wild-caught South Carolina shrimps vs. farm-raised imported shrimps. Can. J. Microbiol. 2007, 53, 919–924. [Google Scholar] [CrossRef]
- Brestoff, J.R.; Artis, D. Commensal bacteria at the interface of host metabolism and the immune system. Nat. Immunol. 2013, 14, 676–684. [Google Scholar] [CrossRef]
- Chen, T.; Wu, Q.; Zhou, H.; Deng, K.; Wang, X.; Meng, F.; Yang, S.; Wang, X.; Shah, N.P.; Wei, H. Assessment of commercial probiotic products in China for labelling accuracy and probiotic characterisation of selected isolates. Int. J. Dairy Technol. 2017, 70, 119–126. [Google Scholar] [CrossRef]
- Cherak, Z.; Loucif, L.; Moussi, A.; Rolain, J.-M. Epidemiology of mobile colistin resistance (mcr) genes in aquatic environments. J. Glob. Antimicrob. Resist. 2021, 27, 51–62. [Google Scholar] [CrossRef]
- Coil, D.; Jospin, G.; Darling, A.E. A5-miseq: An updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 2015, 31, 587–589. [Google Scholar] [CrossRef]
- Cosentino, S.; Voldby Larsen, M.; Møller Aarestrup, F.; Lund, O. PathogenFinder-distinguishing friend from foe using bacterial whole genome sequence data. PLoS ONE 2013, 8, e77302. [Google Scholar] [CrossRef]
- Devine, D.A.; Marsh, P.D.; Meade, J. Modulation of host responses by oral commensal bacteria. J. Oral Microbiol. 2015, 7, 26941. [Google Scholar] [CrossRef]
- European Society of Clinical Microbiology and Infectious Diseases. European Committee on Antimicrobial Susceptibility Testing, Breakpoint Tables for Interpretation of MICs and Zone Diameters; European Society of Clinical Microbiology and Infectious Diseases: Basel, Switzerland, 2015. [Google Scholar]
- FAO. The State of World Fisheries and Aquaculture 2016: Contributing to Food Security and Nutrition for All; Food and Agriculture Organization of the United Nations: Rome, Italy, 2016. [Google Scholar]
- Fey, P.D.; Safranek, T.J.; Rupp, M.E.; Dunne, E.F.; Ribot, E.; Iwen, P.C.; Bradford, P.A.; Angulo, F.J.; Hinrichs, S.H. Ceftriaxone-resistant Salmonella infection acquired by a child from cattle. N. Engl. J. Med. 2000, 342, 1242–1249. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, K.J.; Reyes, A.; Wang, B.; Selleck, E.M.; Sommer, M.O.; Dantas, G. The shared antibiotic resistome of soil bacteria and human pathogens. Science 2012, 337, 1107–1111. [Google Scholar] [CrossRef] [PubMed]
- Haryani, Y.; Halid, N.A.; Guat, G.S.; Nor-Khaizura, M.A.R.; Hatta, M.A.M.; Sabri, S.; Radu, S.; Hasan, H. High prevalence of multiple antibiotic resistance in fermented food-associated lactic acid bacteria in Malaysia. Food Control 2023, 147, 109558. [Google Scholar] [CrossRef]
- Hassan, J.; Eddine, R.Z.; Mann, D.; Li, S.; Deng, X.; Saoud, I.P.; Kassem, I.I. The Mobile Colistin Resistance Gene, mcr-1.1, Is Carried on IncX4 Plasmids in Multidrug Resistant E. coli Isolated from Rainbow Trout Aquaculture. Microorganisms 2020, 8, 1636. [Google Scholar] [CrossRef] [PubMed]
- Hawkey, P.M. The growing burden of antimicrobial resistance. J. Antimicrob. Chemother. 2008, 62, i1–i9. [Google Scholar] [CrossRef] [PubMed]
- Haydushka, I.A.; Markova, N.; Kirina, V.; Atanassova, M. Recurrent sepsis due to Bacillus licheniformis. J. Glob. Infect. Dis. 2012, 4, 82. [Google Scholar] [CrossRef] [PubMed]
- Hemamalini, N.; Shanmugam, S.A.; Kathirvelpandian, A.; Deepak, A.; Kaliyamurthi, V.; Suresh, E.; Ezhilmathi, S. Prevalence, Antimicrobial Susceptibility and Resistance Gene Detection in Bacteria Isolated from Goldfish and Tiger Barb from Ornamental Fish Farms of Tamil Nadu. Indian J. Microbiol. 2022, 62, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Hemamalini, N.; Shanmugam, S.A.; Kathirvelpandian, A.; Deepak, A.; Kaliyamurthi, V.; Suresh, E. A critical review on the antimicrobial resistance, antibiotic residue and metagenomics-assisted antimicrobial resistance gene detection in freshwater aquaculture environment. Aquac. Res. 2022, 53, 344–366. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, L.; Tiu, L.; Wang, H.H. Characterization of antibiotic resistance in commensal bacteria from an aquaculture ecosystem. Front. Microbiol. 2015, 6, 914. [Google Scholar] [CrossRef]
- Iurlina, M.O.; Saiz, A.I.; Fuselli, S.R.; Fritz, R. Prevalence of Bacillus spp. in different food products collected in Argentina. LWT-Food Sci. Technol. 2006, 39, 105–110. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST. org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Kim, J.S.; Yu, J.K.; Jeon, S.J.; Park, S.-H.; Han, S.; Park, S.H.; Kang, M.; Im Jang, J.; Shin, E.-K.; Kim, J.; et al. Distribution of mcr genes among carbapenem-resistant Enterobacterales clinical isolates: High prevalence of mcr-positive Enterobacter cloacae complex in Seoul, Republic of Korea. Int. J. Antimicrob. Agents 2021, 58, 106418. [Google Scholar] [CrossRef] [PubMed]
- Kitamoto, S.; Nagao-Kitamoto, H.; Kuffa, P.; Kamada, N. Regulation of virulence: The rise and fall of gastrointestinal pathogens. J. Gastroenterol. 2016, 51, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yang, Z.; Hu, J.; Wang, B.; Rong, H.; Li, Z.; Sun, Y.; Wang, Y.; Zhang, X.; Wang, M. Evaluation of culturable ‘last-resort’antibiotic resistant pathogens in hospital wastewater and implications on the risks of nosocomial antimicrobial resistance prevalence. J. Hazard. Mater. 2022, 438, 129477. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Shi, X.; Song, Y.; Yao, S.; Li, K.; Shi, B.; Sun, J.; Liu, Z.; Zhao, W.; Zhao, C.; et al. Genetic diversity, antibiotic resistance, and virulence profiles of Listeria monocytogenes from retail meat and meat processing. Food Res. Int. 2022, 162, 112040. [Google Scholar] [CrossRef]
- Liu, M.; Li, X.; Xie, Y.; Bi, D.; Sun, J.; Li, J.; Tai, C.; Deng, Z.; Ou, H.-Y. ICEberg 2.0: An updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 2019, 47, D660–D665. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Steele, J.C.; Meng, X.-Z. Usage, residue, and human health risk of antibiotics in Chinese aquaculture: A review. Environ. Pollut. 2017, 223, 161–169. [Google Scholar] [CrossRef]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience 2012, 1, 2047–2217X. [Google Scholar] [CrossRef]
- Lv, Z.; Shen, Y.; Liu, W.; Ye, H.; Liu, D.; Liu, J.; Fu, Y.; Peng, C.; Chen, K.; Deng, X.; et al. Prevalence and risk factors of mcr-1-positive volunteers after colistin banning as animal growth promoter in China: A community-based case-control study. Clin. Microbiol. Infect. 2022, 28, 267–272. [Google Scholar] [CrossRef]
- Madslien, E.; Rønning, H.; Lindbäck, T.; Hassel, B.; Andersson, M.; Granum, P.E. Lichenysin is produced by most Bacillus licheniformis strains. J. Appl. Microbiol. 2013, 115, 1068–1080. [Google Scholar] [CrossRef] [PubMed]
- Montes-Horcasitas, C.; Ruiz-Medrano, R.; Magaña-Plaza, I.; Silva, L.G.; Herrera-Martínez, A.; Hernández-Montalvo, L.; Xoconostle-Cázares, B.J.C.m. Efficient transformation of Cellulomonas flavigena by electroporation and conjugation with Bacillus thuringiensis. Curr. Microbiol. 2004, 49, 428–432. [Google Scholar] [CrossRef] [PubMed]
- Musovic, S.; Klümper, U.; Dechesne, A.; Magid, J.; Smets, B.F. Long-term manure exposure increases soil bacterial community potential for plasmid uptake. Environ. Microbiol. Rep. 2014, 6, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Nakamizo, S.; Egawa, G.; Honda, T.; Nakajima, S.; Belkaid, Y.; Kabashima, K. Commensal bacteria and cutaneous immunity. In Seminars in Immunopathology; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Pan, Y.; Zeng, Z.; Niu, H.; Huang, L.; Hu, J.; Li, G.; Li, Y. Whole-genome epidemiology and characterisation of mcr-1-encoding Escherichia coli in aquatic bird farms from the Pearl River Delta, China, 2019–2020. Int. J. Antimicrob. Agents 2022, 59, 106478. [Google Scholar] [CrossRef]
- Raum, E.; Lietzau, S.; von Baum, H.; Marre, R.; Brenner, H. Changes in Escherichia coli resistance patterns during and after antibiotic therapy: A longitudinal study among outpatients in Germany. Clin. Microbiol. Infect. 2008, 14, 41–48. [Google Scholar] [CrossRef]
- Salinas, L.; Loayza, F.; Cardenas, P.; Saraiva, C.; Johnson, T.J.; Amato, H.; Graham, J.P.; Trueba, G. Environmental Spread of Extended Spectrum Beta-Lactamase (ESBL) Producing Escherichia coli and ESBL Genes among Children and Domestic Animals in Ecuador. Environ. Health Perspect. 2021, 129, 27007. [Google Scholar] [CrossRef]
- Salkinoja-Salonen, M.S.; Vuorio, R.; Andersson, M.; Kampfer, P.; Andersson, M.; Honkanen-Buzalski, T.; Scoging, A.J.A. Toxigenic strains of Bacillus licheniformis related to food poisoning. Appl. Environ. Microbiol. 1999, 65, 4637–4645. [Google Scholar] [CrossRef]
- Santini, F.; Borghetti, V.; Amalfitano, G.; Mazzucco, A. Bacillus licheniformis prosthetic aortic valve endocarditis. J. Clin. Microbiol. 1995, 33, 3070–3073. [Google Scholar] [CrossRef]
- Schubert, M.; Lindgreen, S.; Orlando, L. AdapterRemoval v2: Rapid adapter trimming, identification, and read merging. BMC Res. Notes 2016, 9, 88. [Google Scholar] [CrossRef]
- Shen, Y.; Lv, Z.; Yang, L.; Liu, D.; Ou, Y.; Xu, C.; Liu, W.; Yuan, D.; Hao, Y.; He, J.; et al. Integrated aquaculture contributes to the transfer of mcr-1 between animals and humans via the aquaculture supply chain. Environ. Int. 2019, 130, 104708. [Google Scholar] [CrossRef]
- Siguier, P.; Pérochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef] [PubMed]
- International Organization for Standardization. Clinical Laboratory Testing and in Vitro Diagnostic Test Systems-Susceptibility Testing of Infectious Agents and Evaluation of Performance of Antimicrobial Susceptibility Test Devices: Reference Method for Testing the In Vitro Activity of Antimicrobial Agents Against Rapidly Growing Aerobic Bacteria Involved in Infectious Diseases; ISO: Geneva, Switzerland, 2006. [Google Scholar]
- Sun, C.; Wang, Y.; Ma, S.; Zhang, S.; Liu, D.; Wang, Y.; Wu, C. Surveillance of antimicrobial resistance in Escherichia coli and enterococci from food products at retail in Beijing, China. Food Control 2021, 119, 107483. [Google Scholar] [CrossRef]
- Wang, M.; Li, Y.; Lin, X.; Xu, H.; Li, Y.; Xue, R.; Wang, G.; Sun, S.; Li, J.; Lan, Z.; et al. Genetic characterization, mechanisms and dissemination risk of antibiotic resistance of multidrug-resistant Rothia nasimurium. Infect. Genet. Evol. 2021, 90, 104770. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.E. Benefit and mischief from commensal bacteria. J. Clin. Pathol. 1973, 26, 811. [Google Scholar] [CrossRef] [PubMed]
- Yang, J. Virulence Factors of Pathogenic Bacteria. 2016. Available online: http://www.mgc.ac.cn/VFs/ (accessed on 24 October 2022).
- Ye, L.; Lu, Z.; Li, X.; Shi, L.; Huang, Y.; Wang, H.H. Antibiotic-resistant bacteria associated with retail aquaculture products from Guangzhou, China. J. Food Prot. 2013, 76, 295–301. [Google Scholar] [CrossRef]
- Yu, Y.; Tang, M.; Wang, Y.; Liao, M.; Wang, C.; Rong, X.; Li, B.; Ge, J.; Gao, Y.; Dong, X.; et al. Virulence and antimicrobial resistance characteristics assessment of Vibrio isolated from shrimp (Penaeus vannamei) breeding system in south China. Ecotoxicol. Environ. Saf. 2023, 252, 114615. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, T.; Wang, C.; Liang, G.; Lu, Q.; Wen, G.; Guo, Y.; Cheng, Y.; Wang, Z.; Shao, H.; et al. Prevalence of colistin resistance gene mcr-1 in Escherichia coli isolated from chickens in central China, 2014 to 2019. J. Glob. Antimicrob. Resist. 2022, 29, 241–246. [Google Scholar] [CrossRef]
- De Clerck, E.; De Vos, P. Genotypic diversity among Bacillus licheniformis strains from various sources. FEMS Microbiol. Lett. 2004, 231, 91–98. [Google Scholar] [CrossRef]




| Sample Site | Sample Type | Number (%) | Regional Drug Resistance Rate (%) | |
|---|---|---|---|---|
| Isolates Investigated | Resistant Isolates | |||
| China— Guangdong | Penaeus vannamei | 385 | 73 (19.0) | 26.8 |
| Soil | 183 | 62 (33.9) | ||
| Aquafeed | 169 | 45 (26.6) | ||
| Pond water | 274 | 91 (33.2) | ||
| China— Fujian | Penaeus vannamei | 222 | 88 (39.6) | 41.0 |
| Soil | 98 | 26 (26.5) | ||
| Aquafeed | 115 | 62 (53.9) | ||
| Pond water | 106 | 46 (43.4) | ||
| China— Jiangsu | Penaeus vannamei | 92 | 22 (23.9) | 49.2 |
| Soil | 58 | 31 (53.4) | ||
| Aquafeed | 76 | 66 (86.8) | ||
| Pond water | 24 | 4 (16.7) | ||
| China— Shandong | Penaeus vannamei | 120 | 115 (95.8) | 82.7 |
| Soil | 60 | 43 (71.7) | ||
| Aquafeed | 84 | 60 (71.4) | ||
| Pond water | 60 | 50 (83.3) | ||
| Total | 2126 | 884 (41.6) | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Lv, X.; Cao, W.; Zhang, H.; Shi, L.; Bai, W.; Ye, L. Survey of Colistin Resistance in Commensal Bacteria from Penaeus vannamei Farms in China. Foods 2023, 12, 2143. https://doi.org/10.3390/foods12112143
Zhang Y, Lv X, Cao W, Zhang H, Shi L, Bai W, Ye L. Survey of Colistin Resistance in Commensal Bacteria from Penaeus vannamei Farms in China. Foods. 2023; 12(11):2143. https://doi.org/10.3390/foods12112143
Chicago/Turabian StyleZhang, Yilin, Xinrui Lv, Weiwei Cao, Huang Zhang, Lei Shi, Weibin Bai, and Lei Ye. 2023. "Survey of Colistin Resistance in Commensal Bacteria from Penaeus vannamei Farms in China" Foods 12, no. 11: 2143. https://doi.org/10.3390/foods12112143
APA StyleZhang, Y., Lv, X., Cao, W., Zhang, H., Shi, L., Bai, W., & Ye, L. (2023). Survey of Colistin Resistance in Commensal Bacteria from Penaeus vannamei Farms in China. Foods, 12(11), 2143. https://doi.org/10.3390/foods12112143




