Food-Grade Bacteria Combat Pathogens by Blocking AHL-Mediated Quorum Sensing and Biofilm Formation
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Media
2.2. Isolation of New Propionibacterium freudenreichii Strains from Cheese
2.3. Collecting Spent Culture Supernatants from Lactobacilli and Propionibacteria
2.4. Screening Anti-QS and Bactericidal Activities Using C. violaceum Reporters
2.5. HPLC Analyses of Spent Culture Supernatants
2.6. Statistical Analyses
3. Results
3.1. Reporter Grown on LBA Enhances Violacein Production in LBYE
3.2. Violacein Production Is More Efficient in ATCC31532 Grown in PDYT Than LBYE
3.3. High D-Glc Inhibits and High L-Trp Stimulates Violacein Synthesis in C. violaceum
3.4. Violacein Production in ATCC31532 Was Increased by >10-Times in mPDYT
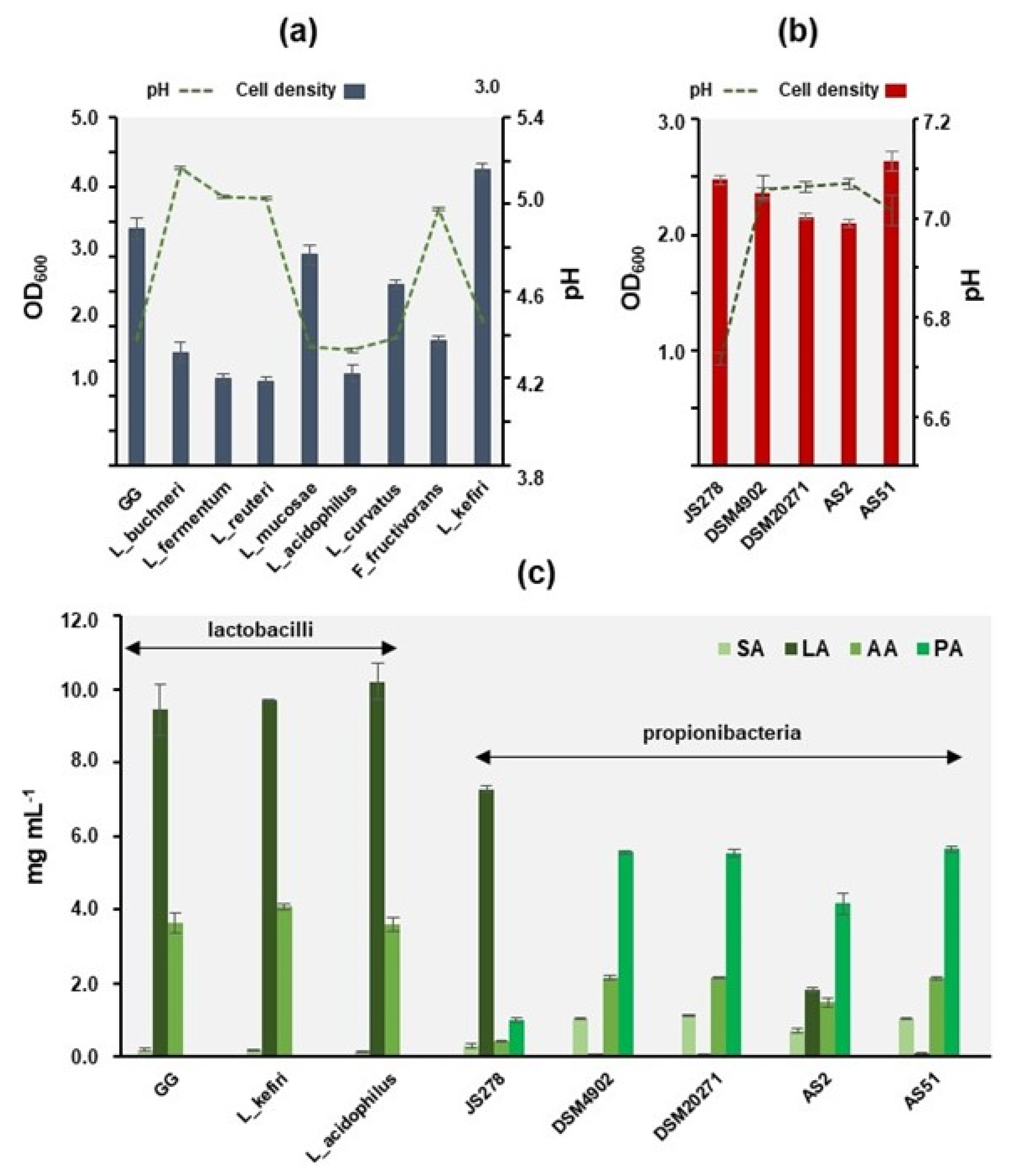
3.5. Lactobacilli Can Exert either QSI or QQ Activities against C. violaceum
3.6. Propionibacteria Interrupt QS Signaling in C. violaceum by QQ
3.7. Acetic and Propionic Acids Can Block the Induction of QS of C. violaceum
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Turkina, M.V.; Vikström, E. Bacteria-Host Crosstalk: Sensing of the quorum in the context of Pseudomonas aeruginosa Infections. J. Innate Immun. 2019, 11, 263–279. [Google Scholar] [CrossRef]
- Mukherjee, S.; Bassler, B.L. Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 2019, 17, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yu, Z.; Ding, T. Quorum-sensing regulation of antimicrobial resistance in bacteria. Microorganisms 2020, 8, 425. [Google Scholar] [CrossRef] [PubMed]
- Haque, S.; Yadav, D.K.; Bisht, S.C.; Yadav, N.; Singh, V.; Dubey, K.K.; Jawed, A.; Wahid, M.; Dar, S.A. Quorum sensing pathways in Gram-positive and -negative bacteria: Potential of their interruption in abating drug resistance. J. Chemother. 2019, 31, 161–187. [Google Scholar] [CrossRef] [PubMed]
- Rajput, A.; Kumar, M. In silico analyses of conservational, functional and phylogenetic distribution of the LuxI and LuxR homologs in Gram-positive bacteria. Sci. Rep. 2017, 7, 6969. [Google Scholar] [CrossRef] [PubMed]
- Defoirdt, T. Quorum-sensing systems as targets for antivirulence therapy. Trends Microbiol. 2018, 26, 313–328. [Google Scholar] [CrossRef]
- Rémy, B.; Mion, S.; Plener, L.; Elias, M.; Chabrière, E.; Daudé, D. Interference in bacterial quorum sensing: A biopharmaceutical perspective. Front. Pharmacol. 2018, 9, 203. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, Y.; Ge, Y.; Zhu, X.; Pan, J. Regulatory mechanisms and promising applications of quorum sensing-inhibiting agents in control of bacterial biofilm formation. Front. Microbiol. 2020, 11, 589640. [Google Scholar] [CrossRef]
- Zhong, S.; He, S. Quorum sensing inhibition or quenching in Acinetobacter baumannii: The novel therapeutic strategies for new drug development. Front. Microbiol. 2021, 12, 558003. [Google Scholar] [CrossRef]
- Khan, F.; Oloketuyi, S.F.; Kim, Y.M. Diversity of bacteria and bacterial products as antibiofilm and antiquorum sensing drugs against pathogenic bacteria. Curr. Drug Targets 2019, 20, 1156–1179. [Google Scholar] [CrossRef]
- Wu, S.; Xu, C.; Liu, J.; Liu, C.; Qiao, J. Vertical and horizontal quorum-sensing-based multicellular communications. Trends Microbiol. 2021, 29, 1130–1142. [Google Scholar] [CrossRef] [PubMed]
- Sikdar, R.; Elias, M. Quorum quenching enzymes and their effects on virulence, biofilm, and microbiomes: A review of recent advances. Expert Rev. Anti Infect. Ther. 2020, 18, 1221–1233. [Google Scholar] [CrossRef] [PubMed]
- Prazdnova, E.V.; Gorovtsov, A.V.; Vasilchenko, N.G.; Kulikov, M.P.; Statsenko, V.N.; Bogdanova, A.A.; Refeld, A.G.; Brislavskiy, Y.A.; Chistyakov, V.A.; Chikindas, M.L. Quorum-sensing inhibition by Gram-positive bacteria. Microorganisms 2022, 10, 350. [Google Scholar] [CrossRef]
- Barzegari, A.; Kheyrolahzadeh, K.; Hosseiniyan Khatibi, S.M.; Sharifi, S.; Memar, M.Y.; Zununi Vahed, S. The Battle of probiotics and their derivatives against biofilms. Infect. Drug Resist. 2020, 13, 659–672. [Google Scholar] [CrossRef]
- Kareb, O.; Aïder, M. Quorum sensing circuits in the communicating mechanisms of bacteria and its implication in the biosynthesis of bacteriocins by lactic acid bacteria. Probiotics Antimicrob. Proteins 2020, 12, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Maldonado-Barragán, A.; West, S.A. The cost and benefit of quorum sensing-controlled bacteriocin production in Lactobacillus plantarum. J. Evol. Biol. 2020, 33, 101–111. [Google Scholar] [CrossRef]
- Patel, M.; Siddiqui, A.J.; Ashraf, S.A.; Surti, M.; Awadelkareem, A.M.; Snoussi, M.; Hamadou, W.S.; Bardakci, F.; Jamal, A.; Jahan, S.; et al. Lactiplantibacillus plantarum-derived biosurfactant attenuates quorum sensing-mediated virulence and biofilm formation in Pseudomonas aeruginosa and Chromobacterium violaceum. Microorganisms 2022, 10, 1026. [Google Scholar] [CrossRef] [PubMed]
- Mathur, H.; Field, D.; Rea, M.C.; Cotter, P.D.; Hill, C.; Ross, R.P. Fighting biofilms with lantibiotics and other groups of bacteriocins. NPJ Biofilms Microb. 2018, 4, 9. [Google Scholar] [CrossRef] [PubMed]
- Rabah, H.; Rosa do Carmo, F.L.; Jan, G. Dairy Propionibacteria: Versatile Probiotics. Microorganisms 2017, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Sarveswari, H.B.; Solomon, A.P. Profile of the intervention potential of the phylum Actinobacteria toward quorum sensing and other microbial virulence strategies. Front. Microbiol. 2019, 10, 2073. [Google Scholar] [CrossRef]
- Jan, G.; Belzacq, A.S.; Haouzi, D.; Rouault, A.; Métivier, D.; Kroemer, G.; Brenner, C. Propionibacteria induce apoptosis of colorectal carcinoma cells via short-chain fatty acids acting on mitochondria. Cell Death Differ. 2002, 9, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Lind, H.; Jonsson, H.; Schnürer, J. Antifungal effect of dairy propionibacteria—Contribution of organic acids. Int. J. Food Microbiol. 2005, 98, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Piwowarek, K.; Lipińska, E.; Hać-Szymańczuk, E.; Kieliszek, M.; Ścibisz, I. Propionibacterium spp.-source of propionic acid, vitamin B12, and other metabolites important for the industry. Appl. Microbiol. Biotechnol. 2018, 102, 515–538. [Google Scholar] [CrossRef]
- Polkade, A.V.; Mantri, S.S.; Patwekar, U.J.; Jangid, K. Quorum sensing: An under-explored phenomenon in the phylum Actinobacteria. Front. Microbiol. 2016, 7, 131. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Durán, N.; Justo, G.Z.; Durán, M.; Brocchi, M.; Cordi, L.; Tasic, L.; Castro, G.R.; Nakazato, G. Advances in Chromobacterium violaceum and properties of violacein-Its main secondary metabolite. Biotechnol. Adv. 2016, 34, 1030–1045. [Google Scholar] [CrossRef]
- Weiland-Bräuer, N. Friends or foes-microbial interactions in nature. Biology 2021, 10, 496. [Google Scholar] [CrossRef]
- Skogman, M.E.; Kanerva, S.; Manner, S.; Vuorela, P.M.; Fallarero, A. Flavones as quorum sensing inhibitors identified by a newly optimized screening platform using Chromobacterium violaceum as reporter bacteria. Molecules 2016, 21, 1211. [Google Scholar] [CrossRef]
- Manner, S.; Fallarero, A. Screening of natural product derivatives identifies two structurally related flavonoids as potent quorum sensing inhibitors against Gram-negative bacteria. Int. J. Mol. Sci. 2018, 19, 1346. [Google Scholar] [CrossRef]
- Ravichandran, V.; Zhong, L.; Wang, H.; Yu, G.; Zhang, Y.; Li, A. Virtual screening and biomolecular interactions of CviR-based quorum sensing inhibitors against Chromobacterium violaceum. Front. Cell. Infect. Microbiol. 2018, 8, 292. [Google Scholar] [CrossRef] [PubMed]
- Beus, M.; Savijoki, K.; Patel, J.Z.; Yli-Kauhaluoma, J.; Fallarero, A.; Zorc, B. Chloroquine fumardiamides as novel quorum sensing inhibitors. Bioorg. Med. Chem. Lett. 2020, 30, 127336. [Google Scholar] [CrossRef]
- Gilbert-Girard, S.; Savijoki, K.; Yli-Kauhaluoma, J.; Fallarero, A. Screening of FDA-approved drugs using a 384-well plate-based biofilm platform: The case of fingolimod. Microorganisms 2020, 8, 1834. [Google Scholar] [CrossRef] [PubMed]
- Venkatramanan, M.; Sankar Ganesh, P.; Senthil, R.; Akshay, J.; Veera Ravi, A.; Langeswaran, K.; Vadivelu, J.; Nagarajan, S.; Rajendran, K.; Shankar, E.M. Inhibition of quorum sensing and biofilm formation in Chromobacterium violaceum by fruit extracts of Passiflora edulis. ACS Omega 2020, 5, 25605–25616. [Google Scholar] [CrossRef]
- Beus, M.; Persoons, L.; Daelemans, D.; Schols, D.; Savijoki, K.; Varmanen, P.; Yli-Kauhaluoma, J.; Pavić, K.; Zorc, B. Anthranilamides with quinoline and β-carboline scaffolds: Design, synthesis, and biological activity. Mol. Divers. 2022, 8, 2595–2612. [Google Scholar] [CrossRef]
- Zore, M.; Gilbert-Girard, S.; San-Martin-Galindo, P.; Reigada, I.; Hanski, L.; Savijoki, K.; Fallarero, A.; Yli-Kauhaluoma, J.; Patel, J.Z. Repurposing the sphingosine-1-phosphate receptor modulator etrasimod as an antibacterial agent against Gram-positive bacteria. Front. Microbiol. 2022, 13, 926170. [Google Scholar] [CrossRef]
- Li, Y.L.; Chu, Z.Y.; Liu, G.M.; Yang, S.Q.; Zeng, H. The derived components of Gnaphalium hypoleucum DC reduce quorum sensing of Chromobacterium violaceum. Molecules 2022, 27, 4881. [Google Scholar] [CrossRef]
- Stauff, D.L.; Bassler, B.L. Quorum sensing in Chromobacterium violaceum: DNA recognition and gene regulation by the CviR receptor. J. Bacteriol. 2011, 193, 3871–3878. [Google Scholar] [CrossRef] [PubMed]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef] [PubMed]
- McClean, K.H.; Winson, M.K.; Fish, L.; Taylor, A.; Chhabra, S.R.; Camara, M.; Daykin, M.; Lamb, J.H.; Swift, S.; Bycroft, B.W.; et al. Quorum sensing and Chromobacterium violaceum: Expression of violacein production and inhibition for the detection of N-acyl homoserine lactones. Microbiology 1997, 143, 3703–3711. [Google Scholar] [CrossRef] [PubMed]
- Mendés, A.S.; De Carvalho, J.E.; Duarte, M.C.T.; Durán, N.; Bruns, R.E. Factorial design and response surface optimization of crude violacein for Chromobacterium violaceum production. Biotechnol. Lett. 2001, 23, 1963–1969. [Google Scholar] [CrossRef]
- Palukurty, M.A.; Pyla, S.P.; Silarapu, S.; Somalanka, S.R. Analyzing alternative nutrient supplements and optimization of production parameters for violacein using central composite design. Int. J. Sci Eng. Res. 2016, 7, 294–300. [Google Scholar]
- Savijoki, K.; Nyman, T.A.; Kainulainen, V.; Miettinen, I.; Siljamäki, P.; Fallarero, A.; Sandholm, J.; Satokari, R.; Varmanen, P. Growth mode and carbon source impact the surfaceome dynamics of Lactobacillus rhamnosus GG. Front. Microbiol. 2019, 10, 1272. [Google Scholar] [CrossRef]
- Jan, G.; Rouault, A.; Maubois, J.-L. Acid stress susceptibility and acid adaptation of Propionibacterium freudenreichii subsp. shermanii. Le Lait 2000, 80, 325–336. [Google Scholar] [CrossRef]
- Kankainen, M.; Paulin, L.; Tynkkynen, S.; von Ossowski, I.; Reunanen, J.; Partanen, P.; Satokari, R.; Vesterlund, S.; Hendrickx, A.P.; Lebeer, S.; et al. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human- mucus binding protein. Proc. Natl. Acad. Sci. USA 2009, 106, 17193–17198. [Google Scholar] [CrossRef]
- Naghmouchi, K.; Belguesmia, Y.; Bendali, F.; Spano, G.; Seal, B.S.; Drider, D. Lactobacillus fermentum: A bacterial species with potential for food preservation and biomedical applications. Crit. Rev. Food Sci. Nutr. 2020, 60, 3387–3399. [Google Scholar] [CrossRef] [PubMed]
- Mu, Q.; Tavella, V.J.; Luo, X.M. Role of Lactobacillus reuteri in human health and diseases. Front. Microbiol. 2018, 9, 757. [Google Scholar] [CrossRef]
- Chen, Y.; Yu, L.; Qiao, N.; Xiao, Y.; Tian, F.; Zhao, J.; Zhang, H.; Chen, W.; Zhai, Q. Latilactobacillus curvatus: A candidate probiotic with excellent fermentation properties and health benefits. Foods 2020, 9, 1366. [Google Scholar] [CrossRef] [PubMed]
- Roos, S.; Karner, F.; Axelsson, L.; Jonsson, H. Lactobacillus mucosae sp. nov., a new species with in vitro mucus-binding activity isolated from pig intestine. Int. J. Syst. Evol. Microbiol. 2000, 50, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Anjum, N.; Maqsood, S.; Masud, T.; Ahmad, A.; Sohail, A.; Momin, A. Lactobacillus acidophilus: Characterization of the species and application in food production. Crit. Rev. Food. Sci. Nutr. 2014, 54, 1241–1251. [Google Scholar] [CrossRef]
- Nam, S.H.; Choi, S.H.; Kang, A.; Lee, K.S.; Kim, D.W.; Kim, R.N.; Kim, D.S.; Park, H.S. Genome sequence of Lactobacillus fructivorans KCTC 3543. J. Bacteriol. 2012, 194, 2111–2112. [Google Scholar] [CrossRef][Green Version]
- Toscano, M.; De Grandi, R.; Miniello, V.L.; Mattina, R.; Drago, L. Ability of Lactobacillus kefiri LKF01 (DSM32079) to colonize the intestinal environment and modify the gut microbiota composition of healthy individuals. Dig. Liver Dis. 2017, 49, 261–267. [Google Scholar] [CrossRef]
- Deptula, P.; Smolander, O.P.; Laine, P.; Roberts, R.J.; Edelmann, M.; Peltola, P.; Piironen, V.; Paulin, L.; Storgårds, E.; Savijoki, K.; et al. Acidipropionibacterium virtanenii sp. nov., isolated from malted barley. Int. J. Syst. Evol. Microbiol. 2018, 68, 3175–3183. [Google Scholar] [CrossRef]
- Falentin, H.; Deutsch, S.M.; Loux, V.; Hammani, A.; Buratti, J.; Parayre, S.; Chuat, V.; Barbe, V.; Aury, J.M.; Jan, G.; et al. Permanent draft genome sequence of the probiotic strain Propionibacterium freudenreichii CIRM-BIA 129 (ITG P20). Stand. Genom. Sci. 2016, 11, 6. [Google Scholar] [CrossRef]
- Koskinen, P.; Deptula, P.; Smolander, O.-P.; Tamene, F.; Kammonen, J.; Savijoki, K.; Paulin, L.; Piironen, V.; Auvinen, P.; Varmanen, P. Complete genome sequence of Propionibacterium freudenreichii DSM 20271(T). Stand. Genom. Sci. 2015, 10, 83. [Google Scholar] [CrossRef]
- Deptula, P.; Kylli, P.; Chamlagain, B.; Holm, L.; Kostiainen, R.; Piironen, V.; Savijoki, K.; Varmanen, P. BluB/CobT2 fusion enzyme activity reveals mechanisms responsible for production of active form of vitamin B12 by Propionibacterium freudenreichii. Microb. Cell Fact. 2015, 14, 186. [Google Scholar] [CrossRef] [PubMed]
- Guerin, T.F.; Mondido, M.; McClenn, B.; Peasley, B. Application of resazurin for estimating abundance of contaminant-degrading micro-organisms. Lett. Appl. Microbiol. 2001, 32, 340–345. [Google Scholar] [CrossRef]
- Sandberg, M.E.; Schellmann, D.; Brunhofer, G.; Erker, T.; Busygin, I.; Leino, R.; Vuorela, P.M.; Fallarero, A. Pros and cons of using resazurin staining for quantification of viable Staphylococcus aureus biofilms in a screening assay. J. Microbiol. Methods 2009, 78, 104–106. [Google Scholar] [CrossRef]
- Xie, C.; Coda, R.; Chamlagain, B.; Edelmann, M.; Deptula, P.; Varmanen, P.; Piironen, V.; Katina, K. In situ fortification of vitamin B12 in wheat flour and wheat bran by fermentation with Propionibacterium freudenreichii. J. Cereal Sci. 2018, 81, 133–139. [Google Scholar] [CrossRef]
- Zhang, J.H.; Chung, T.D.; Oldenburg, K.R. A Simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen 1999, 4, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Bollini, S.; Herbst, J.J.; Gaughan, G.T.; Verdoorn, T.A.; Ditta, J.; Dubowchik, G.M.; Vinitsky, A. High-throughput fluorescence polarization method for identification of FKBP12 ligands. J. Biomol. Screen 2002, 7, 526–530. [Google Scholar] [CrossRef] [PubMed]
- Iversen, P.W.; Eastwood, B.J.; Sittampalam, G.S.; Cox, K.L. A comparison of assay performance measures in screening assays: Signal Window, Z’ Factor, and Assay Variability Ratio. J. Biomol. Screen 2006, 11, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Kivisaar, M. Stationary phase mutagenesis: Mechanisms that accelerate adaptation of microbial populations under environmental stress. Environ. Microbiol. 2003, 5, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Jin, Z.W.; Li, X.; Wang, Z. Effect of yeast extract on L-tryptophan production in batch culture of Escherichia coli. Adv. Mat. Res. 2013, 807–809, 2009–2013. [Google Scholar] [CrossRef]
- Füller, J.J.; Röpke, R.; Krausze, J.; Rennhack, K.E.; Daniel, N.P.; Blankenfeldt, W.; Schulz, S.; Jahn, D.; Moser, J. Biosynthesis of violacein, structure and function of l-tryptophan oxidase VioA from Chromobacterium violaceum. J. Biol. Chem. 2016, 291, 20068–20084. [Google Scholar] [CrossRef]
- Antônio, R.V.; Creczynski-Pasa, T.B. Genetic analysis of violacein biosynthesis by Chromobacterium violaceum. Genet. Mol. Res. 2004, 3, 85–91. [Google Scholar]
- Rana, S.; Bhawal, S.; Kumari, A.; Kapila, S.; Kapila, R. pH-dependent inhibition of AHL-mediated quorum sensing by cell-free supernatant of lactic acid bacteria in Pseudomonas aeruginosa PAO1. Microb. Pathog. 2020, 42, 104–105. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhong, X.; Liu, X.; Wang, X.; Gao, X. Therapeutic and improving function of lactobacilli in the prevention and treatment of cardiovascular-related diseases: A novel perspective from gut microbiota. Front. Nutr. 2021, 8, 693412. [Google Scholar] [CrossRef] [PubMed]
- Kheiri, F.; Kermanshahi, R.K.; Feizabadi, M.M. The inhibitory effects of Lactobacillus supernatants and their metabolites on the growth and biofilm formation of Klebsiella pneumoniae. Infect. Disord. Drug Targets 2020, 20, 902–912. [Google Scholar] [CrossRef]
- Kim, A.-L.; Park, S.-Y.; Lee, C.-H.; Lee, C.-H.; Lee, J.-K. Quorum quenching bacteria isolated from the sludge of a wastewater treatment plant and their application for controlling biofilm formation. J. Microbiol. Biotechnol. 2014, 24, 1574–1582. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Zhang, J.; Qu, J.; Liu, J.; Yin, P.; Zhang, G.; Shang, D. Lactobacillus rhamnosus GG microcapsules inhibit Escherichia coli biofilm formation in coculture. Biotechnol. Lett. 2019, 41, 1007–1014. [Google Scholar] [CrossRef]
- Kusada, H.; Zhang, Y.; Tamaki, H.; Kimura, N.; Kamagata, Y. Novel N-Acyl homoserine lactone-degrading bacteria isolated from penicillin-contaminated environments and their quorum-quenching activities. Front. Microbiol. 2019, 10, 455. [Google Scholar] [CrossRef]
- Cui, T.; Bai, F.; Sun, M.; Lv, X.; Li, X.; Zhang, D.; Du, H. Lactobacillus crustorum ZHG 2-1 as novel quorum-quenching bacteria reducing virulence factors and biofilm formation of Pseudomonas aeruginosa. LWT 2020, 117, 108696. [Google Scholar] [CrossRef]
- Hossain, M.I.; Mizan, M.F.R.; Roy, P.K.; Nahar, S.; Toushik, S.H.; Ashrafudoulla, M.; Jahid, I.K.; Lee, J.; Ha, S.D. Listeria monocytogenes biofilm inhibition on food contact surfaces by application of postbiotics from Lactobacillus curvatus B.67 and Lactobacillus plantarum M.2. Food Res. Int. 2021, 148, 110595. [Google Scholar] [CrossRef] [PubMed]
- Yong, C.C.; Lim, J.; Kim, B.-K.; Park, D.-J.; Oh, S. Suppressive effect of Lactobacillus fermentum Lim2 on Clostridioides Difficile 027 toxin production. Lett. Appl. Microbiol. 2019, 68, 386–393. [Google Scholar] [CrossRef] [PubMed]
- de Rezende Rodovalho, V.; Rodrigues, D.L.N.; Jan, G.; Loir, Y.L.; de Carvalho Azevedo, V.A.; Guédon, E. Propionibacterium freudenreichii: General characteristics and probiotic traits. In Prebiotics and Probiotics—From Food to Health; Robles, E.F., Ed.; IntechOpen: London, UK, 2021. [Google Scholar] [CrossRef]
- Bintsis, T. Lactic acid bacteria as starter cultures: An update in their metabolism and genetics. AIMS Microbiol. 2018, 4, 665–684. [Google Scholar] [CrossRef]
- Alakomi, H.L.; Skyttä, E.; Saarela, M.; Mattila-Sandholm, T.; Latva-Kala, K.; Helander, I.M. Lactic acid permeabilizes Gram-negative bacteria by disrupting the outer membrane. Appl Environ Microbiol. 2000, 66, 2001–2005. [Google Scholar] [CrossRef] [PubMed]
- Almasoud, A.; Hettiarachchy, N.; Rayaprolu, S.; Babu, D.; Kwon, Y.-M.; Mauromoustakos, A. Inhibitory effects of lactic and malic organic acids on autoinducer type 2 (AI-2) quorum sensing of Escherichia coli O157:H7 and Salmonella Typhimurium. LWT Food Sci. Technol. 2016, 66, 560–564. [Google Scholar] [CrossRef]
- Kim, M.; Lee, D.G. Propionic acid induces apoptosis-like death in Escherichia coli O157. J. Basic Microbiol. 2022, 62, 22–34. [Google Scholar] [CrossRef]
- Peh, E.; Kittler, S.; Reich, F.; Kehrenberg, C. Antimicrobial activity of organic acids against Campylobacter spp. and development of combinations-A synergistic effect? PLoS ONE 2020, 15, e0239312. [Google Scholar] [CrossRef]
- Park, T.; Im, J.; Kim, A.R.; Lee, D.; Jeong, S.; Yun, C.-H.; Han, H. Short-chain fatty acids inhibit the biofilm formation of Streptococcus gordonii through negative regulation of competence-stimulating peptide signaling pathway. J. Microbiol. 2021, 59, 1142–1149. [Google Scholar] [CrossRef]
- Nakajima, A.; Sato, H.; Oda, S.; Yokoi, T. Fluoroquinolones and propionic acid derivatives induce inflammatory responses in vitro. Cell Biol. Toxicol. 2018, 34, 65–77. [Google Scholar] [CrossRef]
- Nakamura, K.; O’Neill, A.M.; Williams, M.R.; Cau, L.; Nakatsuji, T.; Horswill, A.R.; Gallo, R.L. Short-chain fatty acids produced by Cutibacterium acnes inhibit biofilm formation by Staphylococcus epidermidis. Sci. Rep. 2020, 10, 21237. [Google Scholar] [CrossRef]
- Wang, Y.; Kuo, S.; Shu, M.; Yu, J.; Huang, S.; Dai, A.; Two, A.; Gallo, R.L.; Huang, C.M. Staphylococcus epidermidis in the human skin microbiome mediates fermentation to inhibit the growth of Propionibacterium acnes: Implications of probiotics in acne vulgaris. Appl. Microbiol. Biotechnol. 2014, 98, 411–424. [Google Scholar] [CrossRef]
- Sauer, K.; Cullen, M.C.; Rickard, A.H.; Zeef, L.A.; Davies, D.G.; Gilbert, P. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J. Bacteriol. 2004, 186, 7312–7326. [Google Scholar] [CrossRef] [PubMed]
- Silva, E.; Monteiro, R.; Grainha, T.; Alves, D.; Pereira, M.O.; Sousa, A.M. Fostering innovation in the treatment of chronic polymicrobial cystic fibrosis-associated infections exploring aspartic acid and succinic acid as ciprofloxacin adjuvants. Front. Cell. Infect. Microbiol. 2020, 10, 441. [Google Scholar] [CrossRef]
- Na, J.I.; Kim, S.Y.; Kim, J.H.; Youn, S.W.; Huh, C.H.; Park, K.C. Indole-3-acetic acid: A potential new photosensitizer for photodynamic therapy of acne vulgaris. Lasers Surg. Med. 2011, 43, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Zhao, H.; Nie, T.; Lu, F.; Zhang, C.; Lu, Y.; Lu, Z. Acetate activates Lactobacillus bacteriocin synthesis by controlling quorum sensing. Appl. Environ. Microbiol. 2021, 87, e0072021. [Google Scholar] [CrossRef] [PubMed]






| Bacterial Species/Strain | Features and Traits | Origin |
|---|---|---|
| Lacticaseibacillus rhamnosus GG | Probiotic paradigm used as a dietary supplement and for the production of fermented foods; cultured at +37 °C | [43] |
| Lactobacillus buchneri | Lactobacillus used as an additive to improve the aerobic stability of silages; cultured at +37 °C | HAMBI69 (a) |
| Lactobacillus fermentum | Probiotic Lactobacillus producing diverse and potent antimicrobial peptides; cultured at +37 °C | HAMBI73, [44] |
| Limosilactobacillus reuteri | Probiotic Lactobacillus used as a dietary supplement and for the production of fermented foods; cultured at +37 °C | HAMBI410, [45] |
| Latilactobacillus curvatus | Probiotic with excellent fermentation and health-promoting properties; cultured at + 30 °C | HAMBI453, [46] |
| Limosilactobacillus mucosae | Potential probiotic Lactobacillus encoding the cell-surface mucus-binding protein; cultured at +37 °C | HAMBI2674, [47] |
| Lactobacillus acidophilus | Probiotic Lactobacillus used as a dietary supplement and for the production of fermented foods; cultured at +37 °C | HAMBI80, [48] |
| Fructilactobacillus fructivorans | Lactobacillus used to produce fermented beverages. Can also act as a spoilage bacterium; cultured at +30 °C | HAMBI1579, [49] |
| Lactobacillus kefiri | Slime-forming probiotic Lactobacillus formed from kefir grains; cultured at +30 °C | HAMBI3070, [50] |
| Acidipropionibacterium virtanenii JS278 | Propionic acid bacterium isolated from malted barley in Finland; cultured at +30 °C | [51] |
| Propionibacterium freudenreichii | Type strain; cultured at +30 °C | DSM4902, [52] |
| Propionibacterium freudenreichii | Type strain; cultured at +30 °C | DSM20271, [53] |
| Propionibacterium freudenreichii AS2 | Propionic acid bacterium isolated from Liechtensteiner cheese (27.07.2020); cultured at +30 °C | This study |
| Propionibacterium freudenreichii AS51 | Propionic acid bacterium isolated from Comte Prestige cheese (21.09.2020); cultured at +30 °C | This study |
| Chromobacterium violaceum | Biofilm-forming QS reporter strain; cultured at +27 °C | ATCC31532, [39] |
| Chromobacterium violaceum CV026 | AHL-negative double miniTn5 disruption mutant of ATCC31532, km; cultured at +27 °C | NCTC13278, [39] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Savijoki, K.; San-Martin-Galindo, P.; Pitkänen, K.; Edelmann, M.; Sillanpää, A.; van der Velde, C.; Miettinen, I.; Patel, J.Z.; Yli-Kauhaluoma, J.; Parikka, M.; et al. Food-Grade Bacteria Combat Pathogens by Blocking AHL-Mediated Quorum Sensing and Biofilm Formation. Foods 2023, 12, 90. https://doi.org/10.3390/foods12010090
Savijoki K, San-Martin-Galindo P, Pitkänen K, Edelmann M, Sillanpää A, van der Velde C, Miettinen I, Patel JZ, Yli-Kauhaluoma J, Parikka M, et al. Food-Grade Bacteria Combat Pathogens by Blocking AHL-Mediated Quorum Sensing and Biofilm Formation. Foods. 2023; 12(1):90. https://doi.org/10.3390/foods12010090
Chicago/Turabian StyleSavijoki, Kirsi, Paola San-Martin-Galindo, Katriina Pitkänen, Minnamari Edelmann, Annika Sillanpää, Cim van der Velde, Ilkka Miettinen, Jayendra Z. Patel, Jari Yli-Kauhaluoma, Mataleena Parikka, and et al. 2023. "Food-Grade Bacteria Combat Pathogens by Blocking AHL-Mediated Quorum Sensing and Biofilm Formation" Foods 12, no. 1: 90. https://doi.org/10.3390/foods12010090
APA StyleSavijoki, K., San-Martin-Galindo, P., Pitkänen, K., Edelmann, M., Sillanpää, A., van der Velde, C., Miettinen, I., Patel, J. Z., Yli-Kauhaluoma, J., Parikka, M., Fallarero, A., & Varmanen, P. (2023). Food-Grade Bacteria Combat Pathogens by Blocking AHL-Mediated Quorum Sensing and Biofilm Formation. Foods, 12(1), 90. https://doi.org/10.3390/foods12010090





